Abstract
Members of the transforming growth factor (TGF)-β superfamily have been shown to play a variety of important roles in embryogenesis, including dorsal and ventral mesoderm induction. The tumor suppressor SMAD4, also known as DPC4, is believed to be an essential factor that mediates TGF-β signals. To explore functions of SMAD4 in development, we have mutated it by truncating its functional C-domain. We show that Smad4 is expressed ubiquitously during murine embryogenesis. Mice heterozygous for the Smad4ex8/+ mutation are developmentally normal, whereas homozygotes die between embryonic day 6.5 (E6.5) and 8.5. All Smad4ex8/ex8 mutants are developmentally delayed at E6 and show little or no elongation in the extraembryonic portion of late egg cylinder stage embryos. Consistent with this, cultured Smad4ex8/ex8 blastocyst outgrowths suffer cellular proliferation defects and fail to undergo endoderm differentiation. Although a portion of mutant embryos at E8.5 show an increase in the embryonic ectoderm and endoderm, morphological and molecular analyses indicate that they do not form mesoderm. Altogether, these data demonstrate that SMAD4-mediated signals are required for epiblast proliferation, egg cylinder formation, and mesoderm induction.
Keywords: gastrulation, gene targeting, transforming growth factor β signaling
The transforming growth factor (TGF)-β superfamily is composed of many secreted signaling molecules that have been found to play important roles in multiple biological systems (reviewed in refs. 1 and 2). TGF-β ligands transduce signals through interaction with the heteromeric complex of two types of transmembrane serine–threonine kinases, known as type I and type II receptors. Ligands first bind to the type II receptor, which then recruits and phosphorylates the type I receptor in the highly conserved GS domain. Once activated, the type I receptor propagates the signal to downstream targets (3–5).
Recently, SMADs have been identified as important components of the signaling pathway of the TGF-β related factors. SMADs constitute a gene family of nine members in vertebrate (6–10) and have highly conserved N-terminal (MH1) and C-terminal (MH2) domains, which are separated by a proline-rich linker region. When directly phosphorylated at a carboxyl-terminal SS(V/M)S consensus site by ligand-activated type I receptor (11, 12), Smad1, Smad2, Smad3, and Smad5 form a stable complex with SMAD4, a common partner of SMADs, and are translocated into nuclei where they regulate target genes in response to TGF-β signals (12, 13). Studies mainly from Xenopus show that Smad1 and Smad5 act downstream of the bone morphogenetic proteins (BMPs) and associate with Smad4 to induce ventral mesoderm formation (12, 13). Smad2 and Smad3 are activated by activin and TGF-β and interact with Smad4 to induce dorsal mesoderm (14–16). Smad6 and Smad7, which lack the carboxyl-terminal phosphorylation site, have been identified as inhibitors of the TGF-β signaling pathway. They can interact with type I receptor but cannot be phosphorylated or released (7, 8, 17).
SMAD4 was first identified as a tumor suppressor gene, Dpc4 (deleted in pancreatic cancer) (18). In humans, mutations in Dpc4/Smad4 have been found in approximately half of all pancreatic carcinomas (18, 19), 30% of colon carcinomas (20), and less than 10% of other cancers (19, 21–23). DPC4/SMAD4 usually exists in a homotrimeric conformation and forms heteromeric complexes with other SMADs in response to TGF-β signals. The C-terminal domain of DPC4/SMAD4 is necessary and sufficient for these interactions (24). The majority of the tumor-derived missense mutations, which are clustered in the C-terminal domain, disrupt the homo-oligomerization both in vitro and in vivo (25).
To investigate the role of SMAD4 during development and tumorigenesis, we created mice carrying a targeted disruption in the Smad4 gene using homologous recombination in embryonic stem (ES) cells. We showed that mice heterozygous for the targeted mutation (Smad4ex8/+) are phenotypically normal. In contrast, homozygotes die at about 6.5–8.5 days during embryonic development (E6.5–8.5). Analysis of the Smad4ex8/ex8 embryos both in in vivo and in vitro indicated that SMAD4 is required for epiblast proliferation, egg cylinder formation, and mesoderm induction. Comparison between Smad4ex8/ex8 embryos with several other mutants in the TGF-β signaling pathway indicated that SMAD4 is the earliest acting factor in the TGF-β signaling pathway found to date, supporting its role as a central mediator of SMAD activities.
MATERIALS AND METHODS
Targeting Construct.
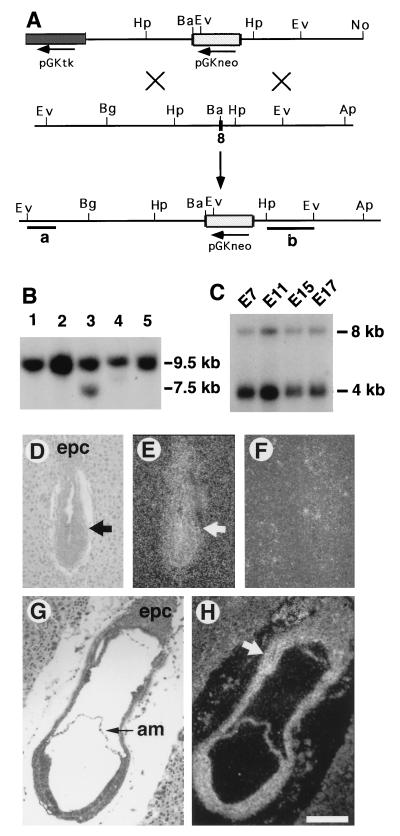
One BAC clone containing genomic DNA of the Smad4 locus was isolated from a 129-mouse genomic library (Genome Systems, St. Louis) by using a human Smad4 cDNA probe (18). A 5-kilobase (kb) BamHI–ApaI fragment that is 3′ to the eighth exon of the Smad4 was subcloned into the HpaI site of the pLoxpneo vector, which is derived from Ppnt (26) by adding a loxp site on the 5′ and 3′ of the neo gene, respectively. The resulting construct was cleaved with BamHI, followed by insertion of a 4.2-kb BglII–BamHI fragment that is 5′ to the eighth exon of the Smad4 gene. The finished construct, pSmad4neo, is shown in Fig. 2A.
Figure 2.
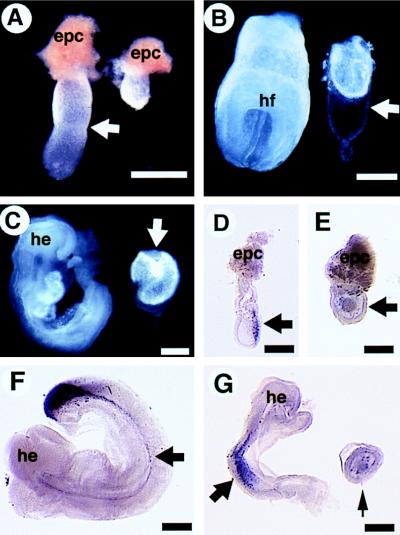
Morphological and molecular analyses of Smad4ex8/ex8 embryos (Right) and their littermate controls (Left). (A) E6.5 embryos. Arrow points to the boundary between extraembryonic and embryonic portions of the control embryo. The mutant embryo is round without such a boundary. epc, ectoplacental cone. (B) E7.5 embryos. Mutant embryos remain small, however, with a relatively large Reichert’s membrane (arrow). (C) E8.5 embryos. Arrow points to mutant embryo. Note the increased sizes of mutant embryos compared with those shown at E6.5 and 7.5. (D–F) Whole mount hybridization showing T expression in E7 normal (D), E8.5 mutant (E), and its littermate control (F) embryos. T transcripts were absent in the mutant (E, arrow) but are detected in the control embryos (D and F, arrows). Lim1 transcripts were also absent in the E8.5 mutant (small arrow in G) but are detected in the control embryos. he, head; hf, headfold. (Bar = 270 μm.)
Homologous Recombination in ES Cells and Generation of Germ-Line Chimeras.
TC1 (27) ES cells were transfected with NotI-digested pSmad4neo and selected with G418 and 1-(2-deoxy-2-fluoro-β-d-avabinofuranosyl-S-iodouracil (FIAU). ES cell colonies that were resistant to both G418 and FIAU were analyzed by Southern blotting for homologous recombination events within the Smad4 locus. Genomic DNAs from these clones and the parental TC1 cell line were digested with EcoRV and then probed with a 5′-flanking probe followed by an internal probe specific to the Smad4 gene. The 5′ probe is a 1-kb EcoRV–XbaI fragment, and the internal probe is a 1.9-kb HpaI–EcoRV fragment. ES cells heterozygous for the targeted mutation were microinjected into C57/B6 blastocysts to obtain germ-line transmission.
Genotype Analysis.
Genotypes were determined by Southern blotting or PCR. For PCR analysis, the wild-type Smad4 allele was detected using a 5′ oligonucleotide (primer 1, 5′-CAGTCGTCCCTTCTTCTTGG-3′) and a 3′ oligonucleotide (primer 2, 5′-CAGCTACTGAATGGAATAGCAG-3′). This primer pair flanks the pLoxpneo insertion site and amplifies a 530-bp fragment from the wild-type Smad4 gene. DNA was also amplified using primer 1 and primer 3 in the pLoxpneo gene (5′-CCAGACTGCCTTGGGAAAAGC-3′) to detect the mutant Smad4 allele. In this case, a 400-bp fragment was detected in mice heterozygous or homozygous for the mutant Smad4 allele, whereas no signal was detected in wild-type mice.
In Situ Hybridization and in Vitro Culture of Blastocysts.
Whole mount and regular in situ hybridization were carried out using standard procedures. Probes used for the hybridization are Lim1 (28), T (29), Mash-2 (30), H19 (31), and Smad4, which is a 0.8-kb fragment containing 594-bp nucleotides that encode the 198 amino acids of the C-domain plus 200 bp of the 3′-untranslated region. The Smad4 probe is also used in Northern blots and for genotyping mutant embryos by in situ hybridization. Because all the sequences contained in this probe are truncated by the neo gene insertion, mutant embryos are stained with significantly reduced intensity compared with wild-type and heterozygous embryos. Conditions for blastocyst culture were as described (29).
RESULTS
Smad4 Expression During Mouse Development.
Smad4 transcripts were detected by Northern blot on poly(A)+ RNA isolated from embryos at E7.0, E11.0, E15.0, and E17.0 (Fig. 1C). At all time points examined, two transcripts of approximately 4.0 and 8.0 kb were detected. The 4-kb transcript was the major form, whereas the 8-kb transcript was expressed at a much lower level. The spatial expression of Smad4 in E6.5–E7.5 embryos was analyzed by in situ hybridization in tissue sections. Smad4 transcripts were detected at these time points at a near ubiquitous pattern except for a slightly stronger signal in the extraembryonic portion of embryos (Fig. 1 D–H). These expression patterns suggest that Smad4 may play an essential role throughout development.
Figure 1.
Smad4 gene expression and targeted disruption. (A) Targeting vector pSmad4neo contains a 9.2-kb Smad4 genomic sequence. The crosshatched box represents PGKneo inserted at a BamHI site in the eighth exon of Smad4, such that a 4.2-kb 5′ and 5-kb 3′ sequence flank the insert. Homologous recombination within Smad4 would replace exon 8 with the neo gene and create a fragment shift from 9.5 to 7.5 kb upon EcoRV digestion. Ap, ApaI; Ba, BamHI; Bg, BglII; Ev, EcoRV; Hp, HpaI; No, NotI. (B) Southern blot analysis of ES DNAs using the 5′-flanking probe (a). The wild-type (9.5 kb) and mutant (7.5 kb) fragments were as indicated. DNA of targeted clones were also analyzed using a 3′-internal probe (b) after digesting with multiple enzymes (not shown). Ten of 185 G418/FIAU double-resistant ES clones were found to be correctly targeted. (C) Northern blot analysis of Smad4 expression in embryos. The filter was purchased from CLONTECH, and RNA samples were evenly loaded as revealed by a probe for glyceraldehyde-3-phosphate dehydrogenase (not shown). (D–H) In situ hybridization of Smad4 expression at E6.5 (D–F) and E7.5 (G–H). D and G are bright-field views, and all others are dark-field views. E and H were hybridized using an antisense probe, and F was hybridized with a sense probe. Arrows point to embryos (D, E) and extraembryonic portion, which has a lightly stronger intensity (H). am, amnion; epc, ectoplacental cone. (Bar = 210 μm in D–H.)
Smad4ex8 Mutation Resulted in Early Embryonic Recessive Lethality.
The Smad4 gene was disrupted in ES cells using the targeting construct pSmad4neo, which contains a 9-kb Smad4 genomic sequence interrupted in exon 8 with a PGKneo cassette (Fig. 1 A and B). The mouse SMAD4 contains 551 amino acids (32) and the neo insertion truncated 198 amino acids from the C-terminal end. It was shown that C-domain truncated SMAD4 was incapable of transmitting either of the activin/TGF-β or BMP signals (13, 14, 18, 19, 21, 38). The targeted truncation we generated should, therefore, create a candidate null mutation of the Smad4 locus.
Mice heterozygous for the Smad4ex8 mutation were phenotypically normal. Over 10 of these mice were monitored for up to 8 months and were completely normal in terms of growth rate, morphology, health, and fertility. The heterozygous mice did not show any type of tumor, but it is still possible that they could develop cancer at older ages. To study the effect of the introduced mutation, Smad4ex8/+ mice were intercrossed to generate homozygous mice, but no homozygotes were found among 56 mice genotyped postnatally (Table 1). This observation suggested that the targeted mutation resulted in a recessive embryonic lethality. To determine the timing of this lethality, we dissected embryos from heterozygous intercrosses at different gestational days. Abnormal or degenerating embryos were recovered between E6.5 and E9.5.
Table 1.
Morphological and histological analysis of offspring from Smad4ex8 heterozygous intercross
| Age | Phenotype
|
Genotype
|
Total | ||||
|---|---|---|---|---|---|---|---|
| Normal | Abnormal | Resorption | +/+ | +/− | −/− | ||
| E3.5 | 116 | 0 | 30 | 62 | 24 | 116 | |
| E5.5 | 9 | 0 | 0 | 3 | 5 | 1 | 9 |
| E6.5 | 21 | 5 | 5 | 9 | 12 | 5 | 31 |
| E7.5 | 23 | 3 | 3 | 8 | 15 | 3 | 29 |
| E8.5 | 38 | 3 | 7 | 12 | 26 | 3 | 48 |
| E9.5 | 29 | 1 | 7 | 12 | 17 | 1 | 37 |
| Postnatal | 56 | 0 | 23 | 33 | 0 | 56 | |
Analyses were performed in both 129/C57BL/6J and 129/Black Swiss background, and comparable phenotypes were found.
At E6.5, about one-quarter of the deciduas contained either resorptions or morphologically abnormal embryos that were genotyped as Smad4ex8 homozygotes (Table 1). The E6.5 Smad4ex8 mutants were only ¼–½ the size of control embryos (wild-type or heterozygous littermates) and showed no apparent boundary between the embryonic and extraembryonic portions (Fig. 2A). At E7.5, the morphologically normal embryos had developed typical structures, including the primitive streak and headfold. However, mutant embryos did not contain any recognizable structures and were much smaller than their littermates (Fig. 2B). Despite their small size, many of the mutant embryos displayed a relatively large Reichert’s membrane (Fig. 2B). At E8.5 (Fig. 2C)–E9.5 (not shown), a small portion of homozygous mutants were still viable and showed an increased size compared with the mutants of E6.5–E7.5 shown in Fig. 2 A and B. But they did not have any structures resembling those of normal embryos (Fig. 2C). Mutant embryos generated from the two independently targeted ES cell clones were analyzed and showed identical phenotypes. These results indicate that loss of SMAD4 results in recessive embryonic lethality during early postimplantation stages.
Smad4ex8/ex8 Embryos Were Developmentally Arrested Before Gastrulation.
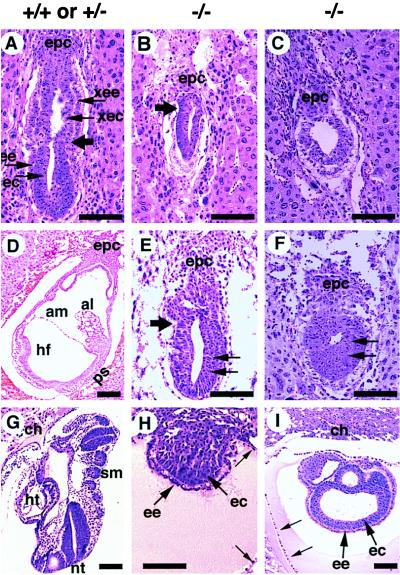
To characterize the structural organization of Smad4ex8/ex8 embryos in detail, we fixed and sectioned intact deciduas of E5.5–E8.5 litters from Smad4ex8 heterozygous intercrosses. Some slides, especially those containing abnormal embryos, were genotyped by in situ hybridization. All E5.5 embryos seemed morphologically normal (not shown). In contrast, E6.5 embryos displayed phenotypic differences. About 75% of these embryos (45/61) exhibited a well organized ectoderm and endoderm. They were well elongated with a clear morphological distinction between the embryonic and extraembryonic portions of the embryos (Fig. 3A). Seven of 61 embryos were significantly retarded in development. They had a small (Fig. 3B) or absent extraembryonic portion of the egg cylinder (Fig. 3C). In the latter case, the embryos appeared round. The remaining nine embryos were completely resorbed, suggesting that these embryos died at earlier stages. At E7.5, all normal embryos examined (n = 64) formed a well defined mesoderm layer (Fig. 3D). The remainder (n = 23) were either abnormal or resorbed (Table 1). Similar to E6.5 mutants, E7.5 Smad4ex8/ex8 embryos exhibited either missing (Fig. 3F) or significantly reduced and distorted extraembryonic egg cylinders (Fig. 3E). Both types of abnormal embryos had larger embryonic portions than mutants at E6.5 (Fig. 3 E and F). E8.5 embryos were also examined. Some abnormal embryos appeared like small solid balls (Fig. 3H), whereas others displayed abnormally enlarged sac-like structures with presumptive embryonic endoderm and ectoderm (Fig. 3I). Despite the relatively larger sizes of these sac-like structures, recognizable structures found in the control embryos (Fig. 3G) were absent. These data indicate that the mutant embryos are developmentally arrested at the onset of gastrulation, although some of them are still alive beyond E6.5.
Figure 3.
Histological sections of embryos generated from crosses between Smad4ex8/+ mice. (A) Sagittal section of a normal E6.5 embryo. Large arrow points to the boundary between extraembryonic and embryonic portions of embryos. epc, ectoplacental cone; ec, embryonic ectoderm; ee, embryonic endoderm; xec, extraembryonic ectoderm; xee, extraembryonic endoderm. (B and C) E6.5 abnormal embryos. Both embryos were developmentally delayed. Arrow in B points to the extraembryonic and embryonic boundary, which is not obvious in the histological section but became apparent by in situ hybridization with a H19 probe, which marks the extraembryonic ectoderm and endoderm. (D–F) E7.5 embryos. (D) Section of a normal embryo. al, allantois; am, amnion; hf, headfold; ps, primitive streak. (E and F) E7.5 abnormal embryos. Compared with the E6.5 mutant embryos, the embryonic portion of the mutant embryos has grown, and the embryonic ectoderm has thickened (arrows). However, the extraembryonic portion was either small (E) or absent (F). (G–I) E8.5 embryos. (G) Section of a normal embryo showing structures, such as chorion (ch), heart (ht), neural tube (nt), and somites (sm), which mutant embryos (H and I) did not have. Some mutant embryos (I) also displayed a sac-like structure with excess amount of presumptive embryonic endoderm and ectoderm. Both mutants (H and I) showed relatively large Reichert’s membranes (small arrow). (Bar, 134 μm in all panels.)
Smad4ex8/ex8 Embryos Showed Abnormal Development of the Egg Cylinder.
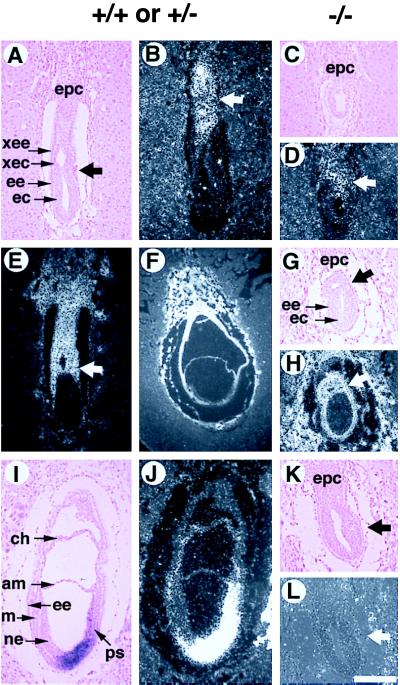
In addition to the reduced dimensions, Smad4ex8/ex8 embryos also displayed abnormalities in egg cylinder formation as revealed by morphological and histological analyses. In normal embryos, the rapid proliferation of epiblasts is accompanied by the elongation and differentiation of the egg cylinder. Beginning at E6, a demarcation is gradually formed that divides the egg cylinder into extraembryonic and embryonic portions. However, Smad4ex8/ex8 embryos did not have a clear boundary that divides the presumptive extraembryonic and embryonic portions (Fig. 2A). Histological analysis indicated that Smad4ex8 embryos contain a reduced (Fig. 3B) or absent extraembryonic portion (Fig. 3C). The expression analysis of the H19 gene, which marks extraembryonic cell types (31 and Fig. 4 E and F), confirmed this observation. The expression domain of H19 within the extraembryonic portion of E6.5 (not shown) and E7.5 (Fig. 4H) Smad4ex8/ex8 embryos was reduced because of the smaller sizes of the extraembryonic portion.
Figure 4.
In situ analysis of Smad4ex8/ex8 embryos. Genotypes of control and presumptive mutant embryos were as indicated. A–E show E6.5 embryos, and F–L show E7.5 embryos. A, C, G, I, and K are bright-field views, and all others are dark-field views. (B and D) Mash-2 expression in the ectoplacental cone (epc) and extraembryonic portion of E6.5 embryos (arrow in B and D). (E–H) H19 expression. The H19 expression domain in E7.5 mutant (H) is significantly reduced as the extraembryonic portion is much smaller compared with the E6.5 (E) and E7.5 (F) normal embryos. Arrows in E, G, and H point to the boundary between extraembryonic and embryonic portions of embryos. (I–L) T gene expression in E7.5 embryos. Transcripts of T gene can be detected in the primitive streak (ps) of normal embryos but cannot be detected in the mutant embryos. Arrows in K and L point to the region where T should normally be expressed. am, amnion; ec, embryonic ectoderm; ee, embryonic endoderm; epc, ectoplacental cone; ps, primitive streak; xec, extraembryonic ectoderm; xee, extraembryonic endoderm. (Bar = 200 μm in all panels.)
Because Smad4ex8 mutants showed abnormal extraembryonic development, we next examined the status of the ectoplacental cone (Epc). Both E6.5 and E7.5 mutants were found to contain histologically distinct Epc whose size was proportional to that of the mutant embryos (Fig. 4 C and K). Smad4ex8/ex8 embryos also expressed Mash-2, which marks Epc and the extraembryonic portion of the embryos (Fig. 4 B and D; ref. 30). The Mash-2 expression domain in mutants (Fig. 4D) was found smaller than their controls (Fig. 4B), but we believed it is because of the overall reduction of the embryos. These data indicate that a major defect of egg cylinder differentiation of Smad4ex8/ex8 embryos is the failure to elongate the extraembryonic portion.
Smad4ex8/ex8 Embryos Did Not Form Mesoderm.
To characterize mesodermal formation in the Smad4ex8 mutants at the molecular level, we examined the expression of brachyury and Lim1, two well established early markers for primitive streak and mesoderm formation, by in situ hybridization. Serial sections of four litters from Smad4ex8/+ intercrosses were examined with those probes. None of the markers were expressed in the E7.5 mutant embryos examined (Fig. 4 K and L), whereas their littermate controls showed characteristic expression patterns (Fig. 4 I and J). Whole mount in situ hybridization using both T and Lim1 (Fig. 2, D–G) confirmed that there was no mesoderm in Smad4ex8/ex8 embryos.
Decreased Embryonic Proliferation in Smad4ex8/ex8 Embryos.
The observation that Smad4ex8/ex8 embryos exhibit diminished size suggested a defect in the ability of cells to proliferate in the mutant embryos. To examine this, we stained three litters of E6.5 embryos from Smad4ex8 heterozygous intercrosses with an antibody that detects the cell proliferation marker, proliferating cell nuclear antigen (PCNA). Nearly all the nuclei of the epiblasts of control embryos exhibited strong staining (Fig. 5A). In contrast, cells of homozygous mutant embryos were stained much weaker (Fig. 5B). These results indicated that the proliferative ability of Smad4ex8/ex8 epiblast cells was decreased at E6.5.
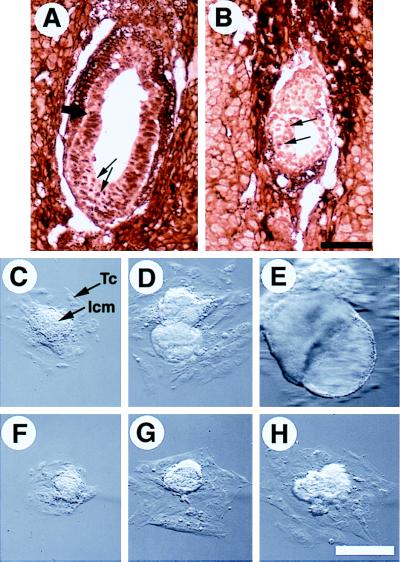
Figure 5.
Proliferation property of Smad4ex8/ex8 embryos. (A and B) Immunohistochemical staining of E6.5 normal and abnormal embryos with an antibody to PCNA. Over 90% of cells in normal embryos were positively stained (arrows in A), whereas the PCNA positive cells (arrows in B) were not only sparse but also stained weaker in the mutant embryo (B). (C–H) Culture of blastocysts in vitro. Blastocyst outgrowths from wild type are shown in C–E, and Smad4ex8/ex8 embryos are shown in F–H. Outgrowths shown in C and F, D and F, and E and G have been cultured for 3, 5, and 7 days in vitro, respectively. Trophoblast cells (Tc) and inner cell mass (Icm) were as indicated. [Bar = 136 μm (A and B); 150 μm (C–H).]
To monitor the growth properties of mutant embryos, blastocysts were isolated from Smad4ex8 heterozygous mating and cultured. Wild-type, heterozygous, and homozygous blastocysts seemed morphologically indistinguishable before culture and were genotyped by PCR in roughly a 1:2:1 ratio at the end of the culture (Table 1). These data indicate that Smad4ex8 mutation did not affect development at preimplantation stages.
Three days after culture, the inner cell mass (ICM) outgrowths of most mutant embryos were found to be slightly smaller than the control embryos (Fig. 5 C and F). After 5 days in culture, a dramatic growth disadvantage of Smad4ex8/ex8 embryos was observed. The ICM outgrowths of mutant embryos were only about ¼–½ size of control embryos (Fig. 5 D and G). The ICM outgrowths of over 70% of control embryos underwent endoderm differentiation and formed cystic structures after 7 days in culture (Fig. 5E). However, none of the mutant ICM outgrowths showed any signs of the differentiation (Fig. 5H).
DISCUSSION
We have shown that Smad4 is expressed at a near ubiquitous pattern in embryos at egg cylinder stages. Consistent with this expression pattern, embryos homozygous for the targeted disruption of the Smad4 gene exhibit abnormal development at egg cylinder stages and die shortly after, without a sign of mesoderm formation. Our data indicate that Smad4 is required for at least three essential functions in early embryogenesis: epiblast proliferation, egg cylinder formation, and mesoderm induction.
SMAD4 and Epiblast Proliferation.
We have found that Smad4ex8 homozygotes are normal at the blastocyst and the early egg cylinder stages. However, at E6.5, the mutant embryos were only about ¼–½ the size of their littermates, revealing an essential function of SMAD4 at stages around E6 of embryogenesis. It has been shown that mouse embryos undergo rapid cell proliferation before gastrulation with the epiblast cells increasing from 120 cells per E5.5 embryo to 660 cells per E6.5 embryo. The cell number increases further to 14,290 per embryo at E7.5 (33). Thus, it is possible that SMAD4 functions as a mediator of mitogenic stimuli for epiblast proliferation. Consistent with this view, Smad4ex8/ex8 embryos exhibited considerably reduced staining with an antibody to PCNA than the control embryos. Moreover, the ICM outgrowth of cultured mutant blastocysts grew much more slowly than that of control blastocysts.
A failure of epiblast proliferation has been previously observed in mouse embryos that are deficient for Bmpr gene, which encodes a BMP-2/4 type I serine–threonine kinase transmembrane receptor (34). Given the extensive studies that show SMAD4 is a downstream mediator of BMP signals, it is no surprise that Smad4ex8/ex8 mutants displayed a phenotype similar to that of BMPr-deficient embryos. However, there are differences between the Bmpr and Smad4ex8 mutants. For example, the cultured BMPr-deficient blastocysts proliferated and differentiated normally, whereas the Smad4ex8 blastocysts displayed a cellular proliferation defect. The more severe phenotype of SMAD4-deficient embryos suggests that SMAD4 may also mediate some other signals in addition to BMPs.
Smad4 and Egg Cylinder Formation.
Concomitant with its rapid proliferation, the egg cylinder, at around E6, undergoes differentiation characterized by the elongation and formation of a boundary between the extraembryonic and embryonic portions. The abnormal development of extraembryonic tissues of Smad4ex8/ex8 embryos suggests a role of Smad4 in this process. However, it is not clear that the effect of Smad4 on egg cylinder differentiation is direct or indirect, as egg cylinder differentiation and proliferation occur simultaneously. Consistent with this notion, growth defects and abnormal egg cylinder development have been found in null mutations of several genes, including BMP4 (35), BMPr (34), Brca1 (36), Fgfr1 (29, 37), Nf2 (38), and others. However, the failure in extraembryonic egg cylinder elongation is unique compared with these mutants. It is possible that the abnormalities in the extraembryonic tissue of Smad4ex8/ex8 embryos is secondary to the proliferation defect. In this case, the cell proliferation in the extraembryonic portion of Smad4ex8/ex8 embryos would depend more on the Smad4 signals than that of the embryonic portion. This notion seems to be consistent with a slightly higher expression of the Smad4 in the extraembryonic tissues. Alternatively, Smad4-mediated signals may play a distinct role during the egg cylinder differentiation and visceral endoderm formation.
Smad4 and Mesoderm Induction.
Members of the TGF-β superfamily have been implicated in mesoderm induction. Studies primarily in Xenopus showed that induction of mesoderm by TGF-βs is mediated by SMADs (14–16, 39, 40). Our data showed that Smad4ex8/ex8 embryos do not form mesoderm by both morphological standards and mesoderm marker analysis. However, these results must be interpreted cautiously because all the mutant embryos were under the influence of a growth defect. It has been suggested that 1,400–1,500 cells must accumulate in the epiblast to initiate gastrulation (41). Thus, the failure of mesoderm formation in the Smad4ex8/ex8 embryos could be primarily because of their diminished sizes. This may indeed be the case for most of the Smad4ex8/ex8 embryos examined before E8. However, at E8.5, a small portion of Smad4ex8/ex8 embryos survived the early proliferation defects, and their embryonic tissues continued to develop. Although their absolute size in the embryonic portion may be larger than those of E6.5–E7 normal embryos, they did not form mesoderm (Fig. 2 E and G), suggesting that without the SMAD4-mediated signals, gastrulation could not be initiated. Thus, this work provides direct genetic evidence that Smad4 is required for mesoderm induction.
Lack of mesoderm formation has been previously observed in BMP4 and BMPr mutant embryos (34, 35). BMP4 embryos homozygous for a targeted mutation died between E6.5 and E9.5 with a variable phenotype partly depending on their genetic background. A portion of Bmp-4tm1blh embryos were developmentally retarded at E6.5 and did not form mesoderm (35). However, some Bmp-4tm1blh embryos were able to advance to E9.5 with apparently normal mesoderm formation. One explanation for the variability in phenotype is that in the absence of BMP4, other ligands, such as BMP2 can provide compensatory function. Consistent with this hypothesis is that the embryos null for BMPr were apparently abnormal at the onset of gastrulation and did not form mesoderm (34). The similarity between Smad4ex8/ex8 embryos and BMP or BMPr null embryos is consistent with the view that SMAD4 is a downstream mediator of BMP signals (12, 13, 40).
Acknowledgments
We thank R. Behringer for Lim1, P. Leder for H19, and J. Rossant for Mash-2 probes. We are grateful to C. Huang for helpful discussion, J. M. Gotay, and M. Weinstein for critical reading of the manuscript, and J. M. Gotay, L. Chen, and S. Shen for technical assistance.
ABBREVIATIONS
- TGF
transforming growth factor
- BMP
bone morphogenetic protein
- ES
embryonic stem
- kb
kilobase(s)
- Epc
ectoplacental cone
- PCNA
proliferating cell nuclear antigen
- ICM
inner cell mass
- E
embryonic day
References
- 1.Kingsley D M. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 2.Massague J. Cell. 1996;85:947–950. doi: 10.1016/s0092-8674(00)81296-9. [DOI] [PubMed] [Google Scholar]
- 3.Wrana J L, Carcamo J, Attisano L, Cheifetz S, Zentella A, Lopez-Casillas F, Massague J. Cold Spring Harbor Symp Quant Biol. 1992;57:81–86. doi: 10.1101/sqb.1992.057.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Letsou A, Arora K, Wrana J L, Simin K, Twombly V, Jamal J, Staehling-Hampton K, Hoffmann F M, Gelbart W M, Massague J, et al. Cell. 1995;80:899–908. doi: 10.1016/0092-8674(95)90293-7. [DOI] [PubMed] [Google Scholar]
- 5.Ruberte E, Marty T, Nellen D, Affolter M, Basler K. Cell. 1995;80:889–897. doi: 10.1016/0092-8674(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 6.Derynck R, Gelbart W M, Harland R M, Heldin C H, Kern S E, Massague J, Melton D A, Mlodzik M, Padgett R W, Roberts A B, et al. Cell. 1996;87:173. doi: 10.1016/s0092-8674(00)81335-5. [DOI] [PubMed] [Google Scholar]
- 7.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Nature (London) 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y Y, Grinnell B W, Richardson M A, Topper J N, Gimbrone M A, Jr, Wrana J L, Falb D. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe T K, Suzuki M, Omori Y, Hishigaki H, Horie M, Kanemoto N, Fujiwara T, Nakamura Y, Takahashi E. Genomics. 1997;42:446–451. doi: 10.1006/geno.1997.4753. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Bhushan A, Vale W. Proc Natl Acad Sci USA. 1997;94:12938–12943. doi: 10.1073/pnas.94.24.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macias-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 12.Kretzschmar M, Liu F, Hata A, Doody J, Massague J. Genes Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- 13.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Feng X, We R, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 15.Graff J M, Bansal A, Melton D A. Cell. 1996;85:479–487. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 16.Baker J C, Harland R M. Curr Opin Genet Dev. 1997;7:467–473. doi: 10.1016/s0959-437x(97)80072-x. [DOI] [PubMed] [Google Scholar]
- 17.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian J L, Heuchel R, Itoh S, Kawabata M, Heldin N E, Heldin C H, ten Dijke P. Nature (London) 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 18.Hahn S A, Schutte M, Hoque A T, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, Kern S E. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 19.Schutte M, Hruban R H, Hedrick L, Cho K R, Nadasdy G M, Weinstein C L, Bova G S, Isaacs W B, Cairns P, Nawroz H, et al. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 20.Thiagalingam S, Lengauer C, Leach F S, Schutte M, Hahn S A, Overhauser J, Willson J K, Markowitz S, Hamilton S R, Kern S E, et al. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 21.Nagatake M, Takagi Y, Osada H, Uchida K, Mitsudomi T, Saji S, Shimokata K, Takahashi T, Takahashi T. Cancer Res. 1996;56:2718–2720. [PubMed] [Google Scholar]
- 22.Lei J, Zou T T, Shi Y Q, Zhou X, Smolinski K N, Yin J, Souza R F, Appel R, Wang S, Cymes K, et al. Oncogene. 1996;13:2459–2462. [PubMed] [Google Scholar]
- 23.Kim S K, Fan Y, Papadimitrakopoulou V, Clayman G, Hittelman W N, Hong W K, Lotan R, Mao L. Cancer Res. 1996;56:2519–2521. [PubMed] [Google Scholar]
- 24.Hata A, Lo R S, Wotton D, Lagna G, Massague J. Nature (London) 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, Hata A, Lo R S, Massague J, Pavletich N P. Nature (London) 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- 26.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 27.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 28.Barnes J D, Crosby J L, Jones C M, Wright C V E, Hogan B L M. Dev Biol. 1994;161:168–178. doi: 10.1006/dbio.1994.1018. [DOI] [PubMed] [Google Scholar]
- 29.Deng C, Wynshaw-boris A, Shen M M, Daugherty C, Ornitz D M, Leder P. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 30.Guillemot F, Nagy A, Rossant J, Joyner A L. Nature (London) 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 31.Poirier F, Chan C T, Timmons P M, Robertson E J, Evans M J, Rigby P W. Development (Cambridge, UK) 1991;113:1105–1114. doi: 10.1242/dev.113.4.1105. [DOI] [PubMed] [Google Scholar]
- 32.Anna C H, Devereux T R. Mamm Genome. 1997;8:443–444. doi: 10.1007/s003359900465. [DOI] [PubMed] [Google Scholar]
- 33.Snow M H L. J Embryol Exp Morphol. 1977;42:293–303. [Google Scholar]
- 34.Mishina Y, Suzuki A, Ueno N, Behringer R R. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 35.Winnier G, Blessing M, Labosky P A, Hogan B L. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 36.Hakem R, de la Pompa J L, Sirard C, Mo R, Woo M, Hakem A, Wakeham A, Potter J, Reitmair A, Billia F, et al. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 37.Yamaguchi T P, Harpal K, Henkemeyer M, Rossant J. Genes Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 38.McClatchey A I, Saotome I, Ramesh V, Gusella J F, Jacks T. Genes Dev. 1997;11:1253–1265. doi: 10.1101/gad.11.10.1253. [DOI] [PubMed] [Google Scholar]
- 39.Liu F, Hata A, Baker J C, Doody J, Carcamo J, Harland R M, Massague J. Nature (London) 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki A, Chang C, Yingling J M, Wang X F, Hemmati-Brivanlou A. Dev Biol. 1997;184:402–405. doi: 10.1006/dbio.1997.8548. [DOI] [PubMed] [Google Scholar]
- 41.Power M A, Tam P P L. Anat Embryol. 1993;187:493–504. doi: 10.1007/BF00174425. [DOI] [PubMed] [Google Scholar]