Abstract
We report the isolation of an empty spiracles class homeodomain-containing gene, Cn-ems, from the hydrozoan Hydractinia symbiolongicarpus, the first gene of this class characterized in a lower metazoan. Cn-ems was found to be expressed in the head of gastrozooids, specifically in endodermal epithelial cells of the taeniolae of the hypostome. Cn-ems is not expressed in gonozooids, which lack taeniolae. Experimental conversion of the posterior region of the planula larva into head structures up-regulates expression of the gene. These findings establish that the association of ems-class genes with head structures preceded the evolution of bilateral symmetry.
The conservation of homeodomain-containing genes and their known roles in specifying positional information in the embryo have reawakened interest in the evolution of development. Of particular note is the finding that many such genes are conserved throughout the animal kingdom, providing legitimate promise that comparative study of patterns of expression may bear on unsolved problems in classical zoology. Here we treat one such problem, the origin of the metazoan head.
Aside from Phylum Porifera, all other metazoans are primitively radially (Cnidaria and Ctenophora) or bilaterally symmetric (all other metazoans) and have a head. If we regard a head as a sensory-rich, mouth-bearing body part placed at or near the anterior pole of the animal, the question of whether the heads of radially symmetric phyla are homologous with those of the bilaterally symmetric phyla is far from obvious. For example, in the radially symmetric Cnidaria, the biradially symmetric Ctenophora, and in the bilaterally symmetric Platyhelminthes, the animal pole of the egg gives rise to the posterior pole of the larva, which in turn gives rise to the mouth-bearing pole of the cnidarian polyp and to the head of the worm (1–6). However, whereas the mouth is positioned at the anterior end of the cnidarian polyp, in many adult flatworms, the mouth is found medially or even at the posterior end of the worm (6, 7). One possibility is that the original polarity of the egg and embryo is conserved but that the position of the mouth and gut has changed relative to this embryonic axis (8). Alternatively, the heads of radially and bilaterally symmetric phyla may have evolved independently. It is, therefore, of particular interest to establish the pattern of expression in radially symmetric organisms of genes whose function has been ascribed to specifying head structures in bilaterally symmetric organisms.
Empty spiracles (ems) is such a gene, first identified in mutant screens in Drosophila (9). Ems mutants display loss of head structures derived from the anterior segments of the fly (10, 11), where ems functions as a gap gene for the head (11, 12). Mouse genes (Emx), identified by sequence similarity with the Drosophila ems homeodomain, also display a head-specific expression in early ontogeny, being expressed in the presumptive cerebral cortex (Emx1 and Emx2) and olfactory regions (Emx2) (13, 14). The association of ems class genes with head structures in both protostome and deuterostome animals led us to characterize its expression in a cnidarian.
MATERIALS AND METHODS
Animals.
Colonies of Hydractinia symbiolongicarpus growing on gastropod shells inhabited by the hermit crab Pagurus longicarpus were obtained from Old Quarry Harbor (Guilford, CT), and Lighthouse Point (New Haven, CT). Fragments of wild-caught colonies containing several polyps were explanted onto glass slides or empty gastropod shells. These stock cultures maintained as a source of eggs, planulae, and tissue were kept in aquaria in artificial seawater (Tropic Marine, Aquarium Systems, Mentor, OH) at 17°C and fed to repletion two to three times per week on brine shrimp (Artemia salina) nauplii. Matings to obtain eggs and planulae were performed as described by Shenk and Buss (15).
Determination of Genomic and Transcript Sequences.
Cn-ems initially was discovered incidentally during a study attempting to identify cnidarian genes homologous to mating-type genes in basidiomycete fungi (16, 17). Total genomic DNA was extracted from H. symbiolongicarpus in urea buffer by using the method of Shure et al. (18). PCR with a mix of degenerate primer sets, including 5′-GTACTGCAGGATCC(AT)C(GT)(AT)GC(AT)(GT)(CT)TAT(GA)AACCA and 5′-GTACTGCAGGATCC(AT)C(GT)(AT)(CT)(GT)(GA)TT(CT)TG(GA)AACCA, amplified a 177-bp fragment of Cn-ems extending from 39 bp upstream of the homeobox to position 138 within the homeobox. This was a purely fortuitous event in that one of these primers bound in the expected region of a homeobox, whereas the other bound nonspecifically upstream of the homeobox. The Cn-ems fragment was among a number of amplification products of varying lengths cloned into pCR2.1 vector (Invitrogen) and manually sequenced (Sequenase Version 2.0 DNA Sequencing Kit, United States Biochemical).
Rapid amplification of cDNA ends (19) was used successfully to obtain the 3′ end of the gene downstream from the initial fragment. Total RNA from H. symbiolongicarpus was extracted in guanidium thiocyanate buffer and purified by ultra centrifugation in cesium trifluoroacetate, and first strand cDNA was prepared as described (ref. 20; see also ref. 21). Gene-specific primers 5′-AGAAACGAAAGCGCCAC and 5′-GCTTTTACACCTACGCAA (positions 829–845 and 852–869 in Fig. 1, respectively) were used in conjunction with rapid amplification of cDNA ends primers Ro and Ri (19), respectively. However, repeated attempts to use ligation-anchored PCR (5′-AmpliFINDER rapid amplification of cDNA ends kit, CLONTECH; see also ref. 22) consistently provided fragments that extended only 25–30 nucleotides upstream of the initial 177-bp PCR fragment.
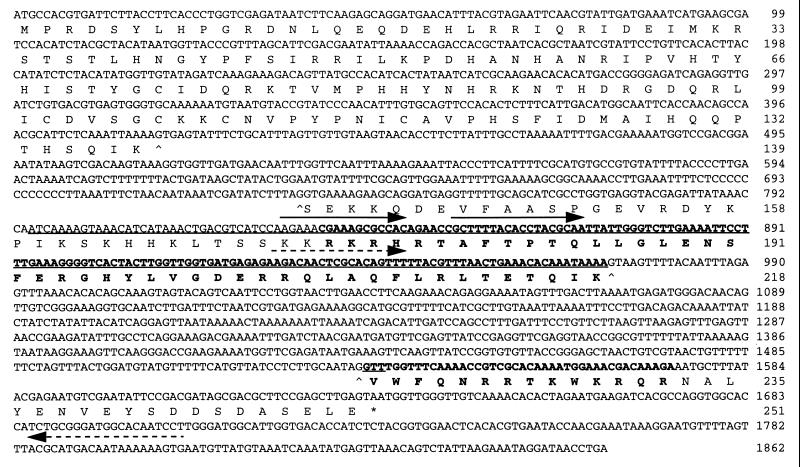
Figure 1.
Nucleotide and predicted amino acid sequence of Cn-ems. Position 1 is the first nucleotide of the putative start codon (see text), and position 1862 is the polyadenylation site. The homeobox and predicted homeodomain are boldfaced. The fragment initially amplified by degenerate primers is underlined. Primers sequences used for 3′ rapid amplification of cDNA ends are overlined by solid arrows, and those used for RT-PCR are overlined by dashed arrows. Exon/intron boundaries are marked (^) beneath the first and last nucleotide of the intron. The stop codon is marked (∗).
An H. symbiolongicarpus genomic library (HindIII digested) was prepared and packaged in λNM1149, host strain ER1647. A Cn-ems cDNA clone of nearly 550 bp, consisting of the homeobox and the 3′ end, was used to generate a radiolabeled probe by random priming (Boehringer Mannheim kit), and 4 × 105 recombinant bacteriophages were screened under stringent conditions at 65°C. A clone containing a 4,100-bp insert was isolated and sequenced from both directions (until an overlap was obtained) on an Applied Biosystems 373A automated sequencer.
Examination of the genomic sequence suggested the position of a methionine start codon. A primer was designed from this region (5′-GTGATTCTTACCTTCACCC, positions 8–26 in Fig. 1) to amplify the 5′ end of Cn-ems from a cDNA pool [prepared by using RNA Isolation kit, Stratagene; mRNA Isolation kit, Dynal (Great Neck, NY); TimeSaver cDNA Synthesis kit, Pharmacia] in conjunction with a primer binding within the homeobox (5′-AGTGACCCCTTTCAAAGG, complement of positions 889–906 in Fig. 1). This 5′ end of Cn-ems cDNA was cloned into pCR2.2 vector (Invitrogen) and sequenced from both directions (completely) on an Applied Biosystems 373A automated sequencer.
Whole Mount in Situ Hybridization.
For use in making riboprobes for in situ hybridization, a 974-bp fragment (henceforth the “full length clone”) extending over the three exon regions from near the putative start codon to near the poly-A tail (positions 8–1859 in Fig. 1) was amplified from a cDNA pool and cloned into pCR2.2 vector (Invitrogen). Aliquots of plasmid DNA were linearized with either NotI or BamHI, and digoxygenin-labeled sense and anti-sense riboprobes were transcribed in vitro by Sp6 or T7 RNA polymerase (Labeling Kit, Boehringer Mannheim). Riboprobes were hydrolyzed to an average length of 150 nucleotides by incubation in 42 mM NaHCO3 and 58 mM Na2CO3 at 60°C for 51 min.
Tissue was fixed for 1 h in 4% paraformaldehyde in 0.1 M Hepes (pH 7.5), 0.42 M NaCl, and 2 mM MgSO4. Whole mount in situ hybridizations were performed as described by Gajewski et al. (23), with one major modification. Instead of a proteinase K treatment (including digestion, digestion-stop, washes, and postdigestion fixation), specimens were heated in PBS/0.1% Tween 20 (PBST) for 5 min at 95°C, chilled on ice, and washed in PBST three times for 5 min at room temperature. The protocol of Gajewski et al. (23) was resumed at the PBST/herring sperm incubation step. Probe concentration was 100–150 ng/ml, and hybridization was carried out at 54°C for 36 h.
Sectioning and Microscopy.
After in situ hybridization and immunochemical visualization, thick transverse sections (≈200 μm) of the hypostome were cut with a microscalpel and mounted in glycerol for light microscopy. Some polyps were embedded in water-soluble JB-4 resin (Polysciences), and thin transverse sections (5–10 μm) were cut with a glass-knife microtome (Reichert-Jung). Alternate thin sections were segregated onto separate slides; one set of slides was stained with standard hematoxylin and eosin to serve as a histological reference for interpreting the hybridization results. Sections were mounted in Permount (Fisher Scientific) and photographed by using a Nikon Optiphot microscope.
Specimens for transmission electron microscopy were fixed in seawater-buffered glutaraldehyde (2.5%), postfixed in 2% OsO4, dehydrated, and embedded in Epon 812. Ultrathin sections (700–900 Å) were prepared by a Reichert-Jung microtome, stained in uranyl acetate (2%) and lead citrate (0.03% in 0.01 N NaOH), and examined in a JEOL 1200-EX electron microscope. Specimens for scanning electron microscopy were fixed and postfixed as above, dehydrated in an alcohol series, and dried by the critical-point method (24).
Experimental Conversion of the Larval Posterior Pole into Head Structures.
Metamorphosin A (MMA) is a member of a naturally occurring family of peptides, the LWamides, known to induce metamorphosis in Hydractinia (23). When exposed to synthetic MMA, Hydractinia planulae have been shown (25) to undergo either complete or partial metamorphosis; in the latter case, the posterior end of the larva is converted into head structures (tentacles, hypostome, and mouth), whereas the anterior end retains the larval form. We used reverse transcriptase (RT)-PCR (see below) to examine the correlation between the time-course of induced head formation and levels of Cn-ems expression. To induce partial metamorphosis, planulae were incubated in 30 μM MMA (in seawater); at this concentration, no complete metamorphoses were observed. From a single batch, 960 planulae were apportioned into four groups of equal number. One group was not induced to metamorphose. Three groups were induced by incubation in 30 μM MMA for 5, 15, and 27 h, respectively, after which mRNA was extracted.
RT-PCR.
For RT-PCR, mRNA was extracted from each of the four groups of planulae described above, as well as from a batch of 650 unfertilized oocytes and a group of 150 gastrozooid polyps from an adult colony (QuickPrep Micro mRNA Purification Kit, Pharmacia), and reverse transcribed with a (dT)12–18 primer (Ready-to-go You Prime First Strand Beads, Pharmacia). Gene-specific primers (5′-AGACAACTCGCACAGTTT and 5′-CACTTTTTTATTGTCATGCGT, positions 927–944 and complement of positions 1785–1805 in Fig. 1, respectively) were designed that would amplify a 318-bp fragment from cDNA and a 879-bp fragment from genomic DNA. PCR amplification was performed on 5 μl (15%) of each cDNA synthesis mix. Reactions were denatured at 94°C, 2 min, cycled 35 times (94°C, 30 s; 47°C, 1 min; 72°C, 30 s), and given a final 10-min extension at 72°C.
We designed actin-specific primers [5′-GACTT(CT)GAACAAGAAATGCA and 5′-TCTTGTTTGGAGATCCACA] from an alignment of published hydroid sequences (Hydra attenuata, GenBank M32364; Podocoryne carnea, GenBank X69058–X69060) for use as a control for uniformity of template concentration and consistency of amplification. These primers were used with equivalent amounts of cDNA template and identical amplification conditions as used in the Cn-ems amplifications.
Amplification products from Cn-ems were separated by electrophoresis on 1% agarose and blotted onto a nylon membrane (ZetaProbe). The membrane was hybridized under stringent conditions, at 65°C, with the full length clone (see above) radiolabeled with 32P by random priming (Boehringer Mannheim Kit).
RESULTS
Genomic Organization and Sequence Analysis of Cn-ems.
Fig. 1 shows the genomic and predicted amino acid sequences of Cn-ems (GenBank Y11836). The translation start site was determined as the first met codon (position 1) within an ORF beginning at position −33 and in frame with the homeobox after intron removal. The extent of the transcription unit in the 5′ direction is uncertain. The coding region is interrupted by two introns, one 317 bp long beginning at position 416 (upstream of the homeobox) and the other 561 bp long beginning at position 972 (toward the 3′ end of the homeobox).
Based on the predicted translation start site, Cn-ems encodes a protein of 251 amino acids with a predicted molecular mass of 29.7 kDa. A homeodomain situated very near the carboxyl-terminal end of the protein shows greatest similarity to ems class homeodomains (Table 1), ranging from 65–70% identity and 80–87% similarity if conservative substitutions are included. The second intron in Cn-ems is close but not identical in position to an intron occurring in human and mouse EMX homeoboxes (26).
Table 1.
Comparison of Cn-ems homeodomain to the most similar known homeodomains
| Gene | Organism* | Homeodomain sequence | Similarity, %† | |
|---|---|---|---|---|
| Antp | RKRGRQTYTRYQTLELEKEFHFNRYLTRRRRIEIAHALCLTERQIKIWFQNRRMKWKKEN | |||
| Cn-ems | Hs | ...H.TAF.PT.L.G..NS.ERGH..VGDE.RQL.QF.R...T...V......T...RQR | ||
| Emx-2 | h, m, zf | P..I.TAFSPS.L.R..HA.EK.H.VVGAE.KQL..S.S...T.V.V......T.F.RQK | 70 | 87 |
| Emx-1 | zf | P..I.TAFSPS.L.R..RA.EK.H.VVGAE.KQL.NG.....T.V.V......T.H.RQK | 70 | 83 |
| Emx-1 | h, m | P..I.TAFSPS.L.R..RA.EK.H.VVGAE.KQL.GS.S.S.T.V.V......T.Y.RQK | 68 | 85 |
| ems | d | P..I.TAFSPS.L.K..HA.ES.Q.VVGAE.KAL.QN.N.S.T.V.V......T.H.RMQ | 65 | 80 |
All sequences are aligned to the Antennapedia homeodomain as a reference sequence. Periods indicate identity with the reference sequence.
Organism abbreviations: d, Drosophila; h, human; Hs, H. symbiolongicarpus; m, mouse; zf, zebra fish.
Similarity of homeodomains to Cn-ems; first column: percentage of identical positions; second column: percentage of identical positions and conservative substitutions. Homeodomains other than Cn-ems were taken from the Swiss-Prot database.
Cn-ems Expression in Polyps.
Cn-ems expression was examined in two polyp types, the gastrozooid and the gonozooid. The head of the gastrozooid is characterized by a dome of tissue, called the hypostome, that extends distally from the zone of tentacle insertion to the mouth (Fig. 2 A and B). The gonozooid is a polyp type specialized for reproduction in which the tentacles are reduced to a ring of buds at the oral end; a hypostome is lacking.
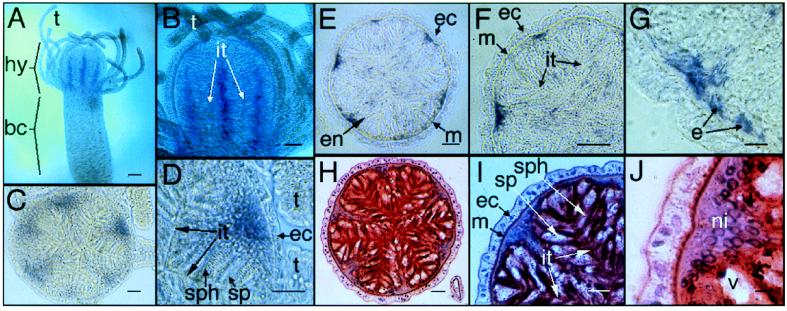
Figure 2.
In situ hybridization of Cn-ems. (A and B) Whole mounts. (A) Longitudinal stripes (purple/blue) representing Cn-ems expression are restricted to the hypostome and are absent from the body-column. (B) Stripes are localized to the center of taeniolae. (C and D) Thick sections. (C) Cross-section of the hypostome area, showing endodermal expression of Cn-ems in the mid-line of taeniolae bases. (D) Close-up of C, showing details of taeniolae. (E–J) Thin sections. (E–G) Immunostaining showing localization of Cn-ems mRNA. (H–J) Hematoxylin and eosin staining. bc, body-column; e, endodermal epithelial cell; ec, ectoderm; en, endoderm; hy, hypostome; it, inter-taeniolae border; m, mesoglea; ni, nuclei; sp, spumeous cell; sph, spherulous cell; t, tentacle; v, vesicle of a spumeous cell. [Magnifications: ×40 (A), ×100 (B), ×200 (C, E, and H), ×400 (D, F, and I), and ×1,000 (G and J); bars = 100 μm (A), 50 μm (B), 20 μm (C–F, H, and I), 5 μm (G and J).]
In gastrozooids, Cn-ems is expressed in endodermal tissues of the hypostome in radially arranged longitudinal stripes, starting just below the tip of the hypostome and increasing in width downward to an abrupt termination at the level of tentacle insertion (Fig. 2 A and B). No expression was seen in the tentacles or in the body-column below the tentacle level. No expression of Cn-ems was observed in gonozooids (data not shown).
The endoderm of the hypostome is populated with three distinct cell types: the endodermal epithelial cells (also referred to as “digestive cells”; refs. 27 and 28) and two types of gland cells, the spumeous and spherulous gland cells (29). Endodermal epithelial cells, which are broad at their basal ends near the mesoglea and narrow to thin processes as they extend inward, line the hypostome with no obvious regional specialization. By contrast, the gland cells, featuring narrow bases near the mesoglea and broadening inwards, occur in a characteristic axial and radial pattern. They are absent in the region directly surrounding the mouth, but, just proximal to the mouth, they are found in a distinctive, radially symmetric arrangement of ridges and furrows. Transverse sections of the hypostome proximal to the mouth reveal this organization (Fig. 2 E–J and Fig. 4 B and C). The ridges, called taeniolae (27, 30, 31), are populated by gland cells alternating with endodermal epithelial cells (27, 28, 32, 33; see Fig. 4 B and C for clarification of cell arrangement within the ridge-and-furrow structure). The furrows between taeniolae are populated by endodermal epithelial cells. The histological complement of taeniolar organization in the hypostome is distinct from that in the body column (28).
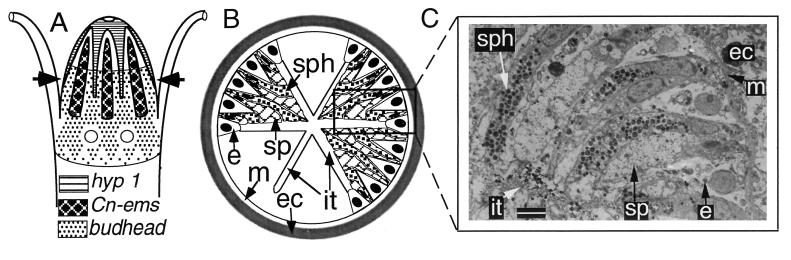
Figure 4.
(A) Hypothetical schematic representation of the relative expression domains of hyp 1, budhead, and Cn-ems in the hydrozoan head, based on descriptions and figures presented independently in the literature for each gene. See text for details. (B) Schematic transverse section of the hypostome at the level indicated by the arrows in A. The inter-taeniolae spaces (it) appear when the polyp feeds (to allow food passage) and are drawn for demonstrative purposes only. (C) Transmission electron microscopy enlargement of the rectangular section indicated in B; (Bar = 3 μm.)
Thick transverse sections (Fig. 2 C and D) show that Cn-ems expression is localized to cells within the basal core of taeniolae. This tissue contains the bases of elongate spumeous and spherulous gland cells as well as endodermal epithelial cells. Thin transverse sections (Fig. 2 E–J) show Cn-ems expression in cells immediately adjacent to the basal gastrodermal boundary and separate from the main cell bodies of differentiated endodermal gland cells. Cn-ems expression appears to be limited to endodermal epithelial cells of taeniolae.
Cn-ems Expression in the Head-Converted Planula Larva.
As described by Leitz et al. (25), treatment with MMA led to the conversion of the posterior end of the planula larvae into head structures. The time course of this conversion is shown in Fig. 3A. The larval posterior begins loss of cilia, axial compression, and thickening by 2-h postinduction. It is markedly shorter and thicker by 4 h and 7 h and almost spherical by 12 h. By 18 h, the posterior has assumed the conical shape typical of the hypostome, and tentacles have appeared at the periphery. The anterior end of the planula shows little overt change during the same period (not shown).
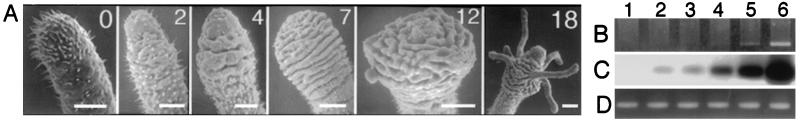
Figure 3.
The onset of Cn-ems expression with respect to metamorphic development and hypostome formation. (A) Scanning electron microscopy photographs of the posterior end of planulae induced to metamorphose by incubation in 30 μM MMA. Incubation time (in hours) is shown in upper-right corner. (Bar = 40 μm.) (B) RT-PCR using Cn-ems-specific primers. (C) Southern hybridization of same gel as in B with the full length clone. (D) RT-PCR using same cDNA pools as in B but actin-specific primers. Source of mRNA: lane 1, 650 unfertilized oocytes; lanes 2–5, groups of 240 planulae each, induced to metamorphose for 0, 5, 15, and 27 h, respectively; and lane 6, 150 polyps from an adult colony.
Expression of Cn-ems was examined by RT-PCR: in oocytes; in planulae undergoing head conversion at 5 and 15 h postinduction; in juveniles at 27 h postconversion; and in mature gastrozooid polyps (Fig. 3 B and C). No expression was detected by autoradiography in the oocytes, and only slight expression was detected in planulae not induced to metamorphose (Fig. 3C). Expression level increased progressively through the 5- to 15-h postinduction stages and the partially metamorphosed planula stage at 27 h and was highest in adult gastrozooids.
Control amplification of ≈420-bp actin gene fragment performed by using amounts of cDNA template from each ontogenetic stage identical to those used to amplify Cn-ems showed uniform levels of amplification product (Fig. 3D). This confirmed that stage-specific differences in amount of Cn-ems amplification product likely were related to differential levels of Cn-ems expression, rather than to variation in efficiency of reverse transcription, total cDNA template amounts, or efficiency of PCR amplification.
DISCUSSION
Sequence similarity in the homeodomain clearly identifies Cn-ems as related to empty spiracles class genes, and its expression in a hydroid head suggests that the association of ems orthologs with head structures preceded the evolution of bilateral symmetry. These findings raise two immediate questions: What role is played by Cn-ems in the organization of the cnidarian head? To what extent do these findings imply that the heads of all eumetazoans are homologous?
Axial and Radial Organization in the Hydrozoan Head Endoderm.
The hydrozoan head is characterized by an axial and radial organization in cell types and head-specific structures. In the endoderm, the axial pattern is manifested by an absence of gland cells at the mouth, below which lies the ridge-and-furrow arrangement of taeniolae, with alternating spherulous and spumeous gland cells restricted to the ridges. Endodermal epithelial cells are evenly distributed radially but display a radially symmetric pattern of cell division, with mitotically active cells limited to the margins of inter-taeniolar furrows (27, 28). An additional radially symmetric feature is the insertion of tentacles at the base of the hypostome, at positions corresponding to inter-taeniolar furrows. The limit of the head is marked both by the basal extent of the tentacle zone and by the anterior boundary of the digestive cavity.
Two genes associated with the axial and radial organization of the hypostome, hyp 1 and budhead, are expressed in endodermal epithelial cells, the cell type that expresses Cn-ems. Together, hyp 1, budhead, and Cn-ems display distinctive patterns of axial and radial organization (Fig. 4). Hyp 1, encoding a product with no significant similarity to any known protein (34), is expressed in a pattern antithetical to that of Cn-ems. Specifically, hyp-1 expression is high at the mouth, where Cn-ems is not expressed, and attenuates aborally, with expression restricted to inter-taeniolar furrows to a limit at the tentacle zone. In contrast, Cn-ems expression attenuates orally and is limited to taeniolar ridges from the upper hypostome into the tentacle zone. Budhead is a recently characterized representative of the forkhead family associated with embryonic “organizers” (35). Budhead is not expressed in the apical part of the hypostome. It is expressed maximally in a circumferential band, spanning both ridges and furrows, just above the zone of tentacles. In the tentacle zone, budhead expression is restricted to the endodermal epithelium of the taeniolae; it is not expressed in those inter-taeniolar regions adjacent to tentacle insertion. Finally, proximal to the tentacle zone, budhead expression resumes its circumferential pattern, although at reduced levels of expression, attenuating in the upper end of the body column. None of these genes is expressed in endoderm of the tentacles.
The association of Cn-ems expression with taeniolate organization is supported further by our finding that gonozooids, which lack taeniolae, do not express Cn-ems. Moreover, Cn-ems expression is enhanced greatly as the axial and radial organization of the head is established during experimental conversion of the posterior region of the larva into head structures. This correlation, coupled with Cn-ems expression restricted to the heads in adult polyps, suggests that Cn-ems also may play a role in developmental patterning of the head. Budhead (35) and hyp 1 (34) both are expressed in regenerating heads, suggesting that these genes may be expressed together with Cn-ems in developing heads. Thus, hyp 1, budhead, and Cn-ems are good candidates for genes responsible for establishing and/or maintaining the axial and radial organization of the hydrozoan head.
Expression patterns of hyp 1, budhead, and Cn-ems are known from two different species of Hydra and one species of Hydractinia, respectively. Amalgamating data from the species provides a composite picture of how these genes may contribute to patterning the hypostome. Verification of this model will require simultaneous examination of expression of these genes in one or more species. The apparent lack of any distinct axial pattern or radial symmetry in the distribution of endodermal epithelial cells in the hydrozoan head belies a distinctive axial and radial organization in expression of hyp 1, budhead, and Cn-ems in these cells. Axial organization, onto which radially symmetrical patterns of expression of particular genes are superimposed, is reflected (Fig. 4) by (i) a zone of expression of only hyp 1 at the top of the hypostome; (ii) a zone below this in which hyp 1 and Cn-ems are expressed in a complementary, nonoverlapping, radially symmetric fashion in hypostomal furrows and ridges, respectively; (iii) a zone just above the tentacles in which budhead is maximally expressed and overlaps both hyp 1 and Cn-ems expression; (iv) the tentacle zone, in which budhead is expressed at low levels, hyp 1 is not expressed, and Cn-ems is expressed in taeniolar ridges; and (v) a zone of low budhead expression extending into, and attenuating in, the upper body column; neither hyp 1 nor Cn-ems is expressed in this region.
Conservation of Head Genes in Radially and Bilaterally Symmetric Metazoa.
Our finding of an association of ems-class genes with head structuring in a cnidarian raises the prospect that the heads of all eumetazoans are homologous. A homology assignment of this order is both premature and exceedingly difficult to make. A statement about homology is a statement about structure, function, and genealogy and often involves statements regarding commonalities in the generative processes (36–39), including common sets of interacting genes. The difficulty lies in determining the appropriate level of homology. A finding that orthologous regulatory genes are both expressed in the head does not alone imply that heads are the appropriate level of homology. Determining the appropriate level of homology entails identifying the morphological features to which the shared genetic determinants correspond. In the case of the heads of radially and bilaterally symmetric taxa, no such obvious candidate presents itself. Subsequent study may reveal such features.
In the case with the cnidarian head, the most progress that currently can be made is to seek to identify additional head-specific genes shared among eumetazoans and to determine their roles. The increasing number of regulatory genes isolated from cnidarians offers several potential candidates. The homeodomain of Cnox-3 from Chlorohydra (40), for example, shows 70% sequence identity to that of Barx1, expressed in part in mouse head (41); 66% identity to the Drosophila brain-specific bsh (42); and 57% identity to Dll, which plays a role in patterning of head structures in Drosophila (43). Budhead, as noted above, is expressed in Hydra from the mid-hypostome to the anterior part of the digestive cavity. In Drosophila, the homolog forkhead is expressed in terminal regions of the embryo that contribute to fore-, hind- and midgut, and in mouse, a homolog HNF3β is expressed in the anterior of the developing gut. Finally, a labial subclass gene recently has been isolated from Hydra and has been shown to display a head-specific expression (H. R. Bode, personal communication). In Drosophila, labial is expressed in both ectodermal and endodermal derivatives in the head region (44–46). The apparent existence of multiple commonalities between head-specific cnidarian genes and genes playing major roles in patterning bilaterian heads supports a hypothesis of homology and suggests that further effort may be rewarded eventually in a clear separation of apomorphy and plesiomorphy.
Acknowledgments
We are grateful to the laboratory of G. Plickert (especially to J. Schloßherr) for guiding us through the intricacies of in situ hybridization of Hydractinia and for kindly providing probes from their research to serve as initial positive controls. Thanks are also due to the laboratory of S. L. Dellaporta (especially to A. Calderon-Urrea and M. Moreno) and to H. R. Bode, A. Grens, and D. E. Martínez for instructive discussions along the research. We thank U. Frank, B. Piekos, Y. Delarea, and V. Wexler for help with microscopy preparations and computer graphics. This work was partly supported by National Science Foundation/Sloan Postdoctoral Fellowship in Molecular Evolution BIR-9510811 (O.M.), National Institutes of Health Training Grant 5T32HD07180-13 (M.D.), and Deutsche Forschungsgemeinschaft Schi 277/10-1 (B.S.).
ABBREVIATIONS
- MMA
metamorphosin A
- RT
reverse transcriptase
Footnotes
Data deposition: The Cn-ems sequence reported in this paper has been deposited in the GenBank database (accession no. Y11836).
References
- 1.Tessier G. Ann Sci Nat Sér Bot Zool. 1931;14:5–60. [Google Scholar]
- 2.Mergner H. In: Experimental Embryology of Marine and Fresh-Water Invertebrates. Reverberi B, editor. Amsterdam: North–Holland; 1971. pp. 1–84. [Google Scholar]
- 3.Freeman G. In: Developmental and Cellular Biology of Coelenterates. Tardent P, Tardent R, editors. Amsterdam: North–Holland; 1980. pp. 97–108. [Google Scholar]
- 4.Skaer R J. In: Experimental Embryology of Marine and Fresh-Water Invertebrates. Reverberi B, editor. Amsterdam: North–Holland; 1971. pp. 104–125. [Google Scholar]
- 5.Rieger R M, Tyler S, Smith J P S, III, Rierger G E. In: Microscopic Anatomy of Invertebrates. Harrison F W, Bogitsh B J, editors. Vol. 3. New York: Wiley–Liss; 1991. pp. 7–141. [Google Scholar]
- 6.Beklemishev W N. Principles of Comparative Anatomy of Invertebrates. 1. Promorphology. trans. MacLennan, J. M. Aberdeen, IL: Univ. Chicago Press; 1969. [Google Scholar]
- 7.Hyman L H. The Invertebrates. III. Platyhelminthes and Rhynchocoela. New York: McGraw–Hill; 1951. [Google Scholar]
- 8.Graff L von. Bronn’s Klassen und Ordnungen des Tierreichs. Leipzig, Germany: C. F. Winter; 1905. [Google Scholar]
- 9.Jürgens G, Wieschaus E, Nüsslein-Volhard C. Roux Arch Dev Biol. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- 10.Dalton D, Chadwick R, McGinnis W. Genes Dev. 1989;3:1940–1956. doi: 10.1101/gad.3.12a.1940. [DOI] [PubMed] [Google Scholar]
- 11.Walldorf U, Gehring W J. EMBO J. 1992;11:2247–2259. doi: 10.1002/j.1460-2075.1992.tb05284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S M, Jürgens G. Nature (London) 1990;346:482–485. doi: 10.1038/346482a0. [DOI] [PubMed] [Google Scholar]
- 13.Simeone A, Gulisano M, Acampora D, Stornaiuolo A, Rambaldi M, Boncinelli E. EMBO J. 1992a;11:2541–2550. doi: 10.1002/j.1460-2075.1992.tb05319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nature (London) 1992b;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- 15.Shenk M A, Buss L W. J Exp Zool. 1991;257:80–86. [Google Scholar]
- 16.Schultz B, Banuett F, Dahl M, Schlesinger R, Schäfer W, Martin T, Herskowitz I, Kahmann R. Cell. 1990;60:295–306. doi: 10.1016/0092-8674(90)90744-y. [DOI] [PubMed] [Google Scholar]
- 17.Gillissen B, Bergemann J, Sandmann C, Schroeer B, Bölker M, Kahmann R. Cell. 1992;68:647–657. doi: 10.1016/0092-8674(92)90141-x. [DOI] [PubMed] [Google Scholar]
- 18.Shure M, Wessler S, Fedoroff N. Cell. 1983;35:225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- 19.Frohman M A, Martin G R. Techniques. 1989;1:165–170. [Google Scholar]
- 20.Dick M H, Buss L W. Mol Phylogenet Evol. 1994;3:146–158. doi: 10.1006/mpev.1994.1017. [DOI] [PubMed] [Google Scholar]
- 21.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 22.Apte A N, Siebert P D. BioTechniques. 1993;15:890–893. [PubMed] [Google Scholar]
- 23.Gajewski M, Leitz T, Schloßherr J, Plickert G. Roux Arch Dev Biol. 1996;205:232–242. doi: 10.1007/BF00365801. [DOI] [PubMed] [Google Scholar]
- 24.Wischnitzer S. Introduction to Electron Microscopy. 3rd Ed. New York: Pergamon; 1981. [Google Scholar]
- 25.Leitz T, Morand K, Mann M. Dev Biol. 1994;163:440–446. doi: 10.1006/dbio.1994.1160. [DOI] [PubMed] [Google Scholar]
- 26.Bürglin T R. In: Guidebook to the Homeobox Genes. Duboule D, editor. New York: Oxford Univ. Press; 1994. pp. 25–72. [Google Scholar]
- 27.Campbell R D. J Embryol Exp Morph. 1967;17:607–616. [PubMed] [Google Scholar]
- 28.Braverman M. J Morph. 1968;126:95–106. doi: 10.1002/jmor.1051260106. [DOI] [PubMed] [Google Scholar]
- 29.Bouillon J. Cahiers Biologie Marine. 1966;7:157–205. [Google Scholar]
- 30.Hamann O. Jena Z Naturw. 1882;15:473–544. [Google Scholar]
- 31.Kühn A. Ergebn Fortschr Zool. 1914;4:1–284. [Google Scholar]
- 32.Hyman L H. The Invertebrates. I. Protozoa through Ctenophora. New York: McGraw–Hill; 1940. [Google Scholar]
- 33.Lentz T L. The Cell Biology of Hydra. Amsterdam: North–Holland; 1966. [Google Scholar]
- 34.Hermans-Borgmeyer I, Schinke B, Schaller H C, Hoffmeister-Ullerich A H. Differentiation. 1996;61:95–101. doi: 10.1046/j.1432-0436.1996.6120095.x. [DOI] [PubMed] [Google Scholar]
- 35.Martínez D E, Dirksen M L, Bode P M, Jamrich M, Steele R E, Bode H R. Dev Biol. 1997;192:523–536. doi: 10.1006/dbio.1997.8715. [DOI] [PubMed] [Google Scholar]
- 36.Roth V L. Biol J Linn Soc. 1984;22:13–29. [Google Scholar]
- 37.Roth V L. In: Ontogeny and Systematics. Humphries C J, editor. New York: Columbia Univ. Press; 1988. pp. 1–26. [Google Scholar]
- 38.Wagner G P. Evolution. 1989;43:1157–1171. doi: 10.1111/j.1558-5646.1989.tb02566.x. [DOI] [PubMed] [Google Scholar]
- 39.Wagner G P. Annu Rev Ecol Syst. 1989;20:51–69. [Google Scholar]
- 40.Schummer M, Scheurlen I, Schaller C, Galliot B. EMBO J. 1992;11:1815–1823. doi: 10.1002/j.1460-2075.1992.tb05233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tissier-Seta J P, Mucchielli M L, Mark M, Mattei M G, Goridis C, Brunet J F. Mech Dev. 1995;51:3–15. doi: 10.1016/0925-4773(94)00343-l. [DOI] [PubMed] [Google Scholar]
- 42.Jones B, McGinnis W. Development. 1993;117:793–806. doi: 10.1242/dev.117.2.793. [DOI] [PubMed] [Google Scholar]
- 43.O’Hara E, Cohen B, Cohen S M, McGinnis W. Development. 1993;117:847–856. doi: 10.1242/dev.117.3.847. [DOI] [PubMed] [Google Scholar]
- 44.Mlodzik M, Fjose A, Gehring W J. EMBO J. 1988;7:2569–2578. doi: 10.1002/j.1460-2075.1988.tb03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merrill V K L, Diederich R J, Turner F R, Kaufman T C. Dev Biol. 1989;135:376–391. doi: 10.1016/0012-1606(89)90187-5. [DOI] [PubMed] [Google Scholar]
- 46.Diederich R J, Merrill V K L, Pultz M A, Kaufman T C. Genes Dev. 1989;3:399–414. doi: 10.1101/gad.3.3.399. [DOI] [PubMed] [Google Scholar]