Abstract
We describe the development, current features, and some directions for future development of the Amber package of computer programs. This package evolved from a program that was constructed in the late 1970s to do Assisted Model Building with Energy Refinement, and now contains a group of programs embodying a number of powerful tools of modern computational chemistry, focused on molecular dynamics and free energy calculations of proteins, nucleic acids, and carbohydrates.
Keywords: Amber, biomolecular simulation programs
Introduction
Molecular dynamics simulations of proteins, which began about 25 years ago, are by now widely used as tools to investigate structure and dynamics under a variety of conditions; these range from studies of ligand binding and enzyme reaction mechanisms to problems of denaturation and protein refolding to analysis of experimental data and refinement of structures. “Amber” is the collective name for a suite of programs that allows users to carry out and analyze molecular dynamics simulations, particularly for proteins, nucleic acids and carbohydrates. None of the individual programs carries this name, but the various parts work reasonably well together, providing a powerful framework for many common calculations.1 The term amber sometimes also refers to the empirical force fields that are implemented here. It should be recognized, however, that the code and force fields are separate; several other computer packages have implemented the amber force fields, and other force fields can be used within the Amber programs.
A history of Amber’s early development was presented about 10 years ago;2 here we give an overview of more recent efforts. Our goal is to provide scientific background for the simulation techniques that are implemented in the Amber programs and to illustrate how certain common tasks are carried out. We cannot be exhaustive here (the Users’ Manual is 310 pages long!), but we do try to give a sense of the trade-offs that are inevitably involved in maintaining a large set of programs as simulation protocols, force fields, and computer power rapidly evolve. There are certain tasks that Amber tries to support well, and these are our focus here. We hope that this account will help potential users decide whether Amber might suit their needs, and to help current users understand why things are implemented the way they are.
It should be clear that this is not a primer on biomolecular simulations, for there are many excellent books that cover this subject at various levels of detail.3-7 More information about Amber itself, including tutorials and a Users’ Manual, is available at http://amber.scripps.edu.
Overview of Amber Simulations
Amber is not a single program, but is rather a collection of codes that are designed to work together. The principal flow of information is shown in Figure 1. There are three main steps, shown top to bottom in the figure: system preparation, simulation, and trajectory analysis. Encoding these operations in separate programs has some important advantages. First, it allows individual pieces to be upgraded or replaced with minimal impact on other parts of the program suite; this has happened several times in Amber’s history. Second, it allows different programs to be written with different coding practices: LEaP is written in C using X-window libraries, ptraj and antechamber are text-based C codes, mm-pbsa is implemented in Perl, and the main simulation programs are coded in Fortran 90. Third, this separation often eases porting to new computing platforms: only the principal simulation codes (sander and pmemd) need to be coded for parallel operation or need to know about optimized (perhaps vendor-supplied) libraries. Typically, the preparation and analysis programs are carried out on local machines on a user’s desktop, whereas time-consuming simulation tasks are sent to a batch system on a remote machine; having stable and well-defined file formats for these interfaces facilitates this mode of operation. Finally, the code separation shown in Figure 1 facilitates interaction with programs written by others. For example, the NAMD simulation program8 can take advantage of Amber tools and force fields by virtue of knowing how to interpret the information in Amber’s prmtop files; similarly, the VMD graphics tools9 can read our trajectory format to prepare animations. The toolkit from the Multiscale Modeling Tools for Structural Biology (MMTSB) project10 can be used to control many aspects of Amber simulations (see Fig. 1). Other users have written tools to improve upon LeAP’s preparation interface, sometimes using a portion of the LEaP code and sometimes bypassing it entirely.
Figure 1.
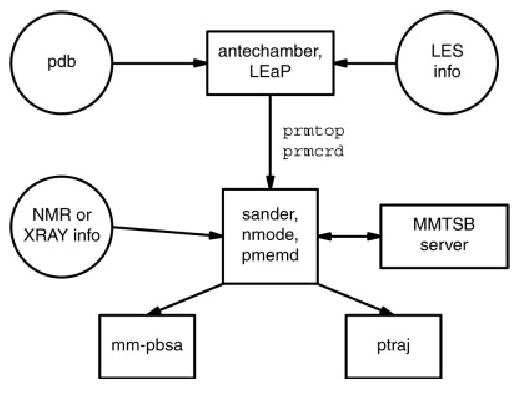
Information flow in the Amber program suite.
Of course, there are also disadvantages in the code fragmentation implied by Figure 1. There is generally no consistent user interface for the various components, which makes it more difficult to learn. Furthermore, there is no (easy) way for code in one section to modify the results of another section. For example, atom types or charges (which are established in the preparation phase) cannot be modified as the simulation proceeds; similarly, the simulation cannot decide which trajectory data to archive based on the sort of analysis to be done, because it does not have any information about that. Despite these limitations, breaking up of the code into distinct pieces has generally served the user and developer communities well.
Preparation Programs
The main preparation programs are antechamber (which assembles force fields for residues or organic molecules that are not part of the standard libraries) and LEaP (which constructs biopolymers from the component residues, solvates the system, and prepares lists of force field terms and their associated parameters). The result of this preparation phase is contained in two text files: a coordinate (prmcrd) file that contains just the Cartesian coordinates of all atoms in the system, and a parameter-topology (prm-top) file that contains all other information needed to compute energies and forces; this includes atom names and masses, force field parameters, lists of bonds, angles, and dihedrals, and additional bookkeeping information. Since version 7, the prmtop file has been written in an extensible format that allows new features or user-supplied information to be included if required.
LEaP incorporates a fairly primitive X-window graphics interface that allows for visual checking of results and for some interactive structure manipulation. Most users, however, find other programs better suited to inspection of PDB files and construction of initial coordinates, and primarily use LEaP in a text mode (tLEaP) to assemble the system from a “clean” PDB-format file that contains acceptable starting coordinates. The nucleic acid builder (NAB) program integrates well with Amber to construct initial models for nucleic acids,11 and the Amber/GLYCAM configurator tool (http://glycam.ccrc.uga.edu/Amber) serves a similar purpose for carbohydrates. Tools for manipulating protein structures (e.g., for constructing homology models) are widespread, and the resulting PDB-format files can generally be processed by LEaP with little or no modification.
Simulation Programs
The main molecular dynamics program is called sander; as with “Amber,” the original acronym is no longer very descriptive. This code is written in Fortran 90, and uses the Fortran namelist syntax to read user-defined parameters as label-value pairs. As one might imagine, there are many possible options, and about 150 possible input variables. Of these, only 32 (identified in boldface in the Users’ Manual) generally need to be changed for most simulations. As much as possible, the default options have been chosen to give good quality simulations.
Sander is a parallel program, using the MPI programing interface to communicate among processors. It uses a replicated data structure, in which each processor “owns” certain atoms, but where all processors know the coordinates of all atoms. At each step, processors compute a portion of the potential energy and corresponding gradients. A binary tree global communication then sums the force vector, so that each processor gets the full force vector components for its “owned” atoms. The processors then perform a molecular dynamics update step for the “owned” atoms, and use a second binary tree to communicate the updated positions to all processors, in preparation for the next molecular dynamics step. Details of this procedure have been given elsewhere.2,12
Because all processors know the positions of all atoms, this model provides a convenient programming environment, in which the division of force-field tasks among the processors can be made in a variety of ways. The main problem is that the communication required at each step is roughly constant with the number of processors, which inhibits parallel scaling. In practice, this communication overhead means that typical explicit solvent molecular dynamics simulations do not scale well beyond about eight processors for a typical cluster with gigabit ethernet, or beyond 16 –32 clusters for machines with more efficient (and expensive) interconnection hardware. Implicit solvent simulations, which have many fewer forces and coordinates to communicate, scale significantly better. For these relatively small numbers of processors, inequities in load-balancing and serial portions of the code are not limiting factors, although more work would have to be done for larger processor counts.
To improve performance, Bob Duke has prepared an extensively revised version of sander, called pmemd, which communicates to each processor only the coordinate information necessary for computing the pieces of the potential energy assigned to it. Many other optimizations were also made to improve single-processor performance. This code does not support all of the options found in sander, but has a significant performance advantage for the most commonly used simulation options. Some pmemd performance numbers may be found at http://amber.scripps.edu/amber8.bench2.html. A similar communications strategy is being added to development versions of sander by Mike Crowley.
The normal mode analysis code, nmode, is by now quite old, but is still useful for certain types of analysis. It is limited to nonperiodic simulations and, as a practical matter, to systems with fewer than 3000 atoms. Its primary use now is to compute estimates of thermodynamic quantities (in particular, vibrational entropies) for configurations extracted from molecular dynamics simulations, as in the mm-pbsa scheme discussed below. Its original purpose, to compute vibrational properties of small molecules used in force-field parameterization, is still relevant as well. The code supports the Amber polarizable potentials, but not (yet) the generalized Born model; second derivatives of GB energies are available in the NAB program (see http://www.scripps.edu/case/nab.html), which has a GB parameterization identical to that of Amber.
Analysis Programs
The task of analyzing MD trajectories faces two main obstacles. First, trajectory files may become very large, and the total time course may need to be assembled from pieces that were computed in different runs of the simulation program. Second, the types of analyses that can be carried out are quite varied, and change with time, as simulation science progresses and as new ideas are developed. The ptraj analysis program (see http://www.chpc.utah.edu/~cheatham/software.html) was designed with these obstacles in mind, although it provides only a partial resolution to them. It can process both Amber and CHARMM trajectories, parsing their respective prmtop or psf files to atom and residue names and connectivity, and can assemble trajectories from partial ones, often stripping out parts (such as solvent) that might not be needed for a particular analysis. After this, a variety of common analysis tasks may be carried out; some of these are illustrated below. The command syntax is designed to allow users to add new tasks, but this requires knowledge of C programming, and is not as straightforward as it might be. But it does provide a powerful framework for adding new commands, and the ptraj repertoire continues to grow; recent additions include a variety of clustering algorithms and variants of principal component analysis.
Our eventual goal is that ptraj will support analyses based on energies as well as structures, but this is not the case in the current codes. Rather, a variety of programs are used to estimate energies and entropies from the snapshots contained within trajectory files. The calculations are organized and spawned by a Perl script, mm-pbsa, which also collects statistics and formats the output in tabular form. As its name suggests, the analysis is primarily based on continuum solvation models.13-15
Overall Strengths and Weaknesses
An overall view of our perception of Amber’s strengths and weaknesses is given in Table 1. In brief, the suite is good at carrying out and analyzing “standard” MD simulations for proteins, nucleic acids, and carbohydrates, using either explicit solvent with periodic boundary conditions or an implicit solvent model. Our aim is to make it easy for users to carry out good-quality simulations, and to keep the codes up to date as standards and expectations evolve. However, some desirable options are missing, and changing the way in which the calculations are performed can require the user to understand and modify a core code that has varying standards of readability. As we prepare new versions of the code, our goal is to ameliorate some of the weaknesses, and to continue to incorporate new simulation techniques, especially those that accelerate convergence of conformational sampling, or which allow us to use force fields that have a greater underlying physical realism.
Table 1.
Strong and Weak Points of the Amber Biomolecular Simulation Programs.
| Strengths | Weaknesses |
|---|---|
| Amber implements efficient simulations with periodic boundary conditions, using the PME method for electrostatic interactions and a continuum model for long-range van der Waals interactions. | One cannot do good simulations of just part of a system, such as the active site of an enzyme: stochastic boundary conditions for the water-continuum interface are missing, as are efficient means for handling long-range electrostatics and a reaction field. |
| Non-periodic simulations are supported, using a generalized Born or numerical Poisson-Boltzmann implicit solvent model. | The component programs lack a consistent user interface; there is only limited scripting capability to support types of calculations not anticipated by the authors. |
| Explicit support is provided for carbohydrate simulations, as well as for proteins, nucleic acids and small organic molecules. | There is limited support for force fields other than those developed by Amber contributors. |
| Free-energy calculations use thermodynamic integration or umbrella sampling techniques, and are not limited to pairwise decomposable potentials. | Missing features include: “dual topology” free energy calculations, reaction-path analysis, Monte Carlo sampling, torsion angle dynamics, and interactive steered molecular dynamics. |
| Convergence acceleration can use locally-enhanced sampling or replica exchange techniques. | QM/MM simulations are limited to semiempirical Hamiltonians, and cannot currently be combined with the PME or generalized Born solvation options. |
| There is a extensive support for trajectory analysis and energetic post-processing. | The codes were written by many authors over many years, and much of it is difficult to understand or modify. |
| Restraints can be very flexible, and can be based on many types of NMR data. | Efficient parallel scaling beyond about a dozen processors may required access to special hardware or the adoption of an implicit solvent model. |
| There is a large and active user community, plus tutorials and a User’s Manual to guide new users. The source code is portable and is available for inspection and modification. | Users are required to compile the programs themselves, and it can be tedious to assemble the needed compilers and libraries. |
No program can hope to do all tasks well, and subjective choices usually need to be made about which capabilities are most needed, and which ones can be supported by the developers. These choices may be expected to change with time, as computers become more powerful, and as algorithms evolve and experience is gained about which sorts of calculations provide the greatest amount of physical realism. Over the years, Amber has discarded a number of features that were once viewed as important parts of its feature set. Some comments about what we no longer support (or support well) may help users better know what to expect from Amber. Some of the features that are now missing are these:
“Vacuum” simulations: Amber is strongly aimed at simulations of biomolecules in water. Although it is still possible to use the codes in situations where there is no solvent environment, we no longer implement this capability in a manner that is as efficient or robust as in earlier versions of the code. The generalized Born (or numerical Poisson–Boltzmann) continuum solvent models allow for more realistic (albeit more expensive) simulations for cases where an explicit treatment of solvation is not appropriate. This lack of full support for solvent-free simulations can be a genuine limitation for cases where coarse-grained, statistical, or other “effective” potentials are appropriate, and future versions of Amber may reintroduce some of this older functionality.
Simulations with nonbonded cutoffs: simulations that ignore long-range nonbonded interactions (particular for electrostatics) often exhibit biased behavior and artifacts that are difficult to predict or correct for. For many types of simulations, it is feasible to include all long-range electrostatics by means of a particle-mesh Ewald (PME) scheme (discussed below), with a simulation cost that is comparable to (or even less than) schemes that do include cutoffs. Given this, Amber neither needs nor implements cutoff-based switching or smoothing functions. In the generalized Born model, electrostatic effects are much less long ranged (especially when a nonzero concentration of added salt is modeled), and Amber can use cutoffs for this situation, although recommended cutoffs are still fairly large (ca. 15 Å).
Free energy simulations for pairwise-decomposable potentials: classical molecular mechanics potentials allow the energy to be written as sums of terms that primarily depend upon two atoms at a time. Free energy calculations that involve changes to only a small part of the system (such as a single amino acid side chain) can exploit this feature by calculating only the small number of terms that change when accumulating free energy differences. This efficiency comes at the expense of some complex bookkeeping, and in any event breaks down when the energy is not written in this pairwise form; the latter, however, is the case for GB or PB theories, for polarizable potentials, and for the “reciprocal space” portion of PME simulations. Because these more modern techniques are the ones we recommend, we have dropped development of the gibbs module that implemented free energy calculations for pairwise potentials. With perhaps less justification, we have also dropped support for free energy perturbation (FEP) calculations, in favor of thermodynamic integration (TI) techniques that are (in most cases) more robust and efficient, and are certainly easier to program and debug.
Force Fields for Biomolecular Simulations
It has long been recognized that the accuracy of the force field model is fundamental to successful application of computational methods. The Amber-related force fields are among the most widely used for biomolecular simulation; the original 1984 article16 is currently the 10th most-cited in the history of the Journal of the American Chemical Society, and the 1995 revision17 has been that journal’s most-cited article published in the last decade. But this widespread use also means that some significant deficiencies are known, especially in terms of the relative stabilities of α-helical vs. extended conformations of peptides.18 -20 There are good recent reviews of the Amber force fields for proteins21 and for nucleic acids.22 These discuss some of the trade-offs that were made in constructing the force fields, and provide comparisons to other potentials in common use. For this reason, we will not summarize this information here. Rather, we will focus on two more recent aspects of the Amber force fields: applications to carbohydrates, and the facilities for defining a model in which part of the system is treated as a quantum subsystem, embedded in a (generally) larger environment described by molecular mechanics force fields.
Amber also supports a more generic force field for organic molecules, called GAFF (the general Amber force field).23 The antechamber program takes a three-dimensional structure as input, and automatically assigns charges, atom types, and force field parameters. Given the wide diversity of functionality in organic molecules, it is not surprising that the resulting descriptions are sometimes not optimal, and “hand-built” force fields are often required for detailed studies. Nevertheless, the tables that drive the antechamber program continue be improved as we gain experience with their weak points. The primary application so far of the GAFF potentials has been to the analysis of complexes of small molecules with proteins or nucleic acids, especially for the automated analysis of diverse libraries of such ligands. The QM/MM facility described below can also be used to describe fairly arbitrary organic molecules in the presence of a receptor described in terms of molecular mechanics potentials.
It is worth noting that the next generation of force fields for proteins and nucleic acids is likely to be significantly more complex than the ones we use today. These coming implementations will certainly have some representation of electronic polarizability,24 -26 and are also likely to include fixed atomic multipoles or off-center charges to provide more realistic descriptions of the electron density. There may also be more complex descriptions of internal distortions (especially for the peptide group27) and terms beyond Lennard–Jones 6 –12 interactions to represent exchange–repulsion and dispersion. A key goal for future development of Amber is to support efficient and parallel simulations that track these new developments.
Applications to Carbohydrates
In eukaryotes, the majority of proteins are glycosylated, that is, they have carbohydrates covalently linked to their surfaces, either through the amido nitrogen atom of asparagine side chains (known as N-linked glycosylation) or through the hydroxyl oxygen atoms of serine or threonine side chains (O-linked). These modifications serve a multitude of roles, spanning the purely structural, such as protecting the protein from proteolytic degradation, to the functional, such as enabling cell adhesion through carbohydrate-protein binding. Computational methods can assist in the interpretation of otherwise insufficient experimental data, and can provide models for the structure of oligosaccharides and insight into the mechanisms of carbohydrate recognition.
Oligosaccharides frequently populate multiple conformational families arising from rotation about the glycosidic linkages (see Fig. 2). As a result, they do not generally exhibit well-defined tertiary structures. A simulation time scale that may be adequate for establishing the performance of a force field for folded proteins cannot be expected to be appropriate for oligosaccharides, whose conformational lifetimes are on the order of 5–10 ns. In this regard, oligo- and polysaccharides behave more like peptides, and force field validation must be based on properties computed from structural ensembles.
Figure 2.
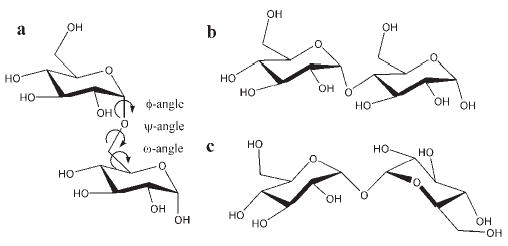
Illustration of 3 of the 10 possible disaccharides generated from linking two α-D-glucopyranosyl residues (α-D-glcp): (a) α-D-glcp-(1→6)-α-D-glcp (iso-maltose), (b) α-D-glcp-(1→4)-α-D-glcp (maltose), and (c) α-D-glcp-(1→1)-α-D-glcp (α,α-trehalose). Conformation-determining glycosidic torsion angles are indicated in (a).
The GLYCAM force field introduced to Amber all of the features that were necessary for carbohydrate conformational simulations, with the key focal points being treatment of glycosidic torsion angles and nonbonded interactions.28 Many valence terms, with the exception of those directly associated with the anomeric carbon atom [notably R(C1–O5), R(C1–O1) and θ(O5–C1–O1)], were taken from the parm94 parameter set, as were all van der Waals terms. The force constants for the newly introduced valence terms were derived by fitting to quantum data at the HF/6-31G* level.
To address the electrostatic properties unique to each monosaccharide, as fully as possible in a nonpolarizable framework, all releases of GLYCAM have employed residue-specific partial atomic charges. In versions of GLYCAM up to and including GLYCAM04, these charges were computed by fitting to the quantum mechanical electrostatic potential computed at the HF/6-31G* level for the methyl glycoside of each residue in each anomeric configuration. For example, the partial charges on the atoms in the glucopyranose methyl α-D-glcp are distinct from those on the atoms in methyl β-D-glcp (see Fig. 3).
Figure 3.
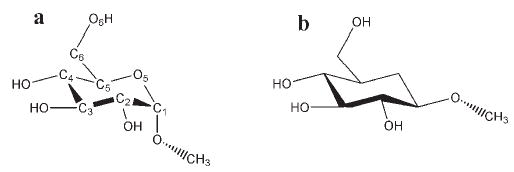
Numbering and anomeric configuration in (a) methyl α- and (b) methyl β-D-glucopyranoside. In GLYCAM, anomeric carbon atom C1 is atom type AC in (a) and atom type EC in (b); in GLYCAM04 both are atom type CG.
Similarly, the structures and partial charges in glucopyranose are distinct from those in mannopyranose (manp) and galactopyranose (galp) etc. Due to the potential for rotations of the hydroxyl groups to influence the electrostatic properties, beginning in 200029 the partial charges were no longer computed from the neutron diffraction structures of the methyl glucosides, but from an ensemble of 100 configurations extracted from solvated MD simulations of the glycosides. The charges were computed at the same quantum level as earlier, consistent with the philosophy of AMBER, but were based on the HF/6-31G* optimized geometries of the ensemble of conformations. It should be noted that the torsion angles of the exocyclic groups were restrained during the geometry optimizations in the solvation preferred orientations. Although the charges were derived at the HF/6-31G* level, the fitting was performed with a larger RESP restraint weight (0.01) than that employed in fitting the charges for the amino acids in AMBER (0.001). Simulations of the crystal lattices of monosaccharides led to the inescapable conclusion that the HF/6-31G* ESP charges were too polar and required the larger damping afforded by the higher restraint weight.30 Although affecting the strengths of direct hydrogen bonds, this damping has negligible effect on molecular dipole moments and ensuing long-range electrostatics.
As in the case of peptides, the interresidue rotational properties (the positions of minima, their relative energies, and interconversion barriers) have a direct influence on the 3D structure and dynamics of an oligosaccharide. In the majority of carbohydrates these rotational properties are associated with only three atomic linkages, emanating from the anomeric carbon atom (Fig. 2).
The first of these is known as the φ-angle, and has been the subject of extensive theoretical and experimental examination because of its unique rotameric properties. In pyranosides, the φ-angle refers to the orientation of the exocyclic O5–C1–O1–Cx sequence. The hyperconjugation associated with the O–C–O sequence imparts a preference for this linkage to adopt gauche orientations, rather than populating all three potential rotamers. This preference is known as the exo-anomeric effect.31 The population of the gauche rotamers differs in α- and β-pyranosides because of further steric effects introduced by the pyran ring system. For this reason, in GLYCAM the rotational properties of the φ-angle were incorporated with a separate torsion term for α-and β-pyranosides. Each term was individually derived by fitting to the rotational energy curves for 2-methoxytetrahydropyran (2-axial serving as a model for α-pyranosides and 2-equatorial for β-pyranosides). All torsion terms associated with the φ-angle, other than those for the O5–C1–O1–Cx sequence, were set to zero. This approach necessitated the introduction of two new atom types for the anomeric carbon in pyranosides, one for use when the linkage was present in an equatorial configuration (atom type EC, generally corresponding to β-pyranosides) and one for axial (AC, generally corresponding to α-).
The second important torsion angle, the ψ-angle, is associated with the C1–O1–Cx–Hx sequence and does not exhibit any profound stereoelectronic properties that are unique to carbohydrates. As such, it was treated with the parameters applicable to ether linkages of this type.
The most problematic carbohydrate-specific torsion term for hexopyranoses is that associated with the exocyclic rotation of the hydroxymethyl group. This O6 –C6 –C5–O5 linkage is known as the ω-angle, and plays a crucial role in defining the 3D structures of any oligosaccharide containing a 16 linkage. The tendency for the O–C–C–O sequence to preferentially adopt a gauche orientation has been associated with the more general gauche effect.32 In the gas phase it arises primarily from hydrogen bonding between the vicinal hydroxyl groups; however, there is a small contribution from hyperconjugation that is further stabilizing. In the condensed phase, its origin is more complex.33 In solution, in pyranosides in which both C6 and O4 are equatorial (most notably in gluco- and mannopyranose), the O6 –C6 –C5–O5 torsion angle adopts nearly exclusively one or the other of the two gauche orientations, known as gauche– gauche (gg) or gauche–trans (gt); labels that refer to the orientation of the O6 –C6 –C5–O5 and O6 –C6 –C5–C4 angles, respectively. Remarkably, when O4 is axial (as in galactopyranose) all three rotamers are populated, with a significant amount of the trans (tg) rotamer being observed.34 To address these issues much computational focus has been given to the O–C–C–O linkage.35-37 In gas phase quantum calculations it is only when intramolecular hydrogen bonding is disallowed that the rotational energy curves agree even qualitatively with the experimental data (see Fig. 4).37 In a combined quantum and simulation study it was concluded that the gauche effect in carbohydrates arose solely from the attenuation of internal hydrogen bonding by interactions with explicit solvent.37
Figure 4.
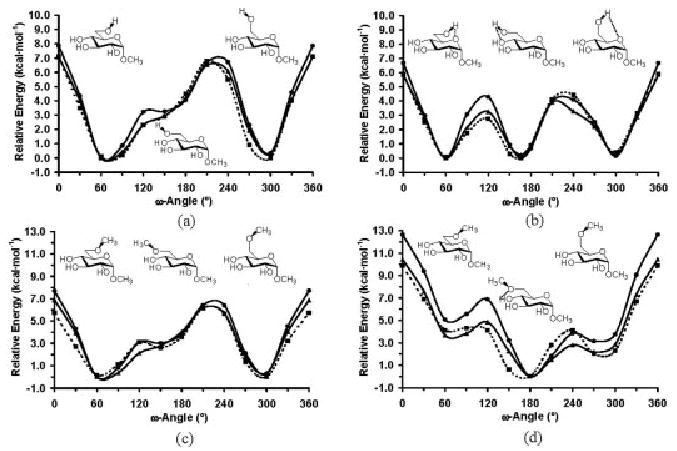
HF/6-31G(d), B3-LYP/6-31++G(2d,2p), and GLYCAM ω-angle rotational curves for methyl α-D-glucopyranoside, without (a) and with (b) internal hydrogen bonding; and methyl 6-O-methyl-α-D-glucopyranoside, without (c) and with (d) internal hydrogen bonding.
To parameterize this property into the GLYCAM force field it was necessary to ensure that the quantum mechanical rotational properties of the O–C–C–O linkage, in the presence and absence of internal hydrogen bonding, could be reproduced. This presented a more serious challenge than might first be expected, largely because of the difficulty in achieving a balance between the strengths of the internal nonbonded energies associated with interactions between O6, O5, and O4. In pyranosides, the O6 . . . O4 interaction is a 1–5 type, while the O6 . . . O5 is a 1– 4. Yet, geometrically, the relevant interatomic distances, both in the presence and absence of internal hydrogen bonding are similar for each case. Thus, the practice in AMBER of damping the magnitude of 1– 4 nonbonded interactions relative to all others by applying a scale factor (of between 0.5– 0.83) introduced an artificial imbalance in the strengths of the O6 . . . O4 and O6 . . . O5 interactions in carbohydrates. It was only when the GLYCAM parameters were derived in the absence of 1– 4 scaling that quantitative agreement with the experimental solution rotamer populations for the ω-angle could be achieved.37
Although adequate for a great variety of carbohydrate systems, the presence in GLYCAM of a unique atom type for the anomeric carbon in α- and β-glycosides meant that it was not possible to simulate processes, such as ring flipping, in which the substituents at the anomeric center changed configuration. This is irrelevant to most pyranosides, which populate only one chair form in solution, but some (like the monosaccharide idopyranose, present in heparin) exist as an equilibrium between the 4C1 and 1C4 chair forms. Further, many carbohydrate processing enzymes are believed to distort the 4C1 chair so as to lower the activation energy. To study these systems it was necessary to derive a parameter set that employed the same atom type for the anomeric carbon in all pyranosides. This was the motivation behind the development of GLYCAM04. Recalling that new anomeric carbon atom types had been introduced to facilitate parameterization of the rotational φ-angle energy curves, it was these parameters that were again the focus of the parameter development. To achieve adequate reproduction of the quantum rotational curves, it was no longer possible to employ only torsion term for the O5–C1–O1–Cx linkage. To employ a single atom type for C1 in GLYCAM04, it was necessary to introduce torsion terms for each of the relevant linkages associated with the φ-angle, namely for the O5–C1–O1–Cx, C2–C1–O1–Cx, and H1–C1–O1–Cx linkages. Unambiguous partitioning of the rotational energies into contributions from each of these sources necessitated a detailed study of the underlying sequences in model structures. This undertaking ultimately led to a completely new set of valence and torsion terms, each fit to quantum data, largely now at the B3LYP/cc-pVTZ level. Partial atomic charges remain as computed earlier. In the course of the development of GLYCAM04, it was decided to remove all default torsion parameters and introduce quantum-derived torsion terms for all linkages found in carbohydrates. This is a paradigm shift that has many advantages; most notable being that it results in a transferable and generalizable force field that enables the introduction of functional groups and chemical modifications into a carbohydrate, without the need to develop a large number of new parameters. However, for a given linkage, the sum of the general torsion terms employed in GLYCAM04 may not be as precise as the explicit fitting employed in GLYCAM. Nevertheless, the accuracy of the GLYCAM04 rotational curves can be rationally tuned, and the parameters extended in ways that were all but impossible in GLYCAM. Whereas the GLYCAM parameters augmented the PARM94 AMBER parameters, the GLYCAM04 parameters are self-contained. To maintain orthogonality with the AMBER protein parameter sets, atom type CT was renamed in GLYCAM04 to CG. In principle, the GLYCAM04 parameters could be extended to generate a completely generalizable force field for proteins.
Currently under development, in both the GLYCAM and GLY-CAM04 formats, are TIP5P compatible parameters that include TIP5P-like lone pairs on the oxygen atoms, and polarizability parameters that employ quantum-derived atomic polarizabilities. An interactive tool (http://glycam.ccrc.uga.edu) has been introduced that greatly facilitates the preparation of the files necessary for running MD simulations with AMBER of proteins, glycoproteins, and oligosaccharides. Further intergration of this facility into the Amber codes is planned, as is continued work on testing and improvement of the GLYCAM force fields.
The QM/MM Approach
Prior to AMBER 8, the QM/MM module was called Roar. This coupled an earlier version of the Amber energy minimization/MD module with Mopac 6.0. As a part of modernization efforts, we wanted to merge more things into the sander program, and we recognized that semiempirical technology had advanced well beyond what was available in Mopac 6.0, particularly in the area of linear-scaling approaches. Because the theory and application of the QM/MM approach38,39 has been extensively reviewed,40 – 42 we only give a brief description of our approach.
The combination of quantum mechanics and molecular mechanics is a natural approach for the study of enzyme reactions and protein–ligand interactions. The active site or binding site is treated by the ab initio density functional theory or semiempirical potentials, whereas the rest of the system is calculated by the force fields based on molecular mechanics. In the current version of sander, one can use the MNDO, AM1, or PM3 semiempirical Hamiltonian for the quantum mechanical region. Interaction between the QM and MM regions includes electrostatics (based on partial charges in the MM part) and Lennard–Jones terms, designed to mimic the exchange-repulsion terms that keep QM and MM atoms from overlapping.
Standard semiempirical molecular orbital methods are widely used in QM/MM applications because they are able to provide fast and reliable QM calculations for energies and molecular properties. However, these methods are still hampered by the need for repeated global matrix diagonalizations in the SCF procedure, resulting in computational expense scaling as O(N3), where N is the number of basis functions. If the QM part is extended to several hundred atoms, it becomes increasingly difficult to apply the standard semiempirical MO method for QM/MM calculation.
This expense can be greatly reduced with a linear scaling approach43,44 based on the density matrix divide and conquer (D&C) method.45 In this approach, global Fock matrix diagonalization and cubic scaling are avoided by decomposing a large-scale electronic structure calculation into a series of relatively inexpensive calculations involving a set of small, overlapping subregions of a system. Each subsystem consists of a core, surrounded by inner and outer buffer regions, and an accurate global description of the system is obtained by combining information from all subsystem density matrices. When the system is large enough, the so-called crossover point is reached, the D&C calculation becomes faster than the standard MO calculation. The accuracy of the D&C approximation can be controlled by the two buffer sizes. According to our experience, the subsetting scheme with the inner and outer buffer layers of 4.0 ± 0.5 Å and 2.0 ± 0.5 Å, respectively appear to be a good compromise between speed and accuracy.46
Figure 5 shows SCF cycle timings for standard and D&C calculations for different QM region sizes studied in the bovine α-chymotrypsin system complexed with 4-fluorobenzylamine. The crossover point occurs at about 170 QM atoms. After this point, the D&C calculation is significantly faster than the standard calculation.
Figure 5.
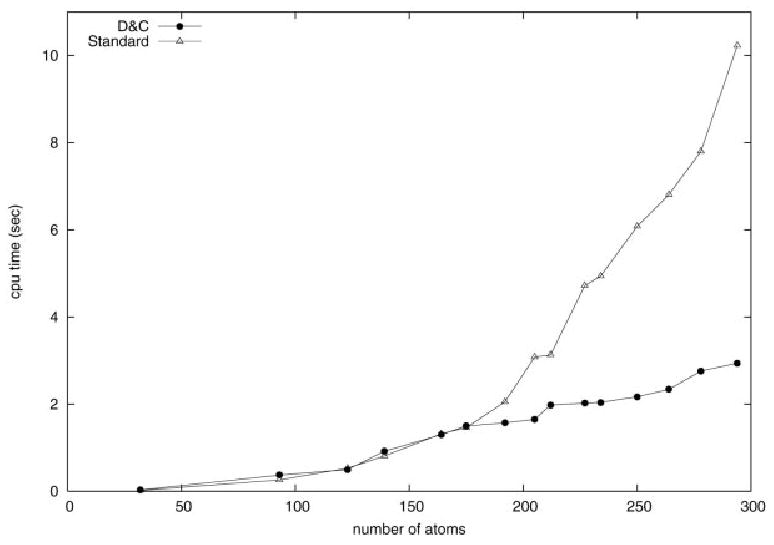
Timings for semiempirical calculations of chymotrypsin, as a function of the size of the QM region.
In some cases, the partitioning of the whole system into QM and MM parts involves the cutting of covalent bonds and raises the question of how to best model the interface between the classical and quantum subsystems. Although many approaches have been proposed to answer this question,47,48 the link atom method is still the most commonly used due to its simplicity. Generally hydrogen atoms are added in the QM part at the covalent bonds cut by the QM/MM interface. Although the link atom approach is subject to the criticism that it partitions the systems in an unphysical manner, results from the link atom approach, if carefully selected, are not so different from other approaches.49 Moreover, because the linear-scaling D&C QM treatment is available, we can readily make the QM part so large that the link atom effect is negligible for the region of interest.
Amber versions under development will allow QM regions to be present when the PME or generalized Born options are chosen, and will be significantly faster, at least for small quantum regions. For protein–ligand complexes, it is natural to treat the ligand and associated residues in the binding site as the QM part, and the rest as the MM part,50 and this facility will be further integrated into the codes.
Treating Solvent Effects
Explicit Solvent Models
Amber provides support for the TIP3P,51 TIP4P and TIP4P-Ew,52-54 TIP5P,55 SPC/E,56 and POL357 models for water, as well as solvent models for methanol, chloroform, N-methylacetamide, and urea/water mixtures. General triclinic unit cells can be used, although some of the analysis and visualization tools are limited to the most common (rectangular and truncated octahedron) shapes; there is no support for symmetry elements (such as screw axes) that involve rotations. By default, electrostatic interactions are handled by a particle-mesh Ewald (PME) procedure, and long-range Lennard–Jones attractions are treated by a continuum model. This gives densities and cohesive energies of simple liquids that are in excellent agreement with more elaborate methods,54 at a computational cost that is often less than that of cutoff based simulations.
The PME is, as the name suggests, a modified form of Ewald summation that is inspired by and closely related to the original Hockney–Eastwood PPPM method.58 Ewald summation is a method to efficiently calculate the infinite range Coulomb interaction under periodic boundary conditions (PBC), and PME is a modification to accelerate the Ewald reciprocal sum to near linear scaling, using the three dimensional fast Fourier transform (3DFFT).
Because the Coulomb interaction has infinite range, under PBC particle i within the unit cell interacts electrostatically with all other particles j within the cell, as well as with all the periodic images of j. It also interacts with all of its own periodic images. The electrostatic energy of the unit cell, and related quantities such as forces on individual particles, are found by summing the resulting infinite series. This latter converges (slowly) to a finite limit only if the unit cell is electrically neutral, and furthermore, the limit is found to depend on the order of summation (conditional rather than absolute convergence). Ewald59 applied a Jacobi theta transform to convert this slowly, conditionally convergent series to a pair of rapidly, absolutely convergent series, called the Ewald direct and reciprocal sums. The conditional convergence of the original series is expressed in a third term60 as a quadratic function of the dipole moment of the unit cell, whose form depends on the order of summation. It is standard to assume that the whole assembly of unit cells is immersed in an external dielectric. Most commonly this dielectric is assumed to be fully conducting (“tin-foil” boundary conditions), in which case the third term vanishes and the order of summation becomes irrelevant. A more elementary derivation of the Ewald sum, using compensating Gaussian charge densities together with Poisson’s equation under PBC in place of the Jacobi theta transform, can be found in the appendix to Kittel.61
The Ewald direct sum resembles the standard Coulomb interaction, but with the term qiqj/rij, representing the Coulomb energy of interaction between particles i and j, replaced by qiqj erfc(βrij)/rij, where β is the so-called Ewald convergence parameter. This latter term involving erfc converges rapidly to zero as a function of the interparticle distance rij, allowing the use of a finite cutoff. In the sander and pmemd programs the Ewald direct sum is calculated together with the van der Waals interactions. The default value of the direct sum cutoff is 8 Å, independent of system size. Accordingly, β is chosen to be ≈0.35 Å−1, leading to a relative RMS force error due to truncation below 5 × 10−4.
The Ewald reciprocal sum is the sum, over all reciprocal lattice vectors m, (m ≠ 0), of a Gaussian-like weight factor exp(−π2m2/β2)/(2πm2) multiplied by |S(m)|,2 where the so-called structure factor S(m) is given by the sum of qjexp(2πim · rj) over all particles j in the unit cell (rj is the Cartesian coordinate vector of particle j). A cutoff can also be applied to the reciprocal sum. With the above choice of β, the number of reciprocal vectors needed so that the relative RMS force error due to truncation is below 5 × 10−4 is typically several times the number of particles in the unit cell. Because the computational cost of calculating S(m) is of order N for each such vector m, the cost of the reciprocal sum is thus of order N.2 Unfortunately, this cost becomes prohibitive for systems containing tens of thousands of particles as is typical today.
What the PME algorithm does is to accurately approximate the structure factors S(m) using the 3DFFT. The essential idea is to first note that exp(2πim · rj) can be factored into three one-dimensional trigonometric terms (even in triclinic unit cells). One can then simply apply table lookup to these terms, approximating the trigonometric functions (evaluated at the crystallographic fractional coordinates of particle j) in terms of their values at nearby grid points. By this means the structure factors are approximated as sums over regular grid points, that is as a discrete Fourier transform that can be rapidly calculated using the 3DFFT, delivering all the needed structure factors at order Nlog(N) computational cost.
The original version62 of the PME utilized Lagrange interpolation to do the table lookup of trigonometric functions. The Ewald reciprocal sum forces were approximated separately from the energy. The smooth PME (SPME)63 replaced Lagrange polynomials with cardinal B-splines, which are better behaved at higher order, and which can be differentiated via recursion, allowing forces to be derived analytically. Thus, in SPME, the forces are obtained from the gradient of the energy, unlike in the original PME. It is also possible to use B-splines to separately approximate the forces, and used in this mode, PME becomes essentially identical to PPPM as implemented by Pollock and Glosli64 (originally we used B-spline interpolation, but least-squares B-spline approximation, advocated by Pollock and Glosli, yields superior accuracy, so we adopted it beginning with version 6 of Amber65). Separate force approximation has the advantage that the resulting forces satisfy Newton’s 2nd law, thus conserving momentum. The accuracy is also better than that of SPME. However, the discrepancy in momentum conservation with SPME is of the order of the force error, which so far appears to be adequate for typical biomolecular simulations (there may yet be situations in which more precise momentum conservation is important). Furthermore, SPME requires half as many 3DFFTs, and thus when used with the standard defaults in sander or pmemd is more efficient than the method of separate force approximation. Finally, the SPME approach of differentiating the approximate reciprocal sum electrostatic potential using the properties of B-splines is critical for efficient approximation of Ewald sums involving atomic dipoles and multipoles (see below). For these reasons we have pursued the SPME approach in recent sander versions and in pmemd.
Over the last 25–30 years objections have sometimes been raised to the use of Ewald summation in liquid state simulations. In the early 1980s, it was demonstrated that simulations of liquids such as water, using Ewald summation, led to calculated properties in quantitative agreement with those of similar simulations using reaction field boundary conditions.60 This alleviated many of the early concerns. More recently, Hummer et al.66 demonstrated that ionic charging free energies could be calculated accurately using Ewald summation, and that the finite size effects, due to limited unit cells, could be accounted for. The picture emerged that simulations of biomolecules in water, using Ewald summation in sufficiently large unit cells, accurately represented the idealized state of the isolated biomolecule in solution. Poisson–Boltzmann calculations under PBC vs. nonperiodic boundary conditions have been used67,68 to estimate the artifacts in conformational free energy due to finite unit cell size (that is, artificial preference for certain conformations due to the imposition of strict PBC within a limited unit cell). From these it is clear that small but nonnegligible finite size artifacts exist, which would be particularly important in protein folding studies and systems containing a low dielectric region (such as lipid bilayers). The cure within Ewald summation (or any modified Ewald method such as PME) is to enlarge the unit cell, which, of course, incurs more computational cost. A number of alternative treatments of long-range electrostatics have been proposed. These typically involve a modified form of cutoff of standard Coulombic interactions, possibly including an approximate treatment of the field due to charges outside the cutoff, as in reaction fields type methods. None of these alternative approaches have been demonstrated to be reliably superior to Ewald summation. Because Ewald summation in a large unit cell is known to be correct, superiority would mean that the alternative method gave the same results as Ewald summation in a large unit cell, using, however, a smaller unit cell. Notably, these alternative treatments all lead to spherically isotropic interaction potentials [An exception, for slab boundary conditions, is the recent method of Wu and Brooks (X.W. Wu, personal communication)]. This spherical isotropy can be expected to be problematic in membrane simulations and other cases involving nonisotropic dielectric media.69,70
Force fields designed for macromolecules have until recently modeled the electrostatic interaction between molecules using fixed atomic point charges. This approximation is thought by many to be the principal limitation in current macromolecular force fields. Recent AMBER forcefields have begun to advance beyond this electrostatic model by introducing off-center charges and inducible atomic dipoles71,72 to account for the nonisotropy of the electron density near atoms and for the inductive response of that density to the local electric field, which changes dynamically as a function of the changing configuration of the system.
The introduction of atomic dipoles necessitates a generalization of the standard Ewald sum, and hence, of the PME algorithm. The generalization of Ewald summation was provided by Smith,73 who gave explicit formulas for Ewald summation of atomic multipoles up to quadrupole level. Smith’s methodology was utilized by Toukmaji et al.74 for performing the Ewald sum involving fixed atomic charges and inducible dipoles, and is implemented in this form into sander. More recently, it was used by Ren and Ponder75 for the AMOEBA force field that includes fixed atomic multipoles up to quadrupole level as well as inducible atomic dipoles using a Thole damping model.76 We have now implemented Ewald summation as well as SPME for atomic Cartesian multipoles up to hexadecapole level,77 using the McMurchie Davidson recursion78 for the Ewald direct sum in place of Smith’s formalism.
Generalizing the SPME to the case of Cartesian multipoles is straightforward. One notes that the Ewald reciprocal sum structure factors now involve a sum of products of Cartesian multipole moments of particle j multiplied by the appropriate derivative, with respect to rj, of the function exp(2πim · rj). Again, these moments and derivatives can all be reexpressed in terms of the crystallographic fractional coordinates of particle j. The function exp(2πim · rj) as well as its successive derivatives with respect to fractional coordinates are approximated in terms of B-splines and their successive derivatives. In practice these derivatives, multiplied by the corresponding transformed moments, are summed onto the PME grid. This grid is then passed to the 3DFFT, modified in reciprocal space, and passed to the inverse 3DFFT precisely as in the standard charge-based SPME algorithm. The reciprocal electrostatic potential is then obtained at an atom i precisely as in the charges only case, and derivatives of this with respect to fractional coordinates of i are multiplied against transformed Cartesian atomic multipole moments of i for all atoms i to get the energy and forces. Due to the above mentioned factorizability of the B-spline approximation, the SPME for atomic multipoles is very efficient. Indeed, for the same grid density and spline order, calculating reciprocal sum interactions up to hexadecapole– hexadecapole order is only twice as expensive as calculating charge– charge interactions (note, however, that the hexadecapole– hexadecapole case generally requires a greater grid density and higher spline order than the charge– charge case). The SPME for atomic multipoles up to quadrupole is currently being implemented into sander and pmemd along with the AMOEBA force field of Ren and Ponder.75,79
Calculating the inductive response of the electron density, modeled by a set of inducible atomic dipoles, usually requires the solution of a set of linear equations relating the electric field to the induced dipoles. In the sander code we have implemented several variants of an iterative scheme for solving these, as well as Car–Parrinello-like extended Lagrangian scheme, wherein the induced dipoles are given a fictitious mass and evolved dynamically along with the atoms. In practice, this latter method is quite efficient, requiring, however, a 1-fs time step and gentle temperature control for stability. We have found that the AMBER ff02 force field, having atomic point charges and induced dipoles leads to stable trajectories using the extended Lagrangian scheme, with some improvement over previous force fields, at least for DNA simulations.80 However, stability problems can occur with force fields involving extra points in addition to inducible dipoles, due to occasional close approaches of the unshielded extra point charges.
Implicit Solvent Models
An accurate description of the aqueous environment is essential for realistic biomolecular simulations, but may become very expensive computationally. For example, an adequate representation of the solvation of a medium-size protein typically requires thousands of discrete water molecules to be placed around it. An alternative replaces the discrete water molecules by “virtual water”—an infinite continuum medium with some of the dielectric and “hydrophobic” properties of water.
These continuum implicit solvent models have several advantages over the explicit water representation, especially in molecular dynamics simulations:
Implicit solvent models are often less expensive, and generally scale better on parallel machines.
There is no need for the lengthy equilibration of water that is typically necessary in explicit water simulations; implicit solvent models correspond to instantaneous solvent dielectric response.
Continuum simulations generally give improved sampling, due to the absence of viscosity associated with the explicit water environment; hence, the macromolecule can more quickly explore the available conformational space.
There are no artifacts of periodic boundary conditions; the continuum model corresponds to solvation in an infinite volume of solvent.
New (and simpler) ways to estimate free energies become feasible; because solvent degrees of freedom are taken into account implicitly, estimating free energies of solvated structures is much more straightforward than with explicit water models.13,14
Implicit models provide a higher degree of algorithm flexibility. For instance, a Monte Carlo move involving a solvent exposed side chain would require nontrivial rearrangement of the nearby water molecules if they were treated explicitly. With an implicit solvent model this complication does not arise.
Most ideas about “landscape characterization” (that is, analysis of local energy minima and the pathways between them) make good physical sense only with an implicit solvent model. Trying to find a minimum energy structure of a system that includes a large number of explicit solvent molecules is both difficult and generally pointless: the enormous number of potential minima that differ only in the arrangement of water molecules might easily overwhelm an attempt to understand the important nature of the protein solute.
Of course, all of these attractive features of the implicit solvent methodology come at a price of making a number of approximations whose effects are often hard, if not impossible, to estimate. Some familiar descriptors of molecular interaction, such as solute–solvent hydrogen bonds, are no longer explicitly present in the model; instead, they come in implicitly, in the mean-field way via a linear dielectric response, and contribute to the overall solvation energy. However, despite the fact that the methodology represents an approximation at a fundamental level, it has in many cases been successful in calculating various macromolecular properties.81– 83
In many molecular modeling applications, and especially in molecular dynamics (MD), the key quantity that needs to be computed is the total energy of the molecule in the presence of solvent. This energy is a function of molecular configuration, its gradients with respect to atomic positions determine the forces on the atoms. The total energy of a solvated molecule can be written as Etot = Evac + ΔGsolv, where Evac represents molecule’s energy in vacuum (gas phase), and ΔGsolv is the free energy of transferring the molecule from vacuum into solvent, that is, solvation free energy. To estimate the total solvation free energy of a molecule, one typically assumes that it can be decomposed into the electrostatic and nonelectrostatic parts:
| (1) |
where ΔGnonel is the free energy of solvating a molecule from which all charges have been removed (i.e., partial charges of every atom are set to zero), and ΔGel is the free energy of first removing all charges in the vacuum, and then adding them back in the presence of a continuum solvent environment. The above decomposition, which is yet another approximation, is the basis of the widely used PB/SA scheme.14 Generally speaking, ΔGnonel comes from the combined effect of two types of interaction: the favorable van der Waals attraction between the solute and solvent molecules, and the unfavorable cost of disturbing the structure of the solvent (water) around the solute. Within the PB/SA, ΔGsurf is taken to be proportional to the total solvent accessible surface area (SA) of the molecule, with a proportionality constant derived from experimental solvation energies of small nonpolar molecules.84,85 Some more complex approaches have been proposed,86,87 but Amber currently follows this simple way to compute ΔGnonel, and uses a fast LCPO algorithm to compute an analytical approximation to the surface-accessible area of the molecule.88 This part is relatively straightforward, and is not the bottleneck of a typical MD simulation. The most time-consuming part is the computation of the electrostatic contribution to the total solvation free energy, mainly because the forces involved are long ranged, and because screening due to the solvent is a complex phenomenon.
Within the framework of the continuum model, a numerically exact way to compute the electrostatic potential φ(r) produced by molecular charge distribution ρm(r), is based on the Poisson-Boltzman (PB) approach in which the following equation (or its equivalent) must be solved; for simplicity we give its linearized form:
| (2) |
Here, ε(r) represents the position-dependent dielectric constant that equals that of bulk water far away from the molecule, and is expected to decrease fairly rapidly across the solute/solvent boundary. The electrostatic screening effects of (monovalent) salt enter via the second term on the right-hand side of eq. (2), where the Debye–Huckel screening parameter κ ≈ 0.1 Å−1 at physiological conditions. Once the potential φ(r) is computed, the electrostatic part of the solvation free energy is
| (3) |
where qi are the partial atomic charges at positions ri that make up the molecular charge density ρm(r) = ∑iδ(r − ri), and φ(ri)|vac is the electrostatic potential computed for the same charge distribution in the absence of the dielectric boundary, for example, in vacuum. Full accounts of this theory are available elsewhere.85,89
Numerical Approaches to the Poisson–Boltzmann Equation
The computational expense and difficulty of applying a Poisson–Boltzmann (PB) model to biomolecules have sparked much interest in developing faster and better numerical methods. Many different numerical methods have been proposed. The finite-difference method (FDPB) is one of the most popular and mature methods for biomolecular applications.90,91 Recently, its numerical efficiency has been dramatically enhanced, especially for dynamics simulations of biomolecules: the overhead of FDPB numerical solver is reduced from higher than explicit water simulations to comparable to vacuum simulations. The Amber package implements this improved FDPB method.92
A typical FDPB method involves the following steps: mapping atomic charges to the finite difference grid points; assigning nonperiodic boundary conditions, (electrostatic potentials on the boundary surfaces of the finite difference grid); and applying a molecular dielectric model to define the boundary between high-dielectric (water) and low-dielectric (molecular interior) regions, sometimes with the help of a dielectric smoothing technique to alleviate the problem in discretization of the dielectric boundary.93 These steps allow the partial differential equation to be converted into a sparse linear system Ax = b, with the electrostatic potential on grid points x as unknowns, the charge distribution on the grid points as the source b, and the dielectric constant on the grid edges and salt-related terms wrapped into the coefficient matrix A, which is a seven-banded symmetric matrix in the usual formulation. The linear system obtained is then solved with an iterative procedure.
In the Amber implementation, a standard trilinear mapping of charges is used. The boundary potentials may use a sum of contributions from individual atom charges,94 from individual residue dipolar charges,95 from molecular dipolar charges,94 or from individual grid charges. Here a grid charge is the net charge at a grid point after the trilinear charge mapping. A previous study shows that the last option (using grid charges) gives the best balance of accuracy and efficiency for electrostatic force calculations when electrostatic focusing is used. The dielectric model requires a definition of molecular volume; the choices offered in the Amber implementation are van der Waals volume and solvent excluded (molecular) volume. (Note that the van der Waals option is based on an improved definition as discussed in detail elsewhere.95 Briefly, the radii of buried atoms have been increased by solvent probe before they are used for van der Waals volume calculation.) The linear system solver adopted in Amber is the Eisenstat’s implementation of preconditioned conjugate gradient algorithm. Care has been taken to utilize electrostatic update in potential calculations for applications to molecular dynamics and minimization. Once the linear system is solved, the solution can be used to compute the electrostatic energies and forces.95
The Amber PB implementation offers two methods to compute electrostatic energy from a FDPB calculation. In the first method, total electrostatic energy is computed as 1/2 ∑iqiφi, where qi is the charge of atom i and φi is the potential at atom i as interpolated from the finite difference grid. This energy includes not only the reaction field energy, but also the finite-difference self energy and the finite-difference Coulombic energy. To compute a reaction field energy in the first method, two runs are usually required: one with solvent region dielectric constant set as its desired value, and one with solvent region dielectric constant set to that in the solute region. The second run is used to compute the finite-difference self-energy and the finite-difference Coulombic energy so that the difference between the total electrostatic energies of the two runs yields the reaction field energy. However, if there is no need to compute reaction field energy directly, for example, as in a molecular dynamics simulation, the finite-difference total electrostatic energy can be corrected to obtain the analytical total electrostatic energy.95 In doing so, the overhead in second FDPB run can be avoided. This approach is the same as that implemented in UHBD. In the second method, FDPB is only used to calculate reaction field energy, which can be expressed as half of the Coulombic energy between solute charges and induced surface charges on the solute/solvent boundary. The induced surface charges can be computed using Gauss’s law with the knowledge of FDPB electrostatic potential. This method is the same as that implemented in Delphi.
The Generalized Born Model
There exists an alternative approach to obtain a reasonable, computationally efficient estimate of the electrostatic contribution to the solvation free energy, to be used in molecular dynamics simulations. The analytic generalized Born (GB) method is an approximate way to calculate ΔGel. The methodology has become popular, especially in MD applications, due to its relative simplicity and computational efficiency, compared to the more standard numerical solution of the Poisson–Boltzmann equation.82,96 Within the GB models currently available in Amber, each atom in a molecule is represented as a sphere of radius ρi with a charge qi at its center; the interior of the atom is assumed to be filled uniformly with material of dielectric constant of 1. The molecule is surrounded by a solvent of a high dielectric εw (78.5 for water at 300 K). The GB model approximates ΔGel by an analytical formula,82,97
| (4) |
where rij is the distance between atoms i and j, Ri is the so-called effective Born radii of atom i, and fGB is a certain smooth function of its arguments (Fig. 6).
Figure 6.
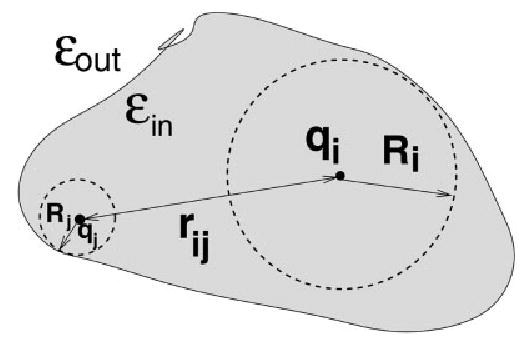
The effective Born radius of an atom reflects the degree of its burial inside the low dielectric region defined by the solvent boundary.
A common choice97 of fGB is
| (5) |
although other expressions have been tried.98,99 The effective Born radius of an atom reflects the degree of its burial inside the molecule: for an isolated ion, Ri is equal to its VDW radius ρi, and eq. (4) takes on a particularly simple form:
| (6) |
where we assumed εw = 78.5 and κ = 0 (pure water). The reader can recognize in eq. (6) the famous expression due to Born of the solvation energy of a single ion. For simple monovalent ions, substituting q = 1 and ρ ~1.5 Å yields ΔGel ~−100 kcal/mol, in reasonable agreement with the experiment. This type of calculation was probably the first success of the implicit solvent model based on continuum electrostatics. The function fGB in eq. (5) is designed to interpolate, in a clever manner, between the limit rij → 0, when atomic spheres merge into one and eq. (6) holds, and the opposite extreme rij → ∞, when the ions can be treated as point charges obeying the Coulomb’s law. In fact,100 without the exponential term in Eq. 5, Eq. 4 corresponds to the exact (in the limit εw→ ∞) solution of the Poisson equation for an arbitrary charge distribution inside a sphere. However, in the case of realistic molecules, the exponential factor in eq. (5) appears to be necessary. This is probably because, compared to a sphere, the electric field lines between a pair of distant charges in a real molecule go more through the high dielectric region, effectively reducing the pairwise interaction. The traditional form of fijGB eq. (5) takes this into account, at least to some extent, by allowing for steeper decay of the interaction with charge– charge distance. For deeply buried atoms, the effective radii are large, Ri >> ρi, and for such atoms one can use a rough estimate Ri ~ L, where L is the distance from the atom to the molecular surface. Closer to the surface, the effective radii become smaller, and for a completely solvent exposed side chain one can expect Ri to approach ρi. Note that the effective radii depend on the molecule’s conformation, and these need to be recomputed every time the conformation changes.
The efficiency of computing the effective radii is therefore a critical issue, and various approximations are normally made that facilitate an effective estimate of ΔGel. In particular, the so-called Coulomb field approximation is often used, which approximates the electric displacement around an atom by the Coulomb field . Within this assumption, the following expression for Ri can be derived:82,101
| (7) |
where the integral is over the solute volume, excluding a sphere of radius ρi around atom i. For a realistic molecule, the solute boundary (molecular surface) is anything but trivial, and so further approximations are often made to obtain a closed-form analytical expression. The Amber programs have adopted the pairwise de-screening approach of Hawkins, Cramer, and Truhlar,102,103 which leads to a GB model termed GBHCT. The electrostatic screening effects of (monovalent) salt are incorporated104 into eq. 4 via the Debye–Huckel screening parameter Note, that within the implicit solvent framework, one does not use explicit counterions unless these are known to remain “fixed” in specific positions around the solute. To set up an MD simulation based on an implicit solvation model, one requires a set of atomic radii {ρi}, which is an extra set of input parameters compared to the explicit solvent case. Over the years, a number of slightly different radii sets have been proposed, each being optimal for some class of problems, for example, for computing solvation free energies of molecules. A good set is expected to perform reasonably well in different types of problems, that is, to be transferable. One example is the Bondi radii set originally proposed in the context of geometrical analysis of macromolecular structures, but later found to be useful in continuum electrostatics models as well.105
Because the GB model shares the same underlying physical approximation— continuum electrostatics—with the Poisson–Boltzmann approach, it is natural, in optimizing the GB performance, to use the PB model as a reference. In particular, it has been shown that a very good agreement between the GB and PB models can be achieved if the effective Born radii match those computed exactly using the PB approach.99 Therefore, by improving the way the effective radii are computed within the analytic generalized Born, one can improve accuracy of the GB model. However, agreement with PB calculations is not the only criterion of optimal GB performance; other tests include comparisons to explicit solvent simulations results, and to the experiment. Also, to ensure computational efficiency in molecular dynamics simulations, the analytical expressions used to compute the effective Born radii, which enter eq. (4), must be simple enough, and the resulting electrostatic energy component ΔGel “well-behaved,” so as not to cause any instabilities in numerical integration of Newton’s equations of motion. The versions of the GB model currently available in Amber reflect various stages of the evolution of this approach; an analysis of these and other popular GB models has recently appeared.106 Historically, the first GB model to appear in Amber was GBHCT. It was later found that the model worked well on small molecules, but not so well on macromolecules, relative to the PB treatment. Note that in the GBHCT, the integral used in the estimation of the effective radii is performed over the VDW spheres of solute atoms, which implies a definition of the solute volume in terms of a set of spheres, rather than the more complex but realistic molecular surface commonly used in the PB calculations. For macromolecules, this approach tends to underestimate the effective radii for buried atoms,101 arguably because the standard integration procedure treats the small vacuum-filled crevices between the VDW spheres of protein atoms as being filled with water, even for structures with large interior.99 This error is expected to be greatest for deeply buried atoms characterized by large effective radii, while for the surface atoms it is largely canceled by the opposing error arising from the Coulomb approximation, which tends82,107,108 to overestimate Ri.
The deficiency of the GBHCT model described above can, to some extent, be corrected by rescaling the effective radii with the empirical parameters that are proportional to the degree of the atom’s burial, as quantified by the value I of the integral in eq. (7). The latter is large for the deeply buried atoms and small for exposed ones. Consequently, one seeks a well-behaved rescaling function, such that for small I, and when I becomes large; here, . One would also want to have a “smooth” upper bound on Ri vs. I to ensure numerical stability, see eq. (8) and Fig. 7). Although there is certainly more than one way to satisfy these constraints, the following simple, infinitely differentiable rescaling function was chosen to replace the GBHCT original expression for the effective radii:
Figure 7.
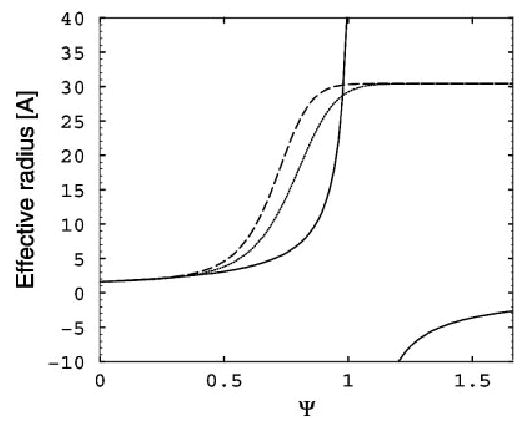
Graphical representation of different expressions used to compute the effective Born radius from the scaled volume Ψ. The broken lines correspond to eq. (8) of the GBOBC model, with parameters corresponding to igb = 2 (dashed) and igb = 5 (dotted). The GBHCT model (corresponding to igb = 1 in Amber) is shown as a solid line. All curves are computed for ρi = 1.7 Å.
| (8) |
where Ψ = I, and α, β, and γ are treated as adjustable dimensionless parameters that were optimized using the guidelines mentioned earlier (primarily agreement with the PB).
Currently, Amber supports two GB models (termed GBOBC) based on eq. (8). These differ by the values of {α, β, γ}, and are invoked by setting igb to either 2 or 5. The details of the optimization procedure and the performance of the GBOBC model relative to the PB treatment and in MD simulations on proteins is described in the original article.109
Enhancing Conformational Sampling
As described above, the use of continuum solvents can improve sampling due to reduced solvent viscosity. This section describes several other methods implemented in Amber to improve conformational sampling. The reader is also encouraged to read articles in a recent special journal issue dedicated to conformational sampling techniques.110
Locally Enchanced Sampling
Locally enhanced sampling (LES)111 is a mean-field approach that has proven useful in improving sampling through a reduction in internal barrier heights. In brief, the LES method provides the opportunity to focus computational resources on the portion of the system of interest by replacing it with multiple copies. The copies do not directly interact, and the system responds to the average force felt through interaction with all of the copies. The mean-field effect obtained from this averaging provides a smoothing effect of the energy landscape,112 improving sampling efficiency through reduction of barrier heights. In this manner, one also obtains multiple trajectories for the region of interest without repeating the entire simulation.
In Amber, the addles module is used to prepare a LES simulation (see Fig. 1). Addles reads files generated by Leap, and outputs the corresponding files with the LES copies as defined by the user in the input. In addition to replicating atoms, the force field parameters are modified; the LES energy function is constructed so that the energy of a LES system when all copies occupy identical positions is the same as the energy of the original system with the same coordinates. This maintains proper balance between interactions between LES and non-LES atoms, and also maintains an exact correspondence between the global minima in the LES and non-LES systems.112 This makes LES particularly useful for global optimization problems such as structure refinement. For example, LES simulations applied to optimization of protein loop conformations are nearly an order of magnitude more efficient than standard MD simulations.113
LES can be employed with several solvent models. Explicit solvent models with PME are supported, and this approach has been used to successfully refine structures of proteins and nucleic acids.114,115 One potential drawback to the use of LES with explicit solvent is that the solvent molecules are typically not placed in the set of atoms that are replicated with LES. Thus, the solvent will interact with all of the copies, and moving the copies apart can require the creation of a larger solvent cavity. This results in a free energy penalty analogous to the hydrophobic effect, and tends to reduce the independence of the copies through an indirect coupling. In addition, the solvent molecules surrounding the group of copies may not be able to simultaneously provide ideal solvation for each of the copies.
Because it is desirable to maximize the independence of the replicas during the simulation to increase both the amount of phase space that is sampled and the magnitude of the mean-field smoothing effect, Amber also supports the use of the GB solvent model in LES simulations. This GB+ LES approach has been described in detail.116 Each LES copy is individually solvated, avoiding the caging effect described above and providing more realistic solvation of the LES atoms. We have observed that GB + LES permits observation of independent transition pathways in a single simulation.
A drawback to the GB + LES approach (in addition to the approximations inherent to the GB models) is that the calculation of effective Born radii becomes more complex, requiring multiple effective radii for each of the non-LES atoms. Increased computational overhead is associated with the use of these extra radii, but this can be significantly reduced through the use of a radii difference threshold that allows atoms to be assigned a single radius if the set of radii differ by less than the threshold. The details of this approximation have been described,116 and a value of 0.01 Å is recommended.
Addles provides flexibility to the definition of LES regions, with the ability to arbitrarily choose the number of LES copies and the atoms included in the LES region. In addition, multiple LES regions can be defined, with the copies in each region interacting in an average manner with all of the copies in other regions. However, this flexibility presents the user with choices that can complicate the application of LES. These options are described in detail elsewhere.113 In general, we have found that the use of three to five copies works well, but the choice of LES region is highly dependent on the type of enhanced motion that is desired. Replication of single residues is sufficient to improve protein side-chain or nucleic acid base rotamer sampling, while at least three to four residues should be included in each LES region to sample alternate backbone conformations. Some experimentation may be necessary to find the optimal choices. In the ideal case, the copies should converge to the same conformation even when the simulation was initiated with different coordinates for each copy. If multiple low-energy states are present, the copies should all interconvert between these states. If the copies occupy different local minima and do not interconvert, then one can assume that they are kinetically trapped. Thus, a single LES simulation has built-in measures of simulation convergence.
Replica Exchange Molecular Dynamics
Another approach to improving conformational sampling is to increase the temperature of the system, providing more kinetic energy to overcome barriers to transitions. Simulated annealing117 can be performed in Amber by changing the target temperature during an MD simulation. A drawback to this approach is that a cooling schedule must be chosen in advance, and this approach is generally limited to locating low-energy states rather than calculation of thermodynamic properties of the system.
In replica exchange molecular dynamics (REMD),118 -121 several noninteracting copies (replicas) are independently and simultaneously simulated at different temperatures. At periodic intervals, conformations of the system being sampled at different temperatures are exchanged based on a Metropolis-type criterion that considers the probability of sampling each conformation at the alternate temperature. If the exchange is accepted, the bath temperatures of these replicas will be swapped, and the velocities will be scaled accordingly. Otherwise, if the exchange is rejected, each replica will continue on its current trajectory with the same bath. In this way, REMD is hampered to a lesser degree by the local minima problem, because the low temperature simulations (replicas) have the potential to escape kinetic traps by jumping to minima that are being sampled by the higher temperature replicas. On the other hand, the high energy regions of conformational basins often sampled by the high-temperature replicas can be relaxed in a way similar to the temperature annealing method. Moreover, the transition probability is constructed such that the canonical ensemble properties are maintained during each simulation, thus providing potentially useful information about conformational probabilities as a function of temperature. Due to these advantages, REMD has been widely applied to studies of peptide and small protein folding.10,18,118,119,122-126
In Amber 8, a single sander run can initiate the entire set of simulations, each with a different bath temperature and set of input/output files that can be specified in a single script. Each replica is run as a separate MD simulation, and multiple MPI communicators are used to enable transfer of data between the simulations. This allows the use of multiple processors for each of the replicas with the same scaling properties that would be obtained for a normal sander MD run. The choice of temperatures requires careful consideration. We typically use a temperature range of approximately 280 – 600 K. The high value is needed to ensure rapid escape from local minima during the simulation. The number of replicas needed to cover this range is determined by the desired probability of accepting the attempted exchanges between replicas. Too large a gap will result in rare exchange and a loss of the advantages of REMD. Obtaining a reasonable acceptance ratio relies on maintaining similarity in the magnitudes of the replica temperature gap and energy fluctuations in each replica simulation; therefore, the acceptable temperature gap between neighboring replicas decreases with larger systems, and more simultaneous simulations are needed to cover the desired temperature range. For large systems with thousands of degrees of freedom, this presents a practical problem. For smaller systems, particularly with implicit solvent models, REMD simulations are more tractable and may require ~10 replicas.
Trajectory Analysis
Molecular dynamics (MD) simulations provide information on molecular mobility on an atomic level, although one has to keep in mind that only recently multiple nanosecond-to-microsecond simulation times can be reached that are comparable to time scales of biologically relevant motions. Information about these relevant motions must then be extracted from the “jiggling and wiggling of atoms,” the time scale of which is governed by the fastest motions occurring in the system (usually on the order of 1 fs).
To consolidate a wide variety of different programs, scripts, and tools written by different people and available for analyzing MD trajectory data created by Amber (in various languages and locations), the ptraj program was created in the early 1990s. A goal was to provide a common interface to the commonly needed analysis tools. Written in C (with some Fortran code), ptraj extended the earlier rdparm program, which reads in an AMBER parameter/topology file (prmtop) and allows a user to interactively inspect the information within, such as lists of bonds, angles, dihedrals, and other parameters. The code in rdparm was not particularly well organized, and inhibited modification and extension of the (minimal) built-in analysis tools. To facilitate reusability of code, to make the program more modular, and to make it more easily extensible and modifiable, completely new code (except for the core prmtop reading routines) evolved. This was primarily accomplished via two significant changes: (1) The development of common/reusable routines, such as routines for parsing command line arguments into keywords and data, routines to select specific atoms (based on the atom selection syntax implemented in Midas127), and (2) the development of a core structure that modularizes the functions. At the time rdparm morphed into ptraj, C++ was not a reliable standard (as different compilers required different code) and therefore the code retained a core C functionality. Documentation was added and a common framework for performing analysis commands and extending the code was added. In addition, although the commands can be run interactively (interpreting commands line by line), the preferred mode of operation was changed from interactive toward parsing of commands from a file. The flow of the program is shown in Figure 8. This highlights the four major operations: (1) reading in (and checking of) coordinates (in various auto-detected formats) a single frame at a time, (2) performing various “actions” on the coordinates a single frame at a time (unless coordinates are buffered internally), (3) output of the modified coordinates, and (4) possible analysis of derived data. Adding a new action routine to perform analysis or modification of coordinates in a frame is relatively straightforward [as described in a prototype action Test() routine]. The intent with all of the action routines is to make them as general as possible with respect to their functionality yet accessible to outside programmers. Overall, the ptraj program has proven widely useful to the Amber user community.
Figure 8.
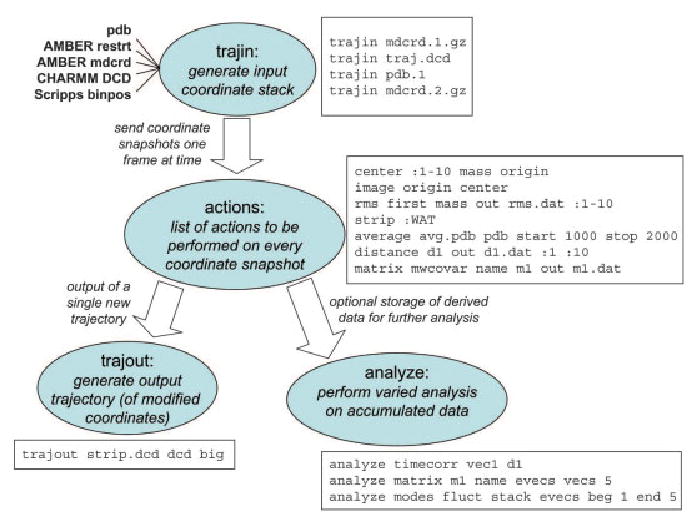
Information flow in ptraj. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
Despite its utility and extensibility there is one notable deficiency with the current implementation, specifically that only a single pass through the coordinate data is supported. This limitation is overcome where necessary by loading parts of, or the entire, trajectory into memory (such as with the 2Drms command, which requires pairwise comparisons between all frames). This, however, is not a general solution as it is an extremely memory intensive task. To overcome this deficiency will require implementation of a means to buffer the data to files or allow (potentially expensive) multiple reads through the raw trajectory data. In addition to overcoming this limitation, future additions to the general release include a general purpose clustering facility (based on pairwise RMSd or distance comparisons), further development of the routines for automated analysis of correlations, and a facility to smooth trajectories via wavelet transform lifting schemes. Other goals include a more direct interface capability to sander for energy analysis, greater support for CHARMM and other file formats, and the ability to convert AMBER parameter/topology formats to other formats.
As an example, in the following sections we describe the analysis of fluctuations and correlations of biomacromolecular motions using the program ptraj. Internal motions are important for the function of biomacromolecules. As such, correlated motions have been suggested to influence enzyme catalysis, and molecular machines like the ribosome or RNA–polymerase show collective hinge-bending or domain rearrangement motions. Furthermore, (macro-)molecular association may change internal motions of the binding partners, which influences binding thermodynamics or plays a role in allosteric regulation. Here, the dynamics of the Ras-binding domain of the protein C-Raf1 will be investigated, which is influenced by binding to the H-Ras protein. For this, snapshots saved at 100 fs intervals along 10 ns production runs for the Ras–Raf complex and unbound Raf will be used.128 For all analyses presented below the trajectories need to be aligned against a reference structure to remove global translational and rotational differences between snapshots, which has been done using the rms command of ptraj. (Note that the choice of the atom set used for fitting can have an important influence on the picture of the internal motion.129,130)
Root-Mean-Square Atomic Fluctuations
To determine the influence of macromolecular association on the magnitude of motions, the root-mean-square amplitude of motion of atoms about their mean positions can be determined from the MD trajectory by the atomicfluct command in ptraj. Results for Cα atoms of unbound Raf (“unbound”) and the Raf structure bound to Ras (“in cplx”) are given in Figure 9. Because in both cases snapshots have been aligned against the starting structures of the trajectories, respectively, the fluctuations for complexed Raf describe not only internal motions of the binding partners, but also include information about (rigid-body) motions of the two proteins with respect to each other. Least-squares fitting of bound Raf only, which has been extracted from the complex structure (“extracted”) provides information solely about internal motions of the protein in the bound state. Not unexpectedly, restrictions of Cα atomic fluctuations upon complex formation are observed for residues in the protein–protein interface (marked by ∇). In contrast, loop L1 of Raf shows considerably increased fluctuations upon formation of a complex. As indicated by the differences in the “in cplx” and “extracted” curves, however, rigid-body motions contribute to these fluctuations. It should be noted that information about calculated atomic fluctuations can be compared to experimentally determined fluctuations (e.g., obtained from crystallographic temperature factors), remembering that the time scales of averaging differ by orders of magnitudes between experiment and MD simulations and that calculated and experimental fluctuations include additional contributions to the atomic motions.
Figure 9.
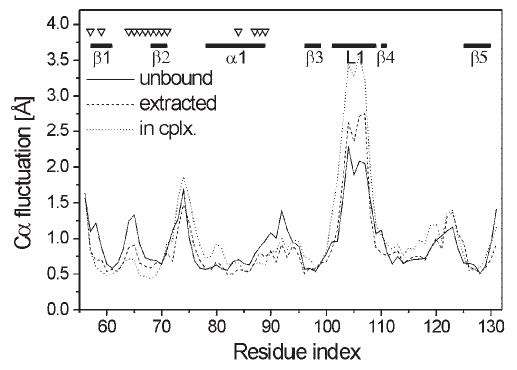
Cα atom root-mean-square atomic fluctuations of Raf in unbound (solid line) and bound (using least-squares fitting of bound, yet extracted, Raf conformations only: dashed line; using least-squares fitting of the whole Ras–Raf complex: dotted line) state. Residues in the interface region are marked with ▽, secondary structure elements are indicated by bold lines.
Equal-Time Crosscorrelations of Atomic Fluctuations
In addition to changes in the magnitude, binding may also influence the direction of motions. This will be reflected in the equal-time crosscorrelations of atomic fluctuations, which are determined by
| (9) |
where 〈. . .〉 indicates an average over the sample period.129 For completely correlated motions, C(i,j) = 1, and for completely anticorrelated motions, C(i,j) = −1. With ptraj, this correlation matrix is obtained by the matrix correl command.
The results are displayed in color-coded form in Figure 10, where dark blue represents C(i,j) = 1 and dark red C(i,j) = −1. The upper left triangle corresponds to motions of Cα atoms of unbound Raf, whereas the lower right triangle shows correlations of motions in the bound but extracted protein. By comparing both parts, changes in internal motions due to complex formation are revealed. As such, orientational correlations become more pronounced in the bound state, as indicated by the more deepened red and blue colors. Particularly intense couplings can be observed for the β-sheet of Raf, where the strands show strongly correlated movements. In turn, loop L1 moves in an anticorrelated fashion with respect to the β-sheet.
Figure 10.
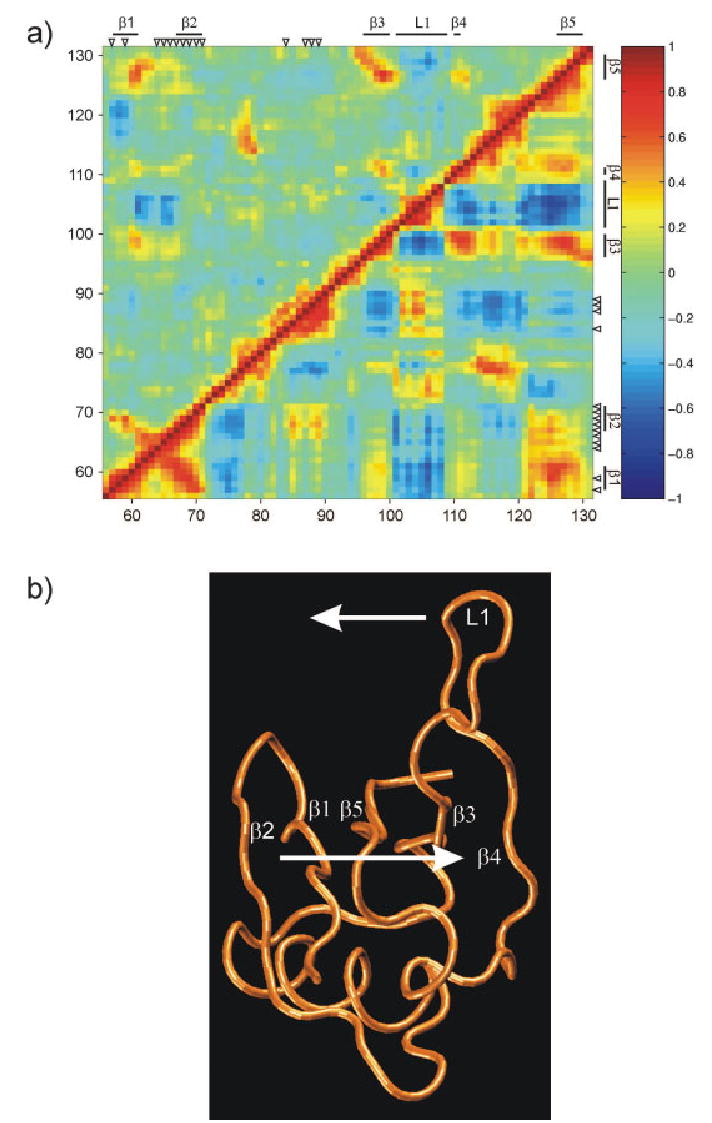
Crosscorrelation map [Eq. (9)] of Cα atomic fluctuations of Raf. (a) The two axes refer to residue indices. Positive correlations are indicated in red, negative in blue. The correlations for the unbound protein are shown in the upper left triangle, the ones for the bound protein in the lower right triangle. Residues in the interface region are marked with ▽, secondary structure elements are indicated by bold lines. Part (b) schematically shows the anticorrelated motion between the β-sheet consisting of β1–β5 and the loop L1 region that occurs in the bound state of Raf.
Quasiharmonic Analysis
From a more coarse-grained perspective, proteins can be considered to be constructed by quasi-rigid bodies (secondary structure elements and domains) that are connected by flexible loops. Hence, the motions of these molecules can be characterized in terms of variables that describe the relative coordinates of these quasi-rigid elements, leading to a considerable reduction in the number of variables compared to that required to describe the dynamics in atomic detail.131 One way to determine these collective variables (also termed “effective modes”) is by quasiharmonic analysis, where effective modes are computed such that the second moments of the amplitude distribution match those found in the MD simulation,132,133 that is, the Hessian (matrix of second derivatives) Hquasi is determined by
| (10) |
Compared to harmonic (normal mode) analysis, quasiharmonic analysis thus includes effects of anharmonic terms in the force field. In ptraj, quasiharmonic analysis is performed by a sequence of two commands. First, the mass-weighted covariance matrix is determined by matrix mwcovar. Subsequently, this matrix is diagonalized by the analyze matrix command, which results in eigenvectors and frequencies Qi and ωi that satisfy
| (11) |
with M being a diagonal matrix of atomic masses. Ptraj also implements principal component analysis (using nonmass-weighted covariance matrices) in Cartesian and distance space134 as well as Isotropically Distributed Ensemble (IDE) analysis.135 The latter is particularly interesting if overall rotational and internal motions of the system cannot be clearly separated, as is the case for molecular systems that exhibit rather unrestricted intramolecular mobility.
Effective modes can then be used to calculate root-mean-square fluctuations, displacements along mode directions in Cartesian coordinates, or dipole– dipole correlation functions, which are all available via the analyze modes command in ptraj. These analyses often show that protein motion is dominated by a relatively small number of effective modes.131 Using the projection modes command, in a subsequent “sweep” through the trajectory snapshots may then be projected onto these “important” modes, which allows determining the distribution of conformational substates in the subspace. An example of this is given in Figure 11, which shows the projection of 104 snapshots onto the two-dimensional space spanned by the first and second effective mode, together with distribution functions of the probability of finding the respective projection value. Interestingly, projections along the first effective mode show a bimodal distribution, indicating that this mode characterizes strongly anharmonic motions. This can be important in helping to establish the range of validity of harmonic or quasiharmonic approximations.136
Figure 11.
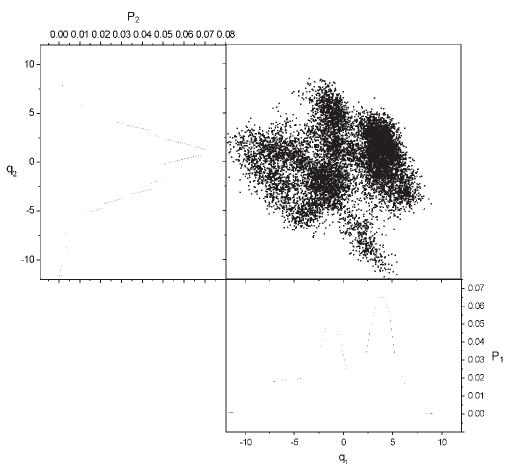
Projection qm of 104 snapshots onto the two-dimensional subspace spanned by the first (m = 1) and second (m = 2) effective modes (a). Probability distribution functions Pm(qm) for the first (b) and second (c) effective mode, respectively, where Pm(qm)dqm represents the probability that the projection is at position qm.
Finally, quasiharmonic analysis can, in principle, be applied to determine absolute rotational and vibrational entropies of biomacromolecules based on classical statistical mechanics.133 With respect to complex formation, changes in the degrees of freedom of the binding partners are then reflected as entropic contributions to the binding affinity. To determine entropies with ptraj, the keyword thermo has to be given in connection with the analyze matrix command mentioned above. Although this route provides an interesting alternative to calculating entropies by normal mode analysis, sampling issues have been found to pose a limitation to its straightforward applicability.15,137,138
Time–Correlation Functions
NMR relaxation experiments provide information on reorientational global and local dynamics on nanosecond and subnanosecond timescales for proteins in solution. This data can be interpreted with the aid of motional models suggested by MD simulations. In particular, time– correlation functions C(τ) determined from MD trajectories characterize the ability of one nucleus (e.g., the amide H) to relax the spin of another (e.g., the amide 15N) to which it is attached. Furthermore, insights into reorientational global motions may be obtained from time– correlation functions even at fairly short times.139 Finally, by applying the “model-free approach” of Lipari and Szabo140 to internal motions that are not coupled to overall tumbling, order-parameters S2 and effective correlation times of the internal motions can be determined from time– correlation functions.141-143
Within ptraj, time– correlation functions for vectors previously determined with the vector command can be calculated with the analyze timecorr command, using an efficient fast Fourier method. As an example, normalized time– correlation functions for three N—H vectors in unbound Raf are shown in Figure 12. Two vectors (Phe6, Leu27) that are located in secondary structure elements of the protein show typical characteristics of relatively rigid parts of the protein (i.e., an initial rapid decay due to vibrational motion followed by a slower decay due to motions of the peptide group as a whole). In contrast, the correlation function of Lys51 located in the loop L1 region decays considerably slower and does not reach a plateau value even after 3 ns, indicating internal motions on the (sub-)nanosecond time scale.
Figure 12.
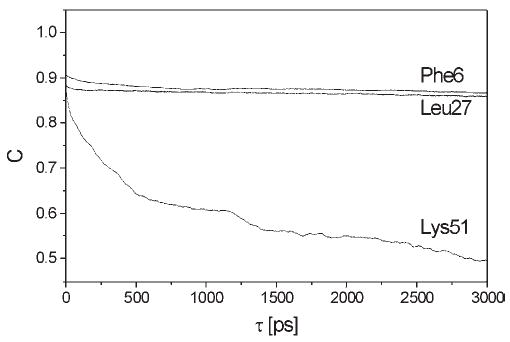
Normalized time– correlation functions for three N—H vectors of unbound Raf after removing overall rotation. Phe6 is located in β-strand 1 of the protein, Leu 27 in the α-helical region, and Lys51 in loop L1.
In addition to autocorrelation functions, ptraj calculates cross-correlation functions (which may be used for comparison to cross-correlated relaxation measurements) as well as time– correlation functions of normal vectors of (least-squares) planes through a series of atoms. Finally, the Isotropic Reorientational Eigenmode Dynamics (IRED) analysis144,145 is available; this model does not require separability of overall tumbling and internal motions and, hence, is applicable to a wide range of systems, such as folded, partially folded, and unfolded biomolecules.
Conclusions
For the past decade, new versions of Amber have been released on a 2-year schedule, and we anticipate that version 9 will become generally available in the Spring of 2006. It is still too early to say much about new features, but current development work is concentrating on improvements in the QM/MM capabilities, implementation of the nudged elastic band approach to finding reaction paths, and the implementation of electrostatic models that include permanent multipoles on the atoms. Also underway are continued improvements in code cleanup, with an eye toward maintainability, portability, and efficiency. Amber is a code that is heavily used by its developers, and reflects their interests, but we are also trying to lower the learning curve for scientists new to the simulation field. The Amber codes, and the entire field, have changed enormously in the 25 years since version 1 was written for a PDP-11 minicomputer; we look forward to seeing it evolve during the next quarter century.
Acknowledgments
Nothing about Amber would have happened without the enthusiastic support and encouragement of Peter Kollman. We also thank the many contributors and friends of Amber; some of them are listed at http://amber.scripps.edu/contributors.html.
References
- 1.Weiner P, Kollman P. J Comput Chem. 1981;2:287. [Google Scholar]
- 2.Pearlman D, Case D, Caldwell J, Ross W, Cheatham T, III, DeBolt S, Ferguson D, Seibel G, Kollman P. Comp Phys Commun. 1995;91:1. [Google Scholar]
- 3.Allen, M.; Tildesley, D. Computer Simulation of Liquids; Clarendon Press: Oxford, 1987.
- 4.Frenkel, D.; Smit, B. Understanding Molecular Simulation: From Algorithms to Applications; Academic Press: San Diego, 1996.
- 5.Becker, O.; MacKerell, A.; Roux, B.; Watanabe, M. Computational Biochemistry and Biophysics; Marcel Dekker: New York, 2001.
- 6.Cramer, C. Essentials of Computational Chemistry: Theories and Models; John Wiley & Sons: New York, 2002.
- 7.Schlick, T. Molecular Modeling and Simulation; Springer-Verlag: New York, 2002.
- 8.Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J Comput Phys. 1999;151:283. [Google Scholar]
- 9.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 10.Feig M, Karanicolas JL, Brooks CL., III J Mol Graph Model. 2004;22:377. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Macke, T.; Case, D. In Molecular Modeling of Nucleic Acids; Leontis, N.; Santa Lucia, J., Eds.; American Chemical Society: Washington, DC, 1998, p. 379.
- 12.Crowley M, Darden T, Cheatham T, III, Deerfield D., II J Supercomput. 1997;11:255. [Google Scholar]
- 13.Srinivasan J, Cheatham TE, III, Kollman P, Case D. J Am Chem Soc. 1998;120:9401. [Google Scholar]
- 14.Kollman P, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case D, Cheatham TE., III Acc Chem Res. 2000;33:889. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 15.Gohlke H, Case D. J Comput Chem. 2004;25:238. doi: 10.1002/jcc.10379. [DOI] [PubMed] [Google Scholar]
- 16.Weiner S, Kollman P, Case D, Singh U, Ghio C, Alagona G, Profeta S, Jr, Weiner P. J Am Chem Soc. 1984;106:765. [Google Scholar]
- 17.Cornell W, Cieplak P, Bayly C, Gould I, Merz K, Jr, Ferguson D, Spellmeyer D, Fox T, Caldwell J, Kollman P. J Am Chem Soc. 1995;117:5179. [Google Scholar]
- 18.García A, Sanbonmatsu K. Proc Natl Acad Sci USA. 2002;99:2782. doi: 10.1073/pnas.042496899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmerling C, Strockbine B, Roitberg AJ. Am Chem Soc. 2002;124:11258. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 20.Okur A, Strockbine B, Hornak V, Simmerling C. J Comput Chem. 2003;24:21. doi: 10.1002/jcc.10184. [DOI] [PubMed] [Google Scholar]
- 21.Ponder J, Case D. Adv Protein Chem. 2003;66:27. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- 22.Cheatham TE, III, Young M. Biopolymers (Nucleic Acid Sci) 2001;56:232. doi: 10.1002/1097-0282(2000)56:4<232::AID-BIP10037>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Wolf R, Caldwell J, Kollman P, Case D. J Comput Chem. 2004;25:1157. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 24.Rick S, Stuart S. Rev Comput Chem. 2002;18:89. [Google Scholar]
- 25.Grossfield A, Ren P, Ponder J. J Am Chem Soc. 2003;125:15671. doi: 10.1021/ja037005r. [DOI] [PubMed] [Google Scholar]
- 26.Kaminski G, Stern H, Berne B, Friesner R. J Phys Chem A. 2004;108:621. [Google Scholar]
- 27.Mannfors B, Mirkin N, Palmo K, Krimm S. J Phys Chem A. 2003;107:1825. [Google Scholar]
- 28.Woods R. J Phys Chem. 1995;99:3832. [Google Scholar]
- 29.Basma M. J Comput Chem. 2000;22:1125. doi: 10.1002/jcc.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woods R, Chappelle R. J Mol Struct. 2000;527:149. doi: 10.1016/S0166-1280(00)00487-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolfe S, Whangbo MH, Mitchell D. Carbohydr Res. 1979;69:1. [Google Scholar]
- 32.Wolfe S. Acc Chem Res. 1972;5:102. [Google Scholar]
- 33.Marchessault R, Perez S. Biopolymers. 1979;18:2369. [Google Scholar]
- 34.Nishida Y, Ohrui H, Meguro H. Tetrahedron Lett. 1984;25:1575. [Google Scholar]
- 35.Kroon–Batenburg L, Krron J. Biopolymers. 1990;29:1243. [Google Scholar]
- 36.Tvaroska I, Carver J. J Phys Chem B. 1997;101:2992. [Google Scholar]
- 37.Kirschner K, Woods R. Proc Natl Acad Sci USA. 2001;98:10541. doi: 10.1073/pnas.191362798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warshel A, Levitt M. J Mol Biol. 1976;103:227. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 39.Singh U, Kollman P. J Comput Chem. 1986;7:718. [Google Scholar]
- 40.Gao J. Rev Comput Chem. 1995;7:119. [Google Scholar]
- 41.Monard G, Merz K. Acc Chem Res. 1999;32:904. [Google Scholar]
- 42.Ryde U. Curr Opin Chem Biol. 2003;7:136. doi: 10.1016/s1367-5931(02)00016-9. [DOI] [PubMed] [Google Scholar]
- 43.Dixon S, Merz K. J Chem Phys. 1996;104:6643. [Google Scholar]
- 44.Dixon S, Merz K. J Chem Phys. 1997;107:879. [Google Scholar]
- 45.Yang W, Lee T. J Chem Phys. 1995;103:5674. [Google Scholar]
- 46.van der Vaart A, Gogonea V, Merz K., Jr J Comput Chem. 2000;21:1494. [Google Scholar]
- 47.Théry V, Rinaldi D, Rivail J, Maigret B, Ferenczy G. J Comput Chem. 1994;15:269. [Google Scholar]
- 48.Gao J, Amara P, Field M. J Phys Chem A. 1998;102:4714. [Google Scholar]
- 49.Reuter N, Dejaegere A, Maigret B, Karplus M. J Phys Chem A. 2000;104:1720. [Google Scholar]
- 50.Wang B, Raha K, Merz K. J Am Chem Soc. 2004;126:11430. doi: 10.1021/ja047695e. [DOI] [PubMed] [Google Scholar]
- 51.Jorgensen W. J Chem Phys. 1982;77:4156. [Google Scholar]
- 52.Jorgensen W, Chandrasekhar J, Madura J, Klein M. J Chem Phys. 1983;79:926. [Google Scholar]
- 53.Jorgensen W, Madura J. Mol Phys. 1985;56:1381. [Google Scholar]
- 54.Horn H, Swope W, Pitera J, Madura J, Dick T, Hura G, Head–Gordon T. J Chem Phys. 2004;120:9665. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 55.Mahoney M, Jorgensen W. J Chem Phys. 2000;112:8910. [Google Scholar]
- 56.Berendsen H, Grigera J, Straatsma T. J Phys Chem. 1987;91:6269. [Google Scholar]
- 57.Caldwell J, Kollman P. J Phys Chem. 1995;99:6208. [Google Scholar]
- 58.Hockney, R.; Eastwood, J. Computer Simulation Using Particles; McGraw–Hill: New York, 1981.
- 59.Ewald P. Ann Phys. 1921;64:253. [Google Scholar]
- 60.DeLeeuw S, Perram J, Smith E. Annu Rev Phys Chem. 1986;37:245. doi: 10.1146/annurev.pc.37.100186.001333. [DOI] [PubMed] [Google Scholar]
- 61.Kittel, C. Introduction to Solid State Physics; John Wiley & Sons: New York, 1986.
- 62.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089. [Google Scholar]
- 63.Essmann U, Perera L, Berkowitz M, Darden T, Lee H, Pedersen L. J Chem Phys. 1995;103:8577. [Google Scholar]
- 64.Pollock E, Glosli J. Comp Phys Commun. 1996;95:93. [Google Scholar]
- 65.Sagui, C.; Darden, T. In Simulation and Theory of Electrostatic Interactions in Solution; Pratt, L.; Hummer, G., Eds.; American Institute of Physics: Melville, NY, 1999, p. 104.
- 66.Hummer G, Pratt L, García A. J Phys Chem. 1996;100:1206. [Google Scholar]
- 67.Hünenberger P, McCammon J. Biophys Chem. 1999;78:69. doi: 10.1016/s0301-4622(99)00007-1. [DOI] [PubMed] [Google Scholar]
- 68.Kastenholz M, Hunenberger P. J Phys Chem B. 2004;108:774. [Google Scholar]
- 69.Feller S, Pastor R, Rojnuckarin A, Bogusz S, Brooks B. J Phys Chem. 1996;100:17011. [Google Scholar]
- 70.Patra M, Karttunen MT, Hyvonen M, Falck E. J Phys Chem B. 2004;108:4485. doi: 10.1063/1.1824033. [DOI] [PubMed] [Google Scholar]
- 71.Dixon R, Kollman P. J Comput Chem. 1997;18:1632. [Google Scholar]
- 72.Cieplak P, Caldwell J, Kollman P. J Comput Chem. 2001;22:1048. [Google Scholar]
- 73.Smith W. CCP5 Inform Q. 1982:4. [Google Scholar]
- 74.Toukmaji A, Sagui C, Board J, Darden T. J Chem Phys. 2000;113:10913. [Google Scholar]
- 75.Ren P, Ponder J. J Phys Chem B. 2003;107:5933. [Google Scholar]
- 76.Thole B. Chem Phys. 1981;59:341. [Google Scholar]
- 77.Sagui C, Pedersen L, Darden T. J Chem Phys. 2004;120:73. doi: 10.1063/1.1630791. [DOI] [PubMed] [Google Scholar]
- 78.McMurchie L, Davidson E. J Comput Phys. 1978;26:218. [Google Scholar]
- 79.Grossfield A, Ren P, Ponder JW. J Am Chem Soc. 2003;125:15671. doi: 10.1021/ja037005r. [DOI] [PubMed] [Google Scholar]
- 80.Baucom J, Transue T, Fuentes–Cabrera M, Krahn J, Darden T, Sagui C. J Chem Phys. 2004;121:6998. doi: 10.1063/1.1788631. [DOI] [PubMed] [Google Scholar]
- 81.Cramer C, Truhlar D. Chem Rev. 1999;99:2161. doi: 10.1021/cr960149m. [DOI] [PubMed] [Google Scholar]
- 82.Bashford D, Case D. Annu Rev Phys Chem. 2000;51:129. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 83.Feig M, Brooks CL., III Curr Opin Struct Biol. 2004;14:217. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 84.Sitkoff D, Sharp K, Honig B. J Phys Chem. 1994;98:1978. [Google Scholar]
- 85.Simonson T. Rep Prog Phys. 2003;66:737. [Google Scholar]
- 86.Levy R, Zhang L, Gallicchio E, Felts A. J Am Chem Soc. 2003;125:9523. doi: 10.1021/ja029833a. [DOI] [PubMed] [Google Scholar]
- 87.Gallicchio E, Levy R. J Comput Chem. 2004;25:479. doi: 10.1002/jcc.10400. [DOI] [PubMed] [Google Scholar]
- 88.Weiser J, Shenkin P, Still W. J Comput Chem. 1999;20:217. doi: 10.1002/(SICI)1096-987X(199905)20:7<688::AID-JCC4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 89.Baker N. Methods Enzymol. 2004;383:94. doi: 10.1016/S0076-6879(04)83005-2. [DOI] [PubMed] [Google Scholar]
- 90.Honig B, Sharp K, Yang AS. J Phys Chem. 1993;97:1101. [Google Scholar]
- 91.Honig B, Nicholls A. Science. 1995;268:1144. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 92.Luo R, David L, Gilson M. J Comput Chem. 2002;23:1244. doi: 10.1002/jcc.10120. [DOI] [PubMed] [Google Scholar]
- 93.Davis M, McCammon J. J Comput Chem. 1991;12:909. [Google Scholar]
- 94.Gilson M, Sharp K, Honig B. J Comp Chem. 1988;9:327. [Google Scholar]
- 95.Lu Q, Luo R. J Chem Phys. 2003;119:11035. [Google Scholar]
- 96.Feig M, Im W, Brooks C. J Chem Phys. 2004;120:903. doi: 10.1063/1.1631258. [DOI] [PubMed] [Google Scholar]
- 97.Still W, Tempczyk A, Hawley R, Hendrickson T. J Am Chem Soc. 1990;112:6127. [Google Scholar]
- 98.Jayaram B, Liu Y, Beveridge D. J Phys Chem. 1998;109:1465. [Google Scholar]
- 99.Onufriev A, Case D, Bashford D. J Comput Chem. 2002;23:1297. doi: 10.1002/jcc.10126. [DOI] [PubMed] [Google Scholar]
- 100.Sigalov G, Scheffe P, Onufriev A. J Chem Phys. 2005;122:094511. doi: 10.1063/1.1857811. [DOI] [PubMed] [Google Scholar]
- 101.Onufriev A, Bashford D, Case D. J Phys Chem B. 2000;104:3712. [Google Scholar]
- 102.Hawkins G, Cramer C, Truhlar D. Chem Phys Lett. 1995;246:122. [Google Scholar]
- 103.Hawkins G, Cramer C, Truhlar D. J Phys Chem. 1996;100:19824. [Google Scholar]
- 104.Srinivasan J, Trevathan M, Beroza P, Case D. Theor Chem Acc. 1999;101:426. [Google Scholar]
- 105.Bondi A. J Chem Phys. 1964;64:441. [Google Scholar]
- 106.Feig M, Onufriev A, Lee M, Im W, Case D, Brooks CL., III J Comput Chem. 2004;25:265. doi: 10.1002/jcc.10378. [DOI] [PubMed] [Google Scholar]
- 107.Schaefer M, Froemmel C. J Mol Biol. 1990;216:1045. doi: 10.1016/S0022-2836(99)80019-9. [DOI] [PubMed] [Google Scholar]
- 108.Schaefer M, Karplus M. J Phys Chem. 1996;100:1578. [Google Scholar]
- 109.Onufriev A, Bashford D, Case D. Proteins. 2004;55:383. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 110.Roitberg A, Simmerling C. Eds J Mol Graph Model. 2004;22:317. doi: 10.1016/j.jmgm.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 111.Elber R, Karplus M. J Am Chem Soc. 1990;112:9161. [Google Scholar]
- 112.Roitberg A, Elber R. J Chem Phys. 1991;95:9277. [Google Scholar]
- 113.Hornak V, Simmerling C. Proteins. 2003;51:577. doi: 10.1002/prot.10363. [DOI] [PubMed] [Google Scholar]
- 114.Simmerling C, Miller J, Kollman P. J Am Chem Soc. 1998;120:7149. [Google Scholar]
- 115.Simmerling C, Lee M, Ortiz A, Kolinski A, Skolnick J, Kollman P. J Am Chem Soc. 2000;122:8392. [Google Scholar]
- 116.Cheng X, Hornak V, Simmerling C. J Phys Chem B. 2004;108:426. doi: 10.1021/jp045437y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kirkpatrick S, Gelatt C, Jr, Vecchi M. Science. 1983;220:671. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 118.Sugita Y, Okamoto Y. Chem Phys Lett. 1999;314:141. [Google Scholar]
- 119.Sugita Y, Okamoto Y. Prog Theor Phys Suppl. 2000;138:402. [Google Scholar]
- 120.Sugita Y, Kitao A, Okamoto Y. J Chem Phys. 2000;113:6042. [Google Scholar]
- 121.Mitsutake A, Sugita Y, Okamoto Y. Biopolymers. 2001;60:96. doi: 10.1002/1097-0282(2001)60:2<96::AID-BIP1007>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 122.Rhee Y, Pande V. Biophys J. 2003;84:775. doi: 10.1016/S0006-3495(03)74897-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pitera J, Swope W. Proc Natl Acad Sci USA. 2003;100:7587. doi: 10.1073/pnas.1330954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Karanicolas J, Brooks C. Proc Natl Acad Sci USA. 2003;100:3954. doi: 10.1073/pnas.0731771100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nymeyer H, Garcia A, Woolf T. Biophys J. 2003;84:381A. [Google Scholar]
- 126.Kinnear B, Jarrold M, Hansmann U. J Mol Graph Model. 2004;22:397. doi: 10.1016/j.jmgm.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 127.Ferrin T, Huang C, Jarvis L, Langridge R. J Mol Graph 1988. 6;13:36. [Google Scholar]
- 128.Gohlke H, Kiel C, Case D. J Mol Biol. 2003;330:891. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 129.Ichiye T, Karplus M. Proteins Struct Funct Genet. 1991;11:205. doi: 10.1002/prot.340110305. [DOI] [PubMed] [Google Scholar]
- 130.Hünenberger P, Mark A, van Gunsteren W. J Mol Biol. 1995;252:492. doi: 10.1006/jmbi.1995.0514. [DOI] [PubMed] [Google Scholar]
- 131.Hayward S, Go N. Annu Rev Phys Chem. 1995;46:223. doi: 10.1146/annurev.pc.46.100195.001255. [DOI] [PubMed] [Google Scholar]
- 132.Karplus M, Kushick J. Macromolecules. 1981;14:325. [Google Scholar]
- 133.Levy R, Karplus M, Kushick J, Perahia D. Macromolecules. 1984;17:1370. [Google Scholar]
- 134.Abseher R, Nilges M. J Mol Biol. 1998;279:911. doi: 10.1006/jmbi.1998.1807. [DOI] [PubMed] [Google Scholar]
- 135.Prompers J, Brüschweiler R. Proteins. 2002;46:177. doi: 10.1002/prot.10025. [DOI] [PubMed] [Google Scholar]
- 136.Amadei A, Linssen A, Berendsen H. Proteins. 1993;17:412. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 137.Schäfer H, Mark A, van Gunsteren W. J Chem Phys. 2000;113:7809. [Google Scholar]
- 138.Schafer H, Daura X, van Gunsteren W. Proteins. 2001;43:45. doi: 10.1002/1097-0134(20010401)43:1<45::aid-prot1016>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 139.Smith P, van Gunsteren W. J Mol Biol. 1994;236:629. doi: 10.1006/jmbi.1994.1172. [DOI] [PubMed] [Google Scholar]
- 140.Lipari G, Szabo A. J Am Chem Soc. 1982;104:4546. [Google Scholar]
- 141.Levy R, Karplus M, Wolynes P. J Am Chem Soc. 1981;103:5998. [Google Scholar]
- 142.Palmer A, Case D. J Am Chem Soc. 1992;114:9059. [Google Scholar]
- 143.Peter C, Daura X, vanGunsteren W. J Biomol NMR. 2001;20:297. doi: 10.1023/a:1011241030461. [DOI] [PubMed] [Google Scholar]
- 144.Prompers J, Brüschweiler R. J Am Chem Soc. 2001;123:7305. doi: 10.1021/ja0107226. [DOI] [PubMed] [Google Scholar]
- 145.Prompers J, Brüschweiler R. J Am Chem Soc. 2002;124:4522. doi: 10.1021/ja012750u. [DOI] [PubMed] [Google Scholar]


