Abstract
The ATP synthase is a nanometric rotary machine that uses a transmembrane electrochemical gradient to form ATP. The structures of most components of the ATP synthase are known, and their organization has been elucidated. However, the supramolecular assembly of ATP synthases in biological membranes remains unknown. Here we show with submolecular resolution the organization of ATP synthases in the yeast mitochondrial inner membranes. The atomic force microscopy images we have obtained show how these molecules form dimers with characteristic 15 nm distance between the axes of their rotors through stereospecific interactions of the membrane embedded portions of their stators. A different interaction surface is responsible for the formation of rows of dimers. Such an organization elucidates the role of the ATP synthase in mitochondrial morphology. Some dimers have a different morphology with 10 nm stalk-to-stalk distance, in line with ATP synthases that are accessible to IF1 inhibition. Rotation torque compensation within ATP synthase dimers stabilizes the ATP synthase structure, in particular the stator-rotor interaction.
INTRODUCTION
The ATP synthase is the major energy transducer protein in all cells (1,2). The structure of the ATP synthase has been well described and can be divided into a soluble F1 part that shows ATPase activity and a membrane-associated FO part that uses the proton motive force to mechanically drive F1. F1 forms a ∼12 nm diameter head comprising three α and three β subunits housing the ADP/ATP binding sites (3) and a central stalk made of subunits γ, δ, and ɛ (4) that provide an elastic coupling between F1 and FO. The FO part forms the rotary machine. In yeast it contains an oligomeric ring of 10 c subunits (5); the number of c subunits varies among species (6,7). This ring is associated with a peripheral stator stalk that is formed by extramembraneous subunits “part of b”, d, F6, and the oligomycin sensitivity conferral protein (the structure of this part is known (8)), and the transmembrane moiety formed by subunits a, “two helices of b”, e, f, g, and A6L. The structure of this last part is the least well known, as only a 32 Å resolution three-dimensional (3D) envelope (in context with the entire enzyme) is available (9). The structure of bovine IF1 inhibitor that binds F1 has been solved isolated (10) and complexed with F1 (11).
A large body of indirect evidence has accumulated showing that the ATP synthase forms dimers and oligomers crucial for mitochondria morphology, structure, and function (12–15). In particular, FO subunits e and g were identified to induce dimerization (12,13). The bovine regulatory protein IF1 was also shown to be associated with F1 dimers (10,11,16). Detergent-solubilized ATP synthase dimers were studied by cryoelectron microscopy (cryo-EM) single particle analysis (17). However direct visualization of ATP synthase dimers and oligomers in the native membrane has remained impossible. To bring light into the issue concerning the supramolecular assembly of ATP synthase holo-enzymes, we have performed an atomic force microscopy (AFM) (18) study of the intermembrane space surface of native mitochondrial inner membranes (MIM) from yeast. The AFM, due to its outstanding signal/noise ratio, has proven to be an invaluable tool for the study of the supramolecular assembly of membrane protein in native membranes (19–21).
METHODS
Membrane preparation
Mitochondria were purified from yeast (Saccharomyces cerevisiae strain W303) cells grown overnight under oxidative conditions in yeast peptone-glycerol media at 30°C. Mitoplasts and outer membrane fragments were prepared as described (22). Mitoplasts were broken by a single passage through a French pressure cell in 1 mM Tris, pH 7.5. The resulting inner mitochondrial membranes were collected and washed by centrifugation. Membranes were stored at 4° for AFM analysis.
Atomic force microscopy
Mica supports were immerged in 40-μl adsorption buffer (10 mM Tris-HCl, pH 7.4, 150 mM KCl, 25 mM MgCl2). Subsequently, 3 μl of membrane solution was injected into the buffer drop. After ∼30 min, the sample was rinsed with recording buffer (10 mM Tris-HCl, pH 7.4, 150 mM KCl). The AFM (18) was operated in contact mode at ambient temperature and pressure. Imaging was performed with a commercial Nanoscope-E AFM (Veeco, Santa Barbara, CA) equipped with a 160-μm scanner (J-scanner) and oxide-sharpened Si3N4 cantilevers (length 100 μm; k = 0.09 N/m; Olympus, Tokyo, Japan). For imaging, minimal loading forces of ∼100 pN were applied at scan frequencies of 4–7 Hz using optimized feedback parameters and manually accounting for force drift.
Data analysis
All image treatment and analysis of AFM topographs were performed using custom-written routines for the ImageJ image processing package (23,24) and IGOR PRO (Wavemetrics, Portland, OR).
RESULTS AND DISCUSSION
Characterization of the mitochondria inner membranes
Before the AFM studies, MIM were characterized by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. The hydrophobicity of the transmembrane proteins alters the migration behavior of the proteins in the gel compared to the expected molecular masses. Therefore, we performed liquid chromatography and mass spectrometry analyses to positively identify the protein bands. These techniques revealed the presence of many different proteins, including ATP synthase, succinate dehydrogenase, cytochrome bc1, succinate-fumarate transporter, phosphate transporter, ATP/ADP transporter, cytochrome oxidase, nicotinamide adenine dinucleotide (NADH) dehydrogenase, and translocase of inner membrane (TIM), as among abundant proteins in the studied membranes (Fig. 1). Furthermore, we have assembled a list of the characteristics of the typical inner mitochondrial membrane proteome (Table 1) that describes the expected size and form of the various complexes that might be found in MIM.
FIGURE 1.
Coomassie brilliant blue stained SDS-PAGE of yeast MIM fractions. Bands labeled and marked with solid lines were identified on the basis of liquid chromatography and mass spectrometry analysis and gel migration behavior. Bands marked with dotted lines were not identified.
TABLE 1.
Inner mitochondrial membrane proteins
| kDa | No. Subunits* | No. TMH† | Soluble domain inter‡/matrix height (Å) | Biological unit | TMD§ dimensions | References | |
|---|---|---|---|---|---|---|---|
| (Complex I) | ∼980 | 46‖ | ∼50 | Matrix (140) | Monomer | 200 Å × 80 Å | (36–38) |
| NADH:ubiquinone oxidoreductase | |||||||
| (Complex II) | ∼130 | 4 | 6 | Matrix (∼80) | Monomer‖ | 45 Å × 45 Å (monomer) | (39–41) |
| Succinate:ubiquinone oxidoreductase | Trimer** | 80 Å × 80 Å (trimer) | |||||
| (Complex III) | ∼250 | 7 | 13 | Matrix (∼75) | Dimer | 70 Å × 90 Å | (42–44) |
| Ubiquinone:cytochrome c oxidoreductase | |||||||
| Cytochrome bc1 complex | |||||||
| (Complex IV) | ∼200 | 12¶ | 28 | Matrix (∼20) | Dimer†† | 200 Å × 100 Å | (45) |
| Cytochrome c oxidase | 13‖ | Inter (∼20) | |||||
| (Complex V) | ∼600 | 21 | 20 (c-ring) | Matrix (∼130) | Monomer* | 60 Å circular (c-ring) | (4,5,8,9,11) |
| ATP synthase | ∼11 ‡‡(FO-stator) | Inter (∼20) | Dimer†† | 50 Å × 70 Å (FO-stator) | |||
| (Mitochondrial carriers: 35 members) | ∼33 | 1 | 6 | Matrix (∼10) | Monomer | 40 Å × 40 Å | (46) |
| ATP/ADP transporter§§ | Inter (∼10) | Dimer†† | |||||
| TIM 22 complex | ∼300 | 5 | 17 | (47,48) | |||
| TIM22 | ∼22 | 1 (4×) | 4 | Matrix | Tetramer | – | |
| TIM54 | ∼54 | 1 | 1–2 | ||||
| TIM 23 complex | ∼340 | 8 | 16** | (47,49) | |||
| TIM23 | ∼23 | 1 (2×) | 4 | Matrix | Dimer | – | |
| TIM17 | ∼17 | 1 (2×) | 4 | Dimer | – | ||
| TIM44 | ∼60 | 1 (2×) | Matrix | Dimer | |||
| Hsp70 | ∼70 | 1 (2×) | Matrix | Dimer | – | ||
| Oxa1 | ∼450 | 1 | 5 | – | Monomer | – | (50) |
Based on x-ray or EM structures or biochemical analysis (see references).
TMH, Transmembrane helices based on x-ray or EM structures or sequence-based structure prediction (see references).
inter, Intermembrane space based on x-ray or EM structures or sequence-based structure prediction (see references).
TMD, Transmembrane domain based on x-ray or EM structures or sequence-based structure prediction (see references).
Yeast mitochondria.
Mammalian mitochondria.
Bacterial homolog.
Evidenced to participate in supercomplex formation in different stoichiometry.
Comprising subunits 6, 8, f, i, 4, and g.
Representative members of the mitochondrial carrier family.
AFM analysis
Yeast MIM were adsorbed on mica and investigated by AFM. Often we found folded membranes that were opened using the AFM tip as a nanodissector (25,26) (Fig. 2 A). These membranes are expected to originate from mitochondrial cristae because of their strong curvature. After opening the folded membrane, the AFM tip can image the protein surfaces of the mitochondrial intermembrane space (Fig. 2 B). Section analysis of low-resolution topographs showed that these membranes had a thickness of ∼17 nm, remaining locally curved. The measured membrane thickness of ∼17 nm corresponds well to the full height of ATP synthases (5). In these membranes densely packed circular complexes with a diameter of ∼8 nm and intercomplex distances of ∼15 nm were observed (Fig. 2, B–E), corresponding well to the observed intercomplex distance of F1 heads (13.6–18 nm) (27).
FIGURE 2.
Overview AFM analysis of MIM. (A) MIM adsorbed on mica. The folded parts of the membrane are marked with white arrows; the membrane area opened with the AFM tip is marked by the red arrow. (B) Topograph of the flattened membrane area shown in A. The AFM tip contours circular complexes of ∼8 nm in diameter and intercomplex distances of ∼15 nm corresponding to c-rings of ATP synthase. (C and D) Overview images reveal arrays of c-ring complexes densely packed. (E) Row of intercalated dimers. The c-ring rotor appears as a ∼1 nm indentation.
At first sight, the circular complexes appeared to be arranged in a hexagonal close packing (Fig. 2, B–D); however, upon closer examination rows of dimers were detected (Fig. 2 E). These rows are formed by dimers at an angle of ∼30° with respect to the row long axis (Figs. 2 E and 3 A). Among typical inner mitochondrial membrane proteomes, only the ATP synthase is expected to have a circular transmembrane domain, the c-ring (Table 1). Furthermore, the ring dimensions correlate well with the size of the c-ring (6,7,28), the interring distances correspond well to the known ATP synthase neighbor spacing in yeast mitochondria (27), and the membrane thickness fits well the total height of the entire ATP synthase (5,9). Therefore, based on the characteristic size and shape of the observed circular complexes and their presence in rows of dimers, we tentatively assign them as a view of the c-ring intermembrane space surface of the ATP synthase.
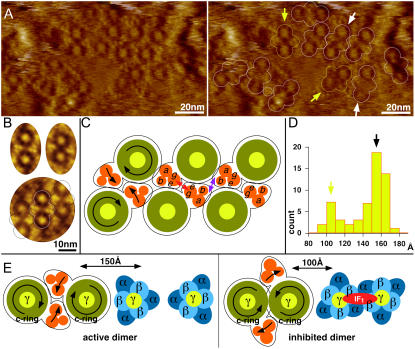
FIGURE 3.
Supramolecular architecture of ATP synthases. (A) High-resolution AFM topograph of MIM. (Right) ATP synthase dimers are outlined. (B) Average topographies. Left: Dimer with 15 nm stalk-to-stalk distance. (Right) Dimer with 10 nm stalk-to-stalk distance. (Bottom) Outlines indicate the molecular packing along a row of dimers. (C) Schematic representation of dimer and oligomer assembly. The positions of the molecules correspond to the average topograph (B, bottom panel). The assignment of subunits e and g at dimer interfaces and the transmembrane portion of b at interfaces along the dimer row are based on Arnold et al. (12) and Paumard et al. (13). (Left) Black arrows indicate rotor and stator rotation torque direction. (D) Stalk-to-stalk distance histogram (n = 58). Two populations of dimers are distinguished; most dimers have 15 nm stalk-to-stalk distance (white arrows in A); a minor fraction has 10 nm stalk-to-stalk distance (yellow arrows in A). (E) Schematic representation of active (left) and IF1-inhibited (right) ATP synthase dimers. The rotation torques stabilize the dimer during ATP synthesis and separate FO stator portions during ATP hydrolysis.
High-resolution raw data images revealed protein topographies in the center of the c-ring and neighboring the c-ring (Fig. 3 A). These central and peripheral proteins protrude ∼1 nm farther than the c-ring and make the rotor appear as an indentation. This may appear counterintuitive but is in line with structural information on the ATP synthase holo-enzyme: first, a cryo-EM analysis of the detergent-solubilized holo-enzyme monomer showed that the stator FO part protrudes farther into the intermembrane space than the rotor (9). This cryo-EM 3D map (9) has proven to be accurate since the atomic structure of the extramembraneous part of the peripheral stalk matches well the EM envelope (8). Second, an EM study of detergent-solubilized ATP synthase dimers revealed a density in the center of the c-ring on the intermembrane space surface (17). Although the protein located at this density remained unassigned, this feature was interpreted as FO bridging important for dimer formation and implicated in cristae biogenesis (17). Third, and equally, the cryo-EM 3D map of the related V-ATP synthase features a well-defined density that “plugs” the c-ring on the surface opposite to F1 (29). Fourth, AFM analysis of purified and reconstituted chloroplast c-rings with associated additional subunits revealed a c-ring plug on one surface (30).
The peripheral protrusions correspond to stator FO transmembrane subunits a, “two transmembrane helices (TMHs) of b”, e, g, and A6L, in agreement with the cryo-EM map (9). Cross correlation averaging showed that these peripheral proteins have three major protruding domains and have a preserved position between ATP synthases within the dimer and between adjacent dimers within the row (Fig. 3 B). Based on our images and a strong body of structural and biochemical data (12,13), we propose that dimerization subunits e, g, and the dimerizing portions of b are positioned at these interfaces (Fig. 3 C), without a precise structural assignment of these subunits to the features observed by AFM. Dimerization over subunits e, g, and b are structurally not equivalent. Both are sufficient to independently induce dimer formation, but are together needed for oligomer formation, in line with the fact that either deletion has drastic effects on mitochondria morphology (13). We propose that the e/g-dimerization surface is located within the dimer, whereas b-dimerization, shown to efficiently cross-link at amino acid positions located in the intermembrane space (13), acts along the dimer row. Notably, we imaged intermembrane space protrusions of neighboring FO close to this interface (Fig. 3, B and C).
Most enzyme dimers reveal a stalk-to-stalk distance of 15 nm; however a second class of ATP synthase shows 10 nm separation (Fig. 3 D; see also Fig. 3, A and B). We propose that these two classes represent active ATP synthase enzyme dimers and ATPase enzyme dimers that are accessible for IF1 inhibition (Fig. 3 E). Two IF1 inhibitor molecules were shown to clamp two F1 together (11). It appears suggestive that the small molecule IF1 requires preformed ATP synthase dimers. Inhibited dimeric F1 are linked together between α and β subunits of each F1. In this assembly the stalks of the two ATP synthases are closer together (∼10 nm) (11) than could be achieved if two of the bulkier subunit edges of each F1 faced each other (∼13 nm). Therefore we suggest that the dimers with 10 nm stalk-to-stalk distance are sterically accessible for IF1 inhibition. The decrease of distance between the two ATP synthases requires that the FO stator moieties are displaced away from their position close to the dimer center (Fig. 3 B). We estimate that both ATP synthases must rotate ∼30° to accomplish this novel dimer conformation, breaking the intramembrane contacts between e/g dimerization surfaces (Fig. 3 B and E, right). We furthermore observe that this conformation appears incompetent for oligomer formation in rows, i.e., the dimerization surface of b subunits are not correctly exposed to neighboring ATP synthases any longer, and thus they do not participate in the oligomers (Fig. 3 A).
Theoretical considerations on the functional role of ATP synthase dimers
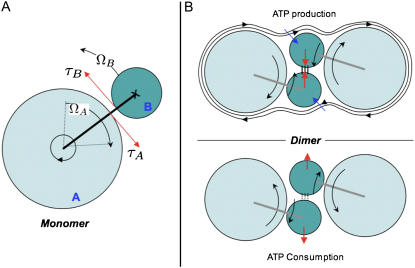
In the ATP synthase, the energy from either proton translocation or ATP hydrolysis is converted into rotary motion. Both processes inject fixed amounts of energy (depending on the ATP/ADP concentration and/or the transmembrane proton potential) into the system, which is converted into mechanical torque on the moving parts of the machine. Because no net force can be produced by the activity of the enzyme, equal but opposite torques  and
and  must be generated on the rotor and the stator at the active site (Fig. 4 A).
must be generated on the rotor and the stator at the active site (Fig. 4 A).
FIGURE 4.
Theoretical considerations about the ATP synthase dimer formation. (A) Monomer of ATP synthase; opposite torques τA and τB are generated on the rotor A and stator B. (B) Functional dimers of ATP synthase. Hydrodynamic (blue arrows) and torque (red arrows) forces stabilize the interaction within the ATP synthase dimer during the ATP production, whereas during the consumption the forces tend to destabilize the dimer.
The energy influx per unit time  is dissipated both by internal friction within the protein and by viscous friction with the membrane environment. Conservation of energy requires that the energy influx be equal to the total dissipation
is dissipated both by internal friction within the protein and by viscous friction with the membrane environment. Conservation of energy requires that the energy influx be equal to the total dissipation
 |
(1) |
where the dissipation has been split into  and
and  which depend on the individual motion of A and B with respect to the membrane, and
which depend on the individual motion of A and B with respect to the membrane, and  which depends on the relative motion of A with respect to B. The latter includes the membrane flow in the vicinity of the A-B interface and the energy converted into internal friction within the protein, both at the proton translocation site (transmembrane interface between rotor and stator) and at the ADP/ATP conversion site (interface between the stator F1(α, β) subunits and the rotor
which depends on the relative motion of A with respect to B. The latter includes the membrane flow in the vicinity of the A-B interface and the energy converted into internal friction within the protein, both at the proton translocation site (transmembrane interface between rotor and stator) and at the ADP/ATP conversion site (interface between the stator F1(α, β) subunits and the rotor  subunit). The internal friction at these sites includes the energy dissipated by pumping protons or synthesizing ATP. Each type of motion is characterized by a friction coefficient
subunit). The internal friction at these sites includes the energy dissipated by pumping protons or synthesizing ATP. Each type of motion is characterized by a friction coefficient  which relates the dissipation to the angular velocities
which relates the dissipation to the angular velocities  and
and  of the rotor and the stator.
of the rotor and the stator.
The torque of A and B can be written  and
and  Torque balance (equal but opposite torques
Torque balance (equal but opposite torques  and
and  ) determines the velocities of the rotor and stator:
) determines the velocities of the rotor and stator:
 |
(2) |
In two dimensions, the hydrodynamic friction force is expected to be proportional to the membrane viscosity and the velocity of the object  (within logarithmic corrections,
(within logarithmic corrections,  the distance from the axis of rotation), giving
the distance from the axis of rotation), giving  where
where  is the membrane viscosity (31). Therefore, we expect the stator to actually move at about the same angular velocity as the rotor for an isolated ATP synthase monomer. The hydrodynamic friction due to the motion of A with respect to B depends on the gradient of the velocity field near the A-B interface and is of the order
is the membrane viscosity (31). Therefore, we expect the stator to actually move at about the same angular velocity as the rotor for an isolated ATP synthase monomer. The hydrodynamic friction due to the motion of A with respect to B depends on the gradient of the velocity field near the A-B interface and is of the order  where
where  characterizes the distance between rotor and stator. The torque produced by ATP hydrolysis is of the order
characterizes the distance between rotor and stator. The torque produced by ATP hydrolysis is of the order  (32), thus if the friction was solely due to hydrodynamics, one would expect an angular velocity of the order 10,000 cycles per s (Hz). The maximal velocity of the F1-
(32), thus if the friction was solely due to hydrodynamics, one would expect an angular velocity of the order 10,000 cycles per s (Hz). The maximal velocity of the F1- part of the protein in solution is ∼50 times smaller (250 Hz) (33), which shows that the internal friction dominates the hydrodynamic drag and must be of the order
part of the protein in solution is ∼50 times smaller (250 Hz) (33), which shows that the internal friction dominates the hydrodynamic drag and must be of the order  The hydrodynamic drag in the membrane is thus much smaller than the internal friction and does not reduce the efficiency of the enzyme. This value correlates well with measurements on the angular velocity of F1-ATPase with attached actin filament as a function of the filament length (33). Similar to Eq. 2, the angular velocity of the actin-bound rotor is given by
The hydrodynamic drag in the membrane is thus much smaller than the internal friction and does not reduce the efficiency of the enzyme. This value correlates well with measurements on the angular velocity of F1-ATPase with attached actin filament as a function of the filament length (33). Similar to Eq. 2, the angular velocity of the actin-bound rotor is given by  where the friction due to the actin
where the friction due to the actin  depends on the actin length
depends on the actin length  and the water viscosity
and the water viscosity  (31). The velocity of the actin-free enzyme (250 Hz) drops 20-fold to 12 Hz when a 1 μm actin filament is attached (33). This leads to the independent estimate
(31). The velocity of the actin-free enzyme (250 Hz) drops 20-fold to 12 Hz when a 1 μm actin filament is attached (33). This leads to the independent estimate 
Our observations show that ATP synthases are functional dimers and provide evidence that dimers undergo a conformational change between the ATP-producing and ATP-consuming state. One may wonder whether rotating objects working in close contact might influence each other through hydrodynamic flow in the membrane. Hydrodynamic forces can be calculated from the viscous stress per unit protein length  (the membrane viscosity multiplied by the gradient of the velocity field). The hydrodynamic force shown (Fig. 4 B, blue arrow) for the ATP-producing state should stabilize the rotor-stator interactions. Nevertheless, its magnitude is quite small (of the order
(the membrane viscosity multiplied by the gradient of the velocity field). The hydrodynamic force shown (Fig. 4 B, blue arrow) for the ATP-producing state should stabilize the rotor-stator interactions. Nevertheless, its magnitude is quite small (of the order  ) and should not have a strong influence on the protein conformation. On the other hand, the torque resulting from the enzyme activity is quite large and leads to forces of the order
) and should not have a strong influence on the protein conformation. On the other hand, the torque resulting from the enzyme activity is quite large and leads to forces of the order  on the stator (Fig. 4 B, red arrows). These forces stabilize the stator-stator interactions within the dimer in the ATP-producing state and destabilize them in the ATP-consuming state. They reduce the energy gain from stator-stator interactions by an amount
on the stator (Fig. 4 B, red arrows). These forces stabilize the stator-stator interactions within the dimer in the ATP-producing state and destabilize them in the ATP-consuming state. They reduce the energy gain from stator-stator interactions by an amount  This is a quite sizeable effect, as these interactions are expected to be fairly small, thus facilitating a conformational change crucial to the inhibition of the enzyme in the ATP consumption state.
This is a quite sizeable effect, as these interactions are expected to be fairly small, thus facilitating a conformational change crucial to the inhibition of the enzyme in the ATP consumption state.
It is worth noting that the lack of viscous losses and the efficiency of the machine rely on its nanometric size. If the machine were 10 times larger (producing 30 ATP/cycle), the hydrodynamic drag (which scales like  ) would dominate the internal friction and the enzyme's efficiency would be appreciably reduced.
) would dominate the internal friction and the enzyme's efficiency would be appreciably reduced.
CONCLUSIONS
Since single molecule measurements have shown that F1 rotates the central stalk counterclockwise upon ATP hydrolysis (32), it is reasonable to assume that the rotation is clockwise during ATP synthesis. In contrast to a previous report (34), the torque force is generated at the interface between the transmembrane portions of the rotor and the stator. As a consequence, whereas the rotor rotates clockwise, the stator, in reaction, rotates counterclockwise around it in a fluid membrane where nothing holds the entire machine in place. This will make the ATP synthase as a whole rotate around an axis approximately at its center of mass and move in the membrane. Such movements dissipate energy. In the oligomeric assembly, in contrast, the torques on the stators of each molecule are compensated and only the rotors will move. Is the blocking of energy dissipation through movement of ATP synthases in the membrane sufficient to explain the increased activity of oligomeric versus monomeric ATP synthases (15)? Apparently not, as the major source of energy dissipation lies inside the ATP synthase, driving ATP synthesis, and the energy provided by proton translocation would produce FO rotation ∼40 times faster than observed (35) if it was only opposed to the viscous friction in the membrane as discussed above.
The energy dissipation through viscous drag is thus negligible compared to friction at both rotor-stator interfaces inside the ATP synthase. Therefore we conclude that oligomer formation has rather a structural role by assuring the integrity of the ATP synthase structure, particularly by stabilizing the weak FO rotor-stator interfaces. Notably, the rotor movement of each ATP synthase will drive the stator of the other toward its rotor. Taken together, the torque during ATP synthesis stabilizes the interfaces within each ATP synthase, within dimers and oligomers, and as a consequence cristae morphology. In contrast ATP hydrolysis destabilizes dimers by pulling the stators apart, allowing the monomers to approach each other and favor IF1 binding. Indeed, in this conformation the rotors come close together (10 nm) and may hinder each other's rotation at their interface (Fig. 3 E, right).
We observed, using high-resolution AFM, rows of ATP synthase dimers in yeast MIM, thus providing first direct evidence, to our knowledge, of the organization of ATP synthase in dimers and oligomers in native membranes. The proposed model describes the interactions within ATP synthase dimers and evidences their functional role. Although atomic structures strongly advanced our knowledge on individual proteins, AFM studies of native membranes visualize the supramolecular assembly of multiple membrane proteins in their native environment (19–21) and provide complementary data for a complete structural description. AFM images the supercomplexes into which structures can be docked (23) and provide insight into the function of these integrated systems.
Acknowledgments
This study was supported by the INSERM (Institut National de la Santé et Recherche Médicale) and INSERM Avenir, CNRS (Centre National de la Recherche Scientifique), and the grant (ANR-06-NANO-023-01).
Editor: Peter Hinterdorfer.
References
- 1.Boyer, P. 1997. The ATP synthase—a splendid molecular machine. Annu. Rev. Biochem. 66:717–749. [DOI] [PubMed] [Google Scholar]
- 2.Stock, D., C. Gibbons, I. Arechaga, A. G. Leslie, and J. E. Walker. 2000. The rotary mechanism of ATP synthase. Curr. Opin. Struct. Biol. 10:672–679. [DOI] [PubMed] [Google Scholar]
- 3.Abrahams, J. P., A. G. W. Leslie, R. Lutter, and J. E. Walker. 1994. Structure at 2.8 angstrom resolution of F1-ATPase from bovine heart mitochondria. Nature. 370:621–628. [DOI] [PubMed] [Google Scholar]
- 4.Gibbons, C., M. G. Montgomery, A. G. Leslie, and J. E. Walker. 2000. The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat. Struct. Biol. 7:1055–1061. [DOI] [PubMed] [Google Scholar]
- 5.Stock, D., A. G. Leslie, and J. E. Walker. 1999. Molecular architecture of the rotary motor in ATP synthase. Science. 286:1700–1705. [DOI] [PubMed] [Google Scholar]
- 6.Seelert, H., A. Poetsch, N. A. Dencher, A. Engel, H. Stahlberg, and D. J. Müller. 2000. Proton powered turbine of a plant motor. Nature. 405:418–419. [DOI] [PubMed] [Google Scholar]
- 7.Meier, T., P. Polzer, K. Diederichs, W. Welte, and P. Dimroth. 2005. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science. 308:659–662. [DOI] [PubMed] [Google Scholar]
- 8.Dickson, V. K., J. A. Silvester, I. M. Fearnley, A. G. Leslie, and J. E. Walker. 2006. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 25:2911–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubinstein, J. L., J. E. Walker, and R. Henderson. 2003. Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J. 22:6182–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabezon, E., M. Runswick, A. Leslie, and J. Walker. 2001. The structure of bovine IF(1), the regulatory subunit of mitochondrial F-ATPase. EMBO J. 20:6990–6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabezon, E., M. G. Montgomery, A. G. W. Leslie, and J. E. Walker. 2003. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat. Struct. Mol. Biol. 10:744–750. [DOI] [PubMed] [Google Scholar]
- 12.Arnold, I., K. Pfeiffer, W. Neupert, R. Stuart, and H. Schagger. 1998. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 17:7170–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paumard, P., J. Vaillier, B. Coulary, J. Schaeffer, V. Soubannier, D. M. Mueller, D. Brèthes, J.-P. di Rago, and J. Velours. 2002. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen, R., C. Schroeder, and A. Fok. 1989. An investigation of mitochondrial inner membranes by rapid-freeze deep-etch techniques. J. Cell Biol. 108:2233–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bornhovd, C., F. Vogel, W. Neupert, and A. S. Reichert. 2006. Mitochondrial membrane potential is dependent on the oligomeric state of F1F0-ATP synthase supracomplexes. J. Biol. Chem. 281:13990–13998. [DOI] [PubMed] [Google Scholar]
- 16.Cabezon, E., I. Arechaga, P. Jonathan, G. Butler, and J. Walker. 2000. Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1. J. Biol. Chem. 275:28353–28355. [DOI] [PubMed] [Google Scholar]
- 17.Minauro-Sanmiguel, F., S. Wilkens, and J. J. Garcia. 2005. Structure of dimeric mitochondrial ATP synthase: novel F0 bridging features and the structural basis of mitochondrial cristae biogenesis. Proc. Natl. Acad. Sci. USA. 102:12356–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binnig, G., C. Gerber, E. Stoll, T. R. Albrecht, and C. F. Quate. 1987. Atomic resolution with atomic force microscopy. Europhys. Lett. 3:1281–1286. [Google Scholar]
- 19.Fotiadis, D., Y. Liang, S. Filipek, D. A. Saperstein, A. Engel, and K. Palczewski. 2003. Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature. 421:127–128. [DOI] [PubMed] [Google Scholar]
- 20.Scheuring, S., and J. N. Sturgis. 2005. Chromatic adaptation of photosynthetic membranes. Science. 309:484–487. [DOI] [PubMed] [Google Scholar]
- 21.Buzhynskyy, N., R. K. Hite, T. Walz, and S. Scheuring. 2007. The supramolecular architecture of junctional microdomains in native lens membranes. EMBO Rep. 8:51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mannella, C. 1982. Structure of the outer mitochondrial membrane: ordered arrays of porelike subunits in outer-membrane fractions from Neurospora crassa mitochondria. J. Cell Biol. 94:680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheuring, S., T. Boudier, and J. N. Sturgis. 2007. From high-resolution AFM topographs to atomic models of supramolecular assemblies. J. Struct. Biol. 159:268–276. [DOI] [PubMed] [Google Scholar]
- 24.Rasband, W. S. 1997. –2005. ImageJ. U.S. National Institutes of Health, Bethesda, MD. http://rsb.info.nih.gov/ij/.
- 25.Fotiadis, D., L. Hasler, D. J. Müller, H. Stahlberg, J. Kistler, and A. Engel. 2000. Surface tongue-and-groove contours on lens MIP facilitate cell-to-cell adherence. J. Mol. Biol. 300:779–789. [DOI] [PubMed] [Google Scholar]
- 26.Scheuring, S., J. N. Sturgis, V. Prima, A. Bernadac, D. Lévy, and J.-L. Rigaud. 2004. Watching the photosynthetic apparatus in native membranes. Proc. Natl. Acad. Sci. USA. 101:11293–11297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicastro, D., A. Frangakis, D. Typke, and W. Baumeister. 2000. Cryo-electron tomography of neurospora mitochondria. J. Struct. Biol. 129:48–56. [DOI] [PubMed] [Google Scholar]
- 28.Stahlberg, H., D. J. Müller, K. Suda, D. Fotiadis, A. Engel, U. Matthey, T. Meier, and P. Dimroth. 2001. Bacterial ATP synthase has an undacemeric rotor. EMBO Rep. 2:229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregorini, M., J. Wang, X.-S. Xie, R. A. Milligan, and A. Engel. 2007. Three-dimensional reconstruction of bovine brain V-ATPase by cryo-electron microscopy and single particle analysis. J. Struct. Biol. In press. [DOI] [PubMed]
- 30.Seelert, H., N. A. Dencher, and D. J. Muller. 2003. Fourteen protomers compose the oligomer III of the proton-rotor in spinach chloroplast ATP synthase. J. Mol. Biol. 333:337–344. [DOI] [PubMed] [Google Scholar]
- 31.Happel, J., and H. Brenner. 1991. Low Reynolds Number Hydrodynamics. Kluwer, Dordrecht, The Netherlands.
- 32.Noji, H., R. Yasuda, M. Yoshida, and K. Kinosita. 1997. Direct observation of the rotation of F1-ATPase. Nature. 386:299–302. [DOI] [PubMed] [Google Scholar]
- 33.Yasuda, R., H. Noji, K. Kinosita, and M. Yoshida. 1998. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell. 93:1117–1124. [DOI] [PubMed] [Google Scholar]
- 34.Rexroth, S., J. Meyer zu Tittingdorf, H. Schwamann, F. Krause, H. Seelert, and N. Dencher. 2004. Dimeric H+-ATP synthase in the chloroplast of Chlamydomonas reinhardtii. Biochim Biophys. Acta. 1658:202–211. [DOI] [PubMed] [Google Scholar]
- 35.Ueno, H., T. Suzuki, K. Kinosita Jr., and M. Yoshida. 2005. ATP-driven stepwise rotation of FoF1-ATP synthase. Proc. Natl. Acad. Sci. USA. 102:1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grigorieff, N. 1998. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 Å in ice. J. Mol. Biol. 277:1033–1046. [DOI] [PubMed] [Google Scholar]
- 37.Abdrakhmanova, A., V. Zickermann, M. Bostina, M. Radermacher, H. Schagger, S. Kerscher, and U. Brandt. 2004. Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica. Biochim. Biophys. Acta. 1658:148–156. [DOI] [PubMed] [Google Scholar]
- 38.Sazanov, L., and P. Hinchliffe. 2006. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 311:1430–1436. [DOI] [PubMed] [Google Scholar]
- 39.Sun, F., X. Huo, Y. Zhai, A. Wang, J. Xu, D. Su, M. Bartlam, and Z. Rao. 2005. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 121:1043–1057. [DOI] [PubMed] [Google Scholar]
- 40.Jormakka, M., S. Tornroth, B. Byrne, and S. Iwata. 2002. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science. 295:1863–1868. [DOI] [PubMed] [Google Scholar]
- 41.Yankovskaya, V., R. Horsefield, S. Tornroth, C. Luna-Chavez, H. Miyoshi, C. Leger, B. Byrne, G. Cecchini, and S. Iwata. 2003. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 299:700–704. [DOI] [PubMed] [Google Scholar]
- 42.Xia, D., C. A. Yu, H. Kim, J. Z. Xia, A. M. Kachurin, L. Zhang, L. Yu, and J. Deisenhofer. 1997. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 277:60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwata, S., J. W. Lee, K. Okada, J. K. Lee, M. Iwata, B. Rasmussen, T. A. Link, S. Ramaswamy, and B. K. Jap. 1998. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 281:64–67. [DOI] [PubMed] [Google Scholar]
- 44.Hunte, C., J. Koepke, C. Lange, T. Rossmanith, and H. Michel. 2000. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 8:669–684. [DOI] [PubMed] [Google Scholar]
- 45.Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono, and S. Yoshikawa. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 272:1136–1144. [DOI] [PubMed] [Google Scholar]
- 46.Pebay-Peyroula, E., C. Dahout-Gonzalez, R. Kahn, V. Trezeguet, G. Lauquin, and G. Brandolin. 2003. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 426:39–44. [DOI] [PubMed] [Google Scholar]
- 47.Bauer, M., S. Hofmann, W. Neupert, and M. Brunner. 2000. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 10:25–31. [DOI] [PubMed] [Google Scholar]
- 48.Koehler, C. M. 2004. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 20:309–335. [DOI] [PubMed] [Google Scholar]
- 49.Pfanner, N., and A. Geissler. 2001. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2:339–349. [DOI] [PubMed] [Google Scholar]
- 50.Reif, S., O. Randelj, G. Domanska, E. Dian, T. Krimmer, C. Motz, and J. Rassow. 2005. Conserved mechanism of Oxa1 insertion into the mitochondrial inner membrane. J. Mol. Biol. 354:520–528. [DOI] [PubMed] [Google Scholar]