Abstract
In the mitochondrial genome of the hemichordate Balanoglossus carnosus, the codon AAA, which is assigned to lysine in most metazoans but to asparagine in echinoderms, is absent. Furthermore, the lysine tRNA gene carries an anticodon substitution that renders its gene product unable to decode AAA codons, whereas the asparagine tRNA gene has not changed to encode a tRNA with the ability to recognize AAA codons. Thus, the hemichordate mitochondrial genome can be regarded as an intermediate in the process of reassignment of mitochondrial AAA codons, where most metazoans represent the ancestral situation and the echinoderms the derived situation. This lends support to the codon capture hypothesis. We also show that the reassignment of the AAA codon is associated with a reduction in the relative abundance of lysine residues in mitochondrial proteins.
Keywords: Balanoglossus carnosus, codon capture, genetic code change, neutral theory, protein evolution
The genetic code, once thought to be “universal,” is now known to vary among several groups of organisms (1). There exist two hypotheses that attempt to explain how changes in the code come about. First, according to the “codon capture hypothesis” (1–4), it would be deleterious for an organism if a codon was assigned to two amino acids (or an amino acid and polypeptide chain termination) simultaneously. Thus, the first step in the change of the genetic code is assumed to be the complete disappearance of a codon from a genome. Subsequently, the tRNA (or release factor) assigned to this codon loses its capacity to recognize it, so that the codon becomes unassigned, and another tRNA acquires this capacity, allowing the codon to reappear at new positions in protein-coding genes. Second, in contrast, the “ambiguous intermediate hypothesis” (5, 6) proposes that the reassignment of a codon takes place via an intermediate stage during which the codon is recognized by two tRNAs assigned to different amino acids (or a tRNA and a release factor). This leads to heterogeneity in the encoded proteins that, according to this hypothesis and experimental results (7, 8), can be tolerated by an organism.
Because a change in the assignment of a codon must occur over relatively long evolutionary periods, it should in principle be possible to find organisms that represent intermediate stages in this process and thus to differentiate between the codon capture and ambiguous intermediate hypotheses. Indeed, unassigned codons have been observed in bacterial and mitochondrial genomes (9–11). Most of these cases are associated with a bias in the base composition. However, none of the codons effected are known to change their amino acid assignment in related evolutionary lineages (1).
The genetic code in echinoderm mitochondria differs from that of other deuterostomes and most other metazoans in that the codon AAA encodes asparagine instead of lysine and the codon AUA isoleucine instead of methionine. We have determined the complete nucleotide sequence of the mitochondrial genome of Balanoglossus carnosus (J.C., G.F., and S.P., unpublished work), a representative of hemichordates, which is the phylum thought to be most closely related to echinoderms (12). We find that while the codon AUA occurs at positions where isoleucine residues are conserved in metazoans, the codon AAA is completely absent from the B. carnosus mitochondrial genome. This, as well as a substitution in an anticodon position in the tRNALys gene, lends strong support to the codon capture hypothesis. Furthermore, we observe that the change in amino acid assignment of the AAA and the AUA codons is associated with a reduction of the relative number of lysine and methionine residues, respectively, in mitochondrial proteins.
METHODS
Complete mitochondrial genome sequences were retrieved from GenBank (13). For nonmetazoans, only mitochondrial genomes containing at least 12 of the 13 protein coding genes found in most metazoans were used. The general features of the mitochondrial genome of the hemichordate B. carnosus are described elsewhere (J.C., G.F., and S.P., unpublished work). The amino acid sequences of the mitochondrially encoded subunits of cytochrome c oxidase (cox1, cox2, and cox3), NADH dehydrogenase (nad1, nad2, nad3, nad4, nad4l, nad5, and nad6), ATP synthase (atp6), and ubiquinol-cytochrome c oxidoreductase (cyt b) were aligned with the program clustalw (14). From every alignment, the conserved regions were selected by a method that allows the definition of a number of pertinent parameters (J.C., unpublished work). In brief, conserved blocks were defined such that: (i) they do not contain more than four contiguous positions where less than 50% of the taxa analyzed have an identical residue, (ii) they are terminated by positions where more than 85% of the taxa have identical residues, and (iii) they have a total length larger than 15 positions. Furthermore, positions with gaps were removed and considered as nonconserved parts of the alignment. The amino acid composition was calculated for the entire sequences of all mitochondrial proteins in a species and, separately, for the conserved and the nonconserved blocks.
RESULTS AND DISCUSSION
Predictions of the Codon Capture Hypothesis.
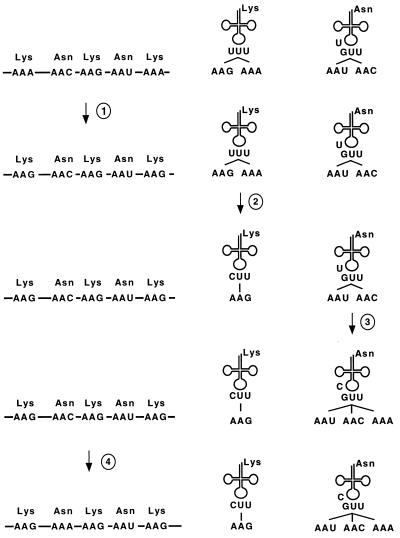
Fig. 1 shows the expected intermediates during the reassignment of AAA codons from lysine to asparagine according to the codon capture hypothesis. A similar scheme was previously proposed to explain the reassignments of such codon (2). The first step is the disappearance of all AAA codons from the protein-coding genes such that lysine residues become encoded exclusively by the codon AAG. This may occur as a consequence of, for example, mutational pressure favoring the increase of genomic GC content. In a second step, the tRNALys changes so that it limits its recognition to the codon AAG. This is expected to occur via a substitution from a T to a C at the first position of the anticodon in the tRNALys gene, because a modified U at the first anticodon position recognizes A and G in the third codon position, whereas a C normally recognizes only G (15, 16). In a third step, tRNAAsn acquires the ability to recognize AAA codons. It is thought that this is achieved in echinoderms by a substitution changing the base preceding the anticodon in the tRNA (position 33) from a U to a C (17). This may result in a changed anticodon loop structure or chemical modification that allows the GUU anticodon to recognize the codon AAA in addition to AAU and AAC. In the final step, AAA codons reappear in the genome at asparagine positions.
Figure 1.
Schematic illustration of the steps predicted for the change of the assignment of the AAA codon from lysine to asparagine by the codon capture hypothesis. Note that the situation after steps 1 and 2 corresponds to the features of the mitochondrial genome of the hemichordate B. carnosus, whereas the features after step 4 correspond to those found in echinoderms.
Lysine Codons in the B. carnosus Mitochondrial DNA.
When the reading frames of all protein-coding genes in the B. carnosus mitochondrial genome are inspected, no AAA codons are found. At positions where most metazoan mitochondrial proteins carry lysine residues, only the AAG codon is found. This is striking because the AAA codon is more common than the AAG codon in most other metazoan mitochondrial genomes (Table 1).
Table 1.
Occurrence of AAN codons in some metazoan mitochondrial genomes
| Species | AAT | AAC | AAA | AAG |
|---|---|---|---|---|
| Nematoda | ||||
| Caenorhabditis elegans | 139 | 12 | 95 (K) | 14 |
| Ascaris suum | 107 | 5 | 29 (K) | 65 |
| Annelida | ||||
| Lumbricus terrestris | 70 | 66 | 70 (K) | 22 |
| Mollusca | ||||
| Cepaea nemoralis | 50 | 37 | 36 (K) | 34 |
| Albinaria coerulea | 106 | 32 | 73 (K) | 13 |
| Katharina tunicata | 110 | 51 | 78 (K) | 18 |
| Arthropoda | ||||
| Artemia franciscana | 93 | 31 | 41 (K) | 40 |
| Drosophila yakuba | 193 | 13 | 76 (K) | 9 |
| Locusta migratoria | 141 | 46 | 71 (K) | 29 |
| Vertebrata | ||||
| Petromyzon marinus | 78 | 67 | 90 (K) | 9 |
| Xenopus laevis | 70 | 80 | 77 (K) | 8 |
| Homo sapiens | 33 | 131 | 85 (K) | 10 |
| Echinodermata | ||||
| Paracentrotus lividus | 44 | 50 | 88 (N) | 54 |
| Arbacia lixula | 46 | 44 | 82 (N) | 52 |
| Asterina pectinifera | 41 | 58 | 112 (N) | 48 |
| Hemichordata | ||||
| B. carnosus | 20 | 98 | 0 | 45 |
AAT and AAC code for asparagine (N), AAG for lysine (K), and AAA for either lysine or asparagine, as shown in parentheses.
Table 2 lists amino acid codons that are absent from complete mitochondrial genomes sequenced in metazoans. Almost all cases involve genomes that have a high AT content (63–85%). The sole exception is the hemichordate mitochondrial genome, where the AT content is 51%. Thus, whereas in most of these cases the absence of codons might be due to AT pressure, the lack of AAA codons in the hemichordate is not due to such pressure, although the original reason for its absence may well be a GC pressure that has since disappeared (4).
Table 2.
Codons absent in metazoan mitochondrial genomes (excluding stop codons)
| Species | Absent codons | % AT |
|---|---|---|
| Nematoda | ||
| C. elegans | CGC | 76 |
| A. suum | TGC, CTC, CCC, CGC | 72 |
| Arthropoda | ||
| Anopheles gambiae | CGC, AGG, GCG | 78 |
| A. quadrimaculatus | CTG, CGC, AGG | 77 |
| D. yakuba | CAG, CGC, AGG | 79 |
| D. melanogaster | CTG, CGC, AGC, AGG | 82 |
| A. mellifera | CTG, CCG, GTG, GCG | 85 |
| TGC, CGC, GGC | ||
| Vertebrata | ||
| P. marinus | GCG | 63 |
| Hemichordata | ||
| B. carnosus | AAA, AGG | 51 |
In addition to AAA codons, only AGG codons are lacking in the B. carnosus mitochondrial genome. The AGG codon is rare in many mitochondrial genomes and it has been reported to be absent in four insect species (Table 2). Thus, its absence in hemichordates may not be very surprising. However, because AGG encodes arginine in cnidarians, serine in most invertebrates (platyhelminths, nematodes, arthropods, mollusks, annelids, and echinoderms), and glycine in urochordates, and is assigned to termination in vertebrates, its tendency to be rare as well as absent may be related to its propensity to change its assignment.
tRNA Genes in the B. carnosus Mitochondrial DNA.
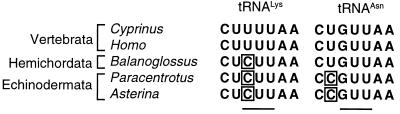
The hemichordate gene for tRNALys that in most metazoans recognizes AAA and AAG codons is similar to echinoderms in having a C at the first anticodon position rather than a T (Fig. 2). This probably limits its codon recognition to AAG (15, 16). It should also be noted that the mitochondrial tRNALys gene of some arthropods carries a CTT anticodon, which in that case is thought to translate both AAA and AAG codons (18, 19). However, because the tRNALys gene carries a TTT anticodon in other protostomes (mollusks, annelids, and nematodes), the CTT anticodon sequence is independently derived in arthropods and, therefore, is of no direct relevance to the situation in hemichordates and echinoderms.
Figure 2.
Alignment of the predicted anticodon loops of tRNALys and tRNAAsn in some deuterostomes. The anticodon region is underlined. Bases suggested to be involved in defining the recognition ability of the respective tRNAs are boxed.
The hemichordate tRNAAsn gene is similar to most other metazoans in carrying a T at position 33 rather than a C as in echinoderms (Fig. 2). Therefore the B. carnosus mitochondrial tRNAAsn is likely to be unable to recognize the AAA codon, which would thus be unassigned.
A Scenario for Codon Reassignments.
In the mitochondrial genome of B. carnosus, the first two steps predicted by the codon capture hypothesis for changes of the genetic code have occurred. First, AAA codons are absent from all protein-coding genes and, second, the gene for tRNALys is similar to echinoderms in having a C at the first anticodon position (Fig. 2), thus limiting its recognition to the codon AAG. However, the “capture” of the unassigned AAA codon by the tRNAAsn has not taken place in hemichordates because its tRNAAsn gene is similar to other metazoans in carrying a T at position 33, which probably limits its gene product to the decodoing of AAU and AAC codons. The B. carnosus mitochondrial genome can thus be seen as representing an intermediate in the process of reassignment of AAA codons, where vertebrates and other metazoan phyla represent the ancestral situation and echinoderms represent the derived situation. This strongly suggests that the codon capture hypothesis correctly predicts how the reassignment of AAA codons from lysine to asparagine has occurred in echinoderms (1, 2).
It is likely that the first two steps of the reassignment occurred in a common ancestor of echinoderms and hemichordates (Fig. 3). Subsequently, the capture of the AAA codon by the tRNAAsn occurred in the echinoderm line, whereas the hemichordate line remained in the stage where no tRNA can recognize AAA codons. If this scenario is correct, the evolutionary line leading to Balanoglossus would have remained devoid of the AAA codon for more than 600 million years (32). However, the determination of nucleotide sequences from the mitochondrial genomes of other hemichordate and echinoderm taxa are desirable to shed further light on the order in which the disappearance of the AAA codon and the change of codon specificity of the tRNALys and tRNAAsn happened.
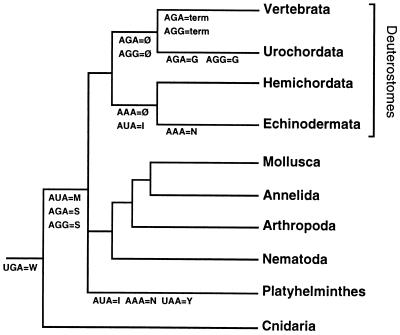
Figure 3.
Phylogenetic tree relating animal phyla where the mitochondrial genetic code is known (15, 20–31). Likely changes in the genetic code are shown. In deuterostomes, also putative unassignment of codons related to codon changes are given (Ø). The phylogenetic relationships among deuterostomes is based on 18S RNA sequences (12). Because the phylogenetic position of Platyhelminthes is uncertain, the change AUA = M may have happened after a putative split of cnidarians and platyhelminths from the other metazoans, thus eliminating the change AUA = I on the line to Platyhelminthes.
The reassignment of the AUA codon from methionine to isoleucine probably happened in a common ancestor of echinoderms and hemichordates (Fig. 3). Furthermore, the absence of the AGG codon from the hemichordate genome could be related to the change of assignment that this codon experienced in vertebrates and urochordates (31). However, because AGG is a rare codon in many metazoan genomes, it is plausible that its disappearance occurred independently in the ancestral lineages of hemichordates and chordates.
Genetic Code Change and Amino Acid Composition.
When the genetic code was still unknown, Sueoka (33) showed that the amino acid composition of proteins is correlated with the GC content of the genomic DNA of an organism. A few years later, when the genetic code had been deciphered, it was found that the number of codons for an amino acid is largely correlated with the relative abundance of the amino acid in proteins, an observation that was adduced by King and Jukes as support of the neutral theory (34). Therefore, it is of interest to investigate whether a change in the genetic code leads to a change in the relative proportions of the affected amino acids. In fact, when a small set of mitochondrial genomes was analyzed, the reassignment of the AUA codon seemed to affect the content of methionine residues in mitochondrial proteins (35).
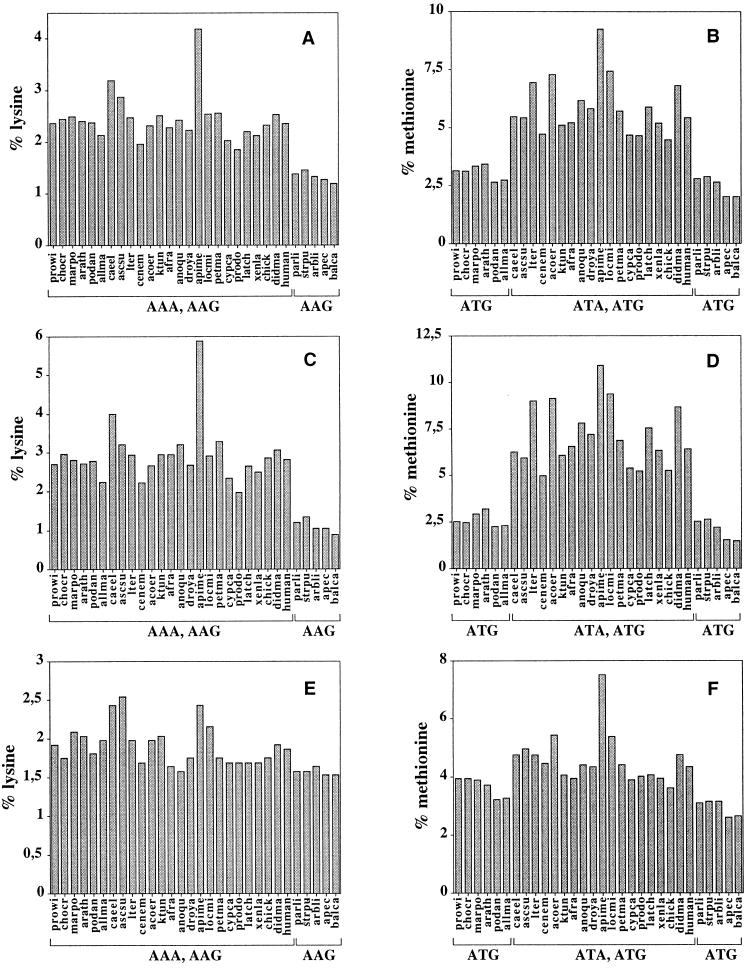
When the amino acid composition of the common set of 12 protein subunits encoded in 30 mitochondrial genomes of plants, fungi, and metazoans are compared, most amino acids are present in similar proportions or exhibit no clear trend among the taxonomic groups. However, lysine shows a decrease in abundance in echinoderms and the hemichordate, and methionine is decreased in non-metazoans, echinoderms and the hemichordate (Fig. 4 A and B). In both cases, these decreases parallel reductions in the number of codons assigned to the amino acids. As discussed above, lysine is encoded by one rather than two codons in echinoderms and the hemichordate, and these two groups show a reduction in the number of lysines. Methionine has experienced an increase in its number of codons in metazoans after the split from cnidarians, where ATA codons changed their assignment from isoleucine to methionine, and a reversal of this change in echinoderms and hemichordates (Fig. 3). Tellingly, methionine content shows a parallel increase in most metazoans that is reversed in echinoderms and hemichordates. When the protein alignments are divided into blocks with a high and a low degree of conservation, the reduction of lysine and methionine content is observed in the protein segments that display a low degree of conservation (Fig. 4 C and D), whereas it is not seen in the conserved protein segments (Fig. 4 E and F).
Figure 4.
Relative numbers of lysines and methionines in mitochondrially encoded proteins in: all 12 proteins common to all groups (A and B), in non-conserved regions of these proteins (2914 positions) (C and D), and in conserved regions (1770 positions) (E and F). Below the species names, the codons that recognize the corresponding amino acid are given. Species names and GenBank accession number follow. For plants: prowi, Prototheca wickerhamii (U02970); chocr, Chondrus crispus (Z47547); marpo, Marchantia polymorpha (M68929); and arath, Arabidopsis thaliana (Y08501). For fungi: podan, Podospora anserina (X55026); and allma, Allomyces macrogynus (U41288); For nematodes: caeel, Caenorhabditis elegans (X54252); and ascsu, Ascaris suum (X54253). For annelids: lter, Lumbricus terrestris (U24570). For molluscs: cenem, Cepaea nemoralis (U23045); acoer, Albinaria coerulea (X83390); and ktun, Katharina tunicata (U09810). For arthropods: afra, Artemia franciscana (X69067); anoqu, Anopheles quadrimaculatus (L04272); droya, Drosophila yakuba (X03240); apime, Apis mellifera (L06178); and locmi, Locusta migratoria (X80245). For vertebrates: petma, Petromyzon marinus (U11880); cypca, Cyprinus carpio (X61010); prodo, Protopterus dolloi (L42813); latch, Latimeria chalumnae (U82228); xenla, Xenopus laevis (M10217); chick, Gallus gallus (X52392); didma, Didelphis virginiana (Z29573); and human, Homo sapiens (X93334). For echinoderms: parli, Paracentrotus lividus (J04815); strpu, Strongylocentrotus purpuratus (X12631); arbli, Arbacia lixula (X80396); and apec, Asterina pectinifera (D16387). For hemichordates: balca, Balanoglossus carnosus (AF051097).
These observations agree with the expectations of the neutral theory of protein evolution (34, 36), according to which a significant proportion of amino acids present in a protein should change by random genetic drift and therefore be influenced by the number of codons assigned to the different amino acids. Furthermore, this effect is more apparent in less conserved parts of proteins than in conserved regions, where purifying selection is expected to be more active. However, it should be noted that other amino acids that have experienced changes in their numbers of codons (arginine, asparagine, isoleucine, serine, and tryptophan) do not show concomitant variations in their abundance in proteins. A possible explanation for this is that when a change in the number of codons is to or from one single codon (lysine, methionine, and tryptophan), it may be easier to detect changes in the amino acid content than when the number of codons change between larger numbers (e.g., between two and three) because in the latter case the relative change in codon number is smaller. Moreover, amino acids such as tryptophan, which are highly conserved in proteins, may be less prone to change their abundance due to changes in codon number. Thus, other factors in addition to the numbers of codons encoding the different amino acids must obviously also influence their occurrence in mitochondrial proteins.
CONCLUSIONS
The mitochondrial genome of B. carnosus provides a remarkable fulfillment of the predictions of the codon capture hypothesis for codon reassignment as proposed by Osawa and Jukes (1, 2, 4). Furthermore, changes in lysine and methionine content in the proteins encoded by the mitochondrial genomes of hemichordate and other organisms upon genetic code changes adhere equally strikingly to the expectations of the neutral theory as outlined by King and Jukes (34) and Kimura (36).
Acknowledgments
We acknowledge a short-term European Molecular Biology Organization fellowship to J.C. and the Deutsche Forschungsgemeinschaft (Pa 452/4-1) for financial support.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF051097).
References
- 1.Osawa S. Evolution of the Genetic Code. Oxford, U.K.: Oxford Univ. Press; 1995. [Google Scholar]
- 2.Jukes T H, Osawa S. In: Evolution of Life. Fossils, Molecules and Culture. Osawa S, Honjo T, editors. Tokyo: Springer; 1991. pp. 79–95. [Google Scholar]
- 3.Jukes T H, Osawa S. J Mol Evol. 1997;45:1–3. doi: 10.1007/pl00006192. [DOI] [PubMed] [Google Scholar]
- 4.Osawa S, Jukes T H. J Mol Evol. 1989;28:271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- 5.Schultz D W, Yarus M. J Mol Biol. 1994;235:1377–1380. doi: 10.1006/jmbi.1994.1094. [DOI] [PubMed] [Google Scholar]
- 6.Yarus M, Schultz D W. J Mol Evol. 1997;45:3–6. doi: 10.1007/pl00006171. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki T, Ueda T, Watanabe K. EMBO J. 1997;16:1122–1134. doi: 10.1093/emboj/16.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuite M F, Santos M A. Biochimie. 1996;78:993–999. doi: 10.1016/s0300-9084(97)86722-3. [DOI] [PubMed] [Google Scholar]
- 9.Kano A, Ohama T, Abe R, Osawa S. J Mol Biol. 1993;230:51–56. doi: 10.1006/jmbi.1993.1125. [DOI] [PubMed] [Google Scholar]
- 10.Oba T, Andachi Y, Muto A, Osawa S. Proc Natl Acad Sci USA. 1991;88:921–925. doi: 10.1073/pnas.88.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolff G, Plante I, Lang B F, Kuck U, Burger G. J Mol Biol. 1994;237:75–86. doi: 10.1006/jmbi.1994.1210. [DOI] [PubMed] [Google Scholar]
- 12.Halanych K M. Mol Phylogenet Evol. 1995;4:72–76. doi: 10.1006/mpev.1995.1007. [DOI] [PubMed] [Google Scholar]
- 13.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohama T, Osawa S, Watanabe K, Jukes T H. J Mol Evol. 1990;30:329–332. doi: 10.1007/BF02101887. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe K, Osawa S. In: tRNA: Structure, Biosynthesis and Function. Söll D, RajBhandary U, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 225–250. [Google Scholar]
- 17.Asakawa S, Himeno H, Miura K, Watanabe K. Genetics. 1995;140:1047–1060. doi: 10.1093/genetics/140.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.HsuChen C C, Cleaves G R, Dubin D T. Nucleic Acids Res. 1983;11:8659–8662. doi: 10.1093/nar/11.24.8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valverde J R, Batuecas B, Moratilla C, Marco R, Garesse R. J Mol Evol. 1994;39:400–408. doi: 10.1007/BF00160272. [DOI] [PubMed] [Google Scholar]
- 20.Anderson S, Bankier A T, Barrell B G, de Bruijn M H, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, et al. Nature (London) 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 21.Barrell B G, Bankier A T, Drouin J. Nature (London) 1979;282:189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- 22.Bessho Y, Ohama T, Osawa S. J Mol Evol. 1992;34:331–335. doi: 10.1007/BF00160240. [DOI] [PubMed] [Google Scholar]
- 23.Boore J L, Brown W M. Genetics. 1995;141:305–319. doi: 10.1093/genetics/141.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clary D O, Wolstenholme D R. J Mol Evol. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- 25.de Bruijn M H L. Nature (London) 1983;304:234–241. doi: 10.1038/304234a0. [DOI] [PubMed] [Google Scholar]
- 26.Garey J R, Wolstenholme D R. J Mol Evol. 1989;28:374–387. doi: 10.1007/BF02603072. [DOI] [PubMed] [Google Scholar]
- 27.Himeno H, Masaki H, Kawai T, Ohta T, Kumagai I, Miura K, Watanabe K. Gene. 1987;56:219–230. doi: 10.1016/0378-1119(87)90139-9. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann R J, Boore J L, Brown W M. Genetics. 1992;131:397–412. doi: 10.1093/genetics/131.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okimoto R, Macfarlane J L, Clary D O, Wolstenholme D R. Genetics. 1992;130:471–498. doi: 10.1093/genetics/130.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pont-Kingdon G A, Beagley C T, Okimoto R, Wolstenholme D R. J Mol Evol. 1994;39:387–399. doi: 10.1007/BF00160271. [DOI] [PubMed] [Google Scholar]
- 31.Yokobori S, Ueda T, Watanabe K. J Mol Evol. 1993;36:1–8. doi: 10.1007/BF02407301. [DOI] [PubMed] [Google Scholar]
- 32.Shu D, Zhang X, Chen L. Nature (London) 1996;380:428–430. [Google Scholar]
- 33.Sueoka N. Proc Natl Acad Sci USA. 1961;47:1141–1149. doi: 10.1073/pnas.47.8.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King J L, Jukes T H. Science. 1969;164:788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- 35.Andersson G E, Kurland C G. Mol Biol Evol. 1991;8:530–544. doi: 10.1093/oxfordjournals.molbev.a040666. [DOI] [PubMed] [Google Scholar]
- 36.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]