Abstract
Several bacterial genomes exhibit preference for G over C on the DNA leading strand extending from the origin of replication to the ter-region in the genomes of Escherichia coli, Mycoplasma genitalium, Bacillus subtilis, and marginally in Haemophilus influenzae, Mycoplasma pneumoniae, and Helicobacter pylori. Strand compositional asymmetry is not observed in the cyanobacterium Synechocystis sp. genome nor in the archaeal genomes of Methanococcus jannaschii, Methanobacterium thermoautotrophicum, and Archaeoglobus fulgidus. A strong strand compositional asymmetry is observed in β-type but not α- or γ-type human herpesviruses featuring G > C downstream of oriL and C > G upstream of oriL. Dinucleotide relative abundances (i.e., dinucleotide representations normalized by the component nucleotide frequencies) are consonant with respect to the leading and lagging strands. Strand compositional asymmetry may reflect on differences in replication synthesis of the leading versus lagging strand, on differences between template and coding strand associated with transcription-coupled repair mechanisms, on differences in gene density between the two strands, on differences in residue and codon biases in relation to gene function, expression level, or operon organization, or on differences in single or context-dependent base mutational rates. The absence of strand asymmetry in the archaeal genomes may reflect the presence of multiple origins of replication.
Active recent studies have focused on the nature and extent of strand compositional asymmetry between the two DNA strands in certain bacterial genomes (1–5). A prevalence of G over C in the leading strand relative to the lagging strand was observed in the genomes of Escherichia coli, of Mycoplasma genitalium, marginally of Haemophilus influenzae, and also manifest about the origin of replication in Bacillus subtilis (2, 3, 5). These observations essentially reflect an excess of G in the leading strand or a deficit of C in the lagging strand extending from the origin of replication to the ter-region of the bacterial genome. The asymmetries are assessed in a given strand via (C−G)/(C + G) counts in a sliding window of 10 kb length and 1 kb displacement; (C − G)/(C + G) = (nC − nG)/(nC + nG) where nC (nG) is the number of C (G) nucleotides in the window at hand. A corresponding asymmetry in (A − T)/(A + T) counts is weak or nonexistent (cf. 2, 3). The bias of the leading strand favoring G over C in the E. coli genome is at variance with the common belief (e.g., ref. 6) that large contigs of each strand in E. coli and most genomes tend to be approximately equal in C and G and approximately equal in A and T. The linear genome of Borrelia burgdorferi (911 kb length) divides into two halves of opposite G–C bias (7). The replication order for the linear B. burgdorferi genome is not yet confirmed. Replication possibly begins at the chromosome termini or from a central origin and proceeds bidirectionally. Guided by the G–C skew, it has been proposed that the origin of replication in the B. burgdorferi chromosome is located about position 450 kb (7).
What are possible sources of strand compositional asymmetry? Is this asymmetry common or special to certain bacterial genomes? To what extent does the asymmetry extend constantly or irregularly over the genome? Why is there the asymmetry of G versus C but generally not of A versus T? Are there compositional asymmetries between the two strands in dinucleotides and higher-order oligonucleotides? Strand compositional asymmetry may be consequent on differences in replication synthesis of the leading versus lagging strand, on differences between template and coding strand associated with transcription-coupled repair mechanisms or deamination events, on differences in promoter and gene density between the two strands, on differences in residue and codon biases depending perhaps on gene function, expression level, or operon organization, or on differences in single-base or context-dependent mutational rates.
In this study, we examine strand compositional asymmetry for 10 complete bacterial genomes and 10 complete herpesvirus and other large viral and phage genomes. Our analysis reveals that the strand compositional asymmetry is found in the several eubacteria mentioned above and weakly in Mycoplasma pneumoniae and Helicobacter pylori but not in the cyanobacterium Synechocystis sp. nor in any of the archaea Methanococcus jannaschii, Methanobacterium thermoautotrophicum, and Archaeoglobus fulgidus.
Lobry (2, 3) associates the basis of strand compositional asymmetry to replication mutational and repair biases different in the leading versus lagging strands. Francino and Ochman (4) emphasize a mutational bias associated with transcription-coupled repair mechanisms and deamination events. The latter occurs in the coding regions where it produces the substitutions C → T more than G → A (4). Compositional asymmetry was asserted by Blattner et al. (5) for both noncoding and coding sequences and at all three codon sites but somewhat less at sites 1 and 2. Possibly also relevant, the Chi octamer GCTGGTGG of E. coli is statistically overabundant and skewed toward the leading strand (5). Biased Okazaki primer sequences conceivably could contribute to the compositional differences of the two strands.
How might different mechanisms influence the genomic DNA sequence? Long-range biases concomitant to replication are expected to be most vigorous in intergenic regions and within gene sequences at codon site 3 because codon sites 1 and 2 must accommodate amino acid requirements. Transcription generally would not discriminate between leading and lagging strands over large stretches of DNA (e.g., >50 kb) except possibly in response to gene density differences between the two strands. Prevalent concordance of orientation of replication and transcription putatively diminishes conflicts between DNA and mRNA polymerase processivity. This correlation is verified near the origin of replication for E. coli (8), for the whole genomes of M. genitalium (9) and of M. pneumoniae but not of H. influenzae (10), and not for E. coli more than 100 kb distant from the origin. Preferences within coding sequences for almost all prokaryotes and eukaryotes are significantly biased featuring base G at codon site 1 and base A at codon site 2 especially reflecting on high usages of acidic amino acids encoded by codons GAN (N stands for any nucleotide). The predominance of G at codon site 1 is further amplified by relatively high glycine, alanine and, valine usages (e.g., ref. 11).
MATERIALS
DNA sequences used in this study include complete genomes of E. coli (5), B. burgdorferi (7), H. influenzae (10), M. genitalium (9), M. pneumoniae (12), M. jannaschii (13), Synechocystis sp. strain PCC6803 (14), B. subtilis (15), Helicobacter pylori (16), Methanobacterium thermoautotrophicum (17), and Archaeoglobus fulgidus (18). The origin of replication has been identified in E. coli, B. subtilis, H. influenzae, M. genitalium, and M. pneumoniae. Also analyzed for potential strand compositional asymmetry are 10 complete genomes of herpesviruses. α-herpesviruses: herpes simplex 1 (HSV1), equine herpesvirus 1 (EHV1), and Varicella–Zoster virus (VZV). β-herpesviruses: human herpesvirus 6 (HHV6), human herpesvirus 7 (HHV7), human cytomegalovirus (HCMV), and mouse cytomegalovirus (MCMV). γ-herpesviruses: Epstein–Barr virus (EBV), equine herpesvirus 2 (EHV2), and herpesvirus saimiri (HVS).
RESULTS
Strand Compositional Asymmetry in Prokaryotic Genomes.
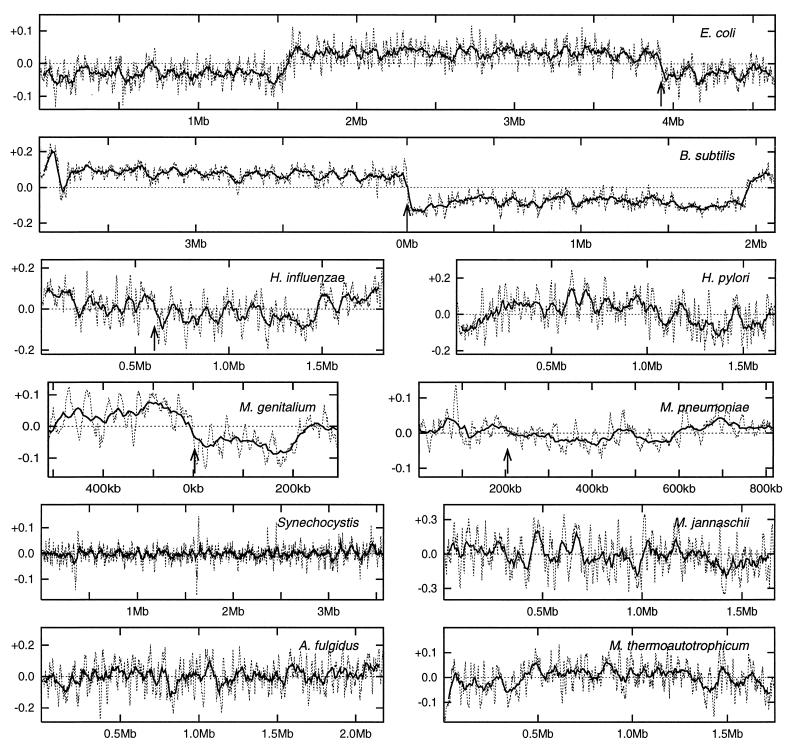
Sliding window (C − G)/(C + G) counts are displayed in Fig. 1 for several complete bacterial genomes. The E. coli genome divides into two parts with predominance of G > C clockwise from oriC and C > G counterclockwise from oriC both extending to the ter-region. The same manifest asymmetry is observed in the genomes of M. genitalium and B. subtilis (2, 3). A marginal asymmetry is observed in the genomes of H. influenzae, H. pylori, and M. pneumoniae with G > C mostly clockwise from the origin and C > G mostly counterclockwise from the origin. The bias is assessed with a 50-kb sliding window but is subject to many fluctuations with a 10-kb sliding window. In the cyanobacterial Synechocystis sp. and in the archaeal M. jannaschii, M. thermoautotrophicum, and A. fulgidus, the sliding window (C − G)/(C + G) counts fluctuate irregularly positively and negatively along the genome revealing no consistent pattern over large portions of the genomes.
Figure 1.
(C − G)/(C + G) counts in sliding windows of 50 kb with 5-kb displacement (solid line) and 10 kb with 1-kb displacement (dashed line) in complete genomes of E. coli, B. subtilis, H. influenzae, H. pylori, M. genitalium, M. pneumoniae, Synechocystis PCC6803, M. jannaschii, M. thermoautotrophicum, and A. fulgidus. The value of (C − G)/(C + G) is assigned to the position at the middle of the window. The arrows indicate locations of the origin of replication (when known).
In seeking to explain the extended strand compositional asymmetry in E. coli, M. genitalium, M. pneumoniae, and H. influenzae we examined intergenic regions and codon site 3 in genes where mutational substitutions are expected to be more pronounced (Table 1). A caveat concerning intergenic regions: these contain regulatory signal sequences (e.g., promoters, cis-acting elements, Shine–Dalgarno sequences, and others) that may be biased. Other highly recurrent sequences (e.g., REP elements in E. coli, HIP in Synechocystis) are symmetric and would not contribute to strand bias. In nonoverlapping 100-kb windows clockwise and counterclockwise of the origin, the B. subtilis intergenic regions show persistent %G > %C by about 3% in the leading strand (i.e., about 300 more G nucleotides than C nucleotides per 10 kb). H. influenzae intergenic regions entail a weak strand asymmetry still favoring %G > %C in the leading strand. Intergenic sequences of E. coli show a gradual compositional change with distance from the origin of replication with C ≈ G near the origin (in the region <100 kb) but unambiguously %G > %C more distant from the origin. The strand compositional asymmetry in M. genitalium intergenic regions is tenuous and is not observed in intergenic regions of M. pneumoniae (Table 1).
Table 1.
Compositional bias in 100-kb windows about the origin of replication at codon site 3 and in intergenic sequences
| Distance from the origin, kb | Overall G − C, %*
|
Aggregate length of genes,† kb
|
Codon site 3 of genes
|
Intergenic sequences§
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G − C difference, %‡
|
A − T difference, %‡
|
Length, kb
|
G − C, %
|
A − T, %
|
||||||||||||
| R | L | R+ | R− | L+ | L− | R | L | R | L | R | L | R | L | R | L | |
| E. coli | ||||||||||||||||
| 0–100 | +2.1 | +1.7 | 61.1 | 22.8 | 63.7 | 25.7 | −0.1 | 0.0 | −3.5 | −1.7 | 12.1 | 10.6 | 0.0 | +0.7 | −0.9 | +0.7 |
| 100–200 | +1.1 | +0.9 | 34.0 | 46.3 | 37.0 | 50.5 | +4.2 | +6.3 | −0.5 | −3.6 | 12.1 | 11.5 | +0.8 | +1.2 | +0.3 | −2.6 |
| 200–300 | +2.5 | +1.5 | 52.1 | 27.9 | 52.5 | 31.4 | +2.1 | +5.5 | −2.2 | 0.0 | 11.0 | 16.0 | +2.1 | +1.4 | −0.6 | −2.0 |
| 300–400 | +1.2 | +1.5 | 37.9 | 44.5 | 47.3 | 42.6 | +7.1 | +3.4 | −2.8 | −2.9 | 13.5 | 9.8 | +0.7 | +0.5 | −0.9 | −1.3 |
| 400–500 | +1.7 | +2.4 | 42.3 | 45.6 | 52.4 | 36.4 | +5.9 | +4.0 | −2.9 | −3.8 | 11.4 | 8.1 | +2.2 | +3.6 | −0.6 | +0.2 |
| 500–600 | +1.1 | +1.4 | 35.9 | 49.9 | 41.3 | 45.2 | +5.8 | +6.4 | −0.3 | −2.1 | 13.8 | 11.8 | +1.9 | +2.1 | 0.0 | +1.5 |
| 600–700 | +1.2 | +1.5 | 36.2 | 48.2 | 44.7 | 45.4 | +5.1 | +3.3 | −1.7 | −1.1 | 16.1 | 11.0 | +2.6 | +2.7 | −0.8 | −0.8 |
| 700–800 | +1.7 | +1.7 | 49.0 | 42.2 | 49.1 | 41.7 | +4.8 | +7.7 | −0.1 | −2.6 | 10.3 | 10.1 | +2.0 | +1.4 | −1.1 | −1.8 |
| 800–900 | +2.7 | +1.9 | 60.0 | 31.9 | 57.1 | 31.5 | +9.2 | +4.8 | −0.5 | −0.9 | 7.5 | 11.1 | +1.7 | +1.6 | −1.2 | −1.4 |
| 900–1,000 | +1.8 | +1.9 | 48.2 | 33.3 | 43.7 | 42.2 | +2.7 | +7.9 | −0.1 | −1.9 | 12.6 | 12.4 | +1.9 | +1.5 | +0.7 | −2.7 |
| 1,000–1,100 | +1.2 | +1.8 | 42.7 | 40.0 | 56.8 | 33.8 | +5.3 | +3.9 | −0.1 | −1.5 | 17.9 | 9.9 | +1.9 | +2.6 | −2.1 | −1.6 |
| 1,100–1,200 | +1.7 | +1.9 | 50.8 | 39.2 | 34.0 | 45.4 | +4.6 | +2.6 | −3.3 | −2.1 | 10.1 | 15.4 | +1.6 | +1.9 | −0.5 | −0.3 |
| 1,200–1,300 | +2.0 | +2.1 | 58.7 | 28.1 | 62.1 | 27.4 | +8.2 | +2.2 | −0.7 | 0.0 | 13.0 | 9.4 | +2.8 | +1.2 | −1.8 | +0.4 |
| 1,300–1,400 | +1.3 | +0.9 | 36.6 | 52.7 | 42.6 | 47.1 | +8.0 | +3.9 | +0.4 | +0.4 | 10.3 | 10.1 | +3.2 | +2.1 | −1.4 | +0.6 |
| 1,400–1,500 | +1.1 | +1.7 | 48.3 | 38.8 | 57.1 | 29.6 | +2.5 | +2.1 | +1.6 | −4.1 | 12.2 | 12.7 | +1.8 | +1.5 | +1.2 | −0.2 |
| 1,500–1,600 | +1.6 | +2.2 | 44.5 | 45.3 | 58.2 | 33.2 | +6.2 | +6.7 | −0.4 | −0.1 | 9.8 | 8.6 | +1.5 | +2.4 | +0.5 | −1.7 |
| 1,600–1,700 | +1.8 | +2.0 | 57.1 | 33.1 | 52.2 | 36.6 | +8.5 | +4.7 | +0.3 | −1.2 | 9.8 | 10.5 | +3.0 | +2.3 | +0.7 | −0.1 |
| H. influenzae | ||||||||||||||||
| 0–100 | +1.3 | +1.7 | 27.7 | 45.5 | 58.1 | 26.7 | +3.1 | −0.2 | −0.6 | −3.4 | 16.6 | 16.0 | +3.1 | +2.0 | −0.1 | −1.6 |
| 100–200 | +1.4 | +0.5 | 38.8 | 38.3 | 41.8 | 47.5 | +0.3 | +2.8 | +4.9 | −2.8 | 18.6 | 10.1 | +2.2 | +0.9 | −0.1 | −1.6 |
| 200–300 | +1.3 | +0.4 | 53.6 | 36.6 | 41.4 | 41.3 | +2.1 | −0.3 | −4.8 | −0.2 | 10.6 | 16.3 | +1.8 | +0.2 | −2.1 | −0.2 |
| 300–400 | +1.0 | +0.5 | 52.9 | 34.1 | 26.1 | 42.7 | +3.6 | +2.7 | −0.0 | −3.5 | 12.2 | 25.8 | +2.4 | +0.8 | −3.0 | −0.5 |
| 400–500 | +0.8 | +2.0 | 45.5 | 38.2 | 38.6 | 37.7 | +2.4 | +5.9 | −0.6 | −3.3 | 15.7 | 17.4 | +0.3 | +1.8 | −2.5 | −0.4 |
| 500–600 | +0.2 | +2.9 | 27.8 | 54.3 | 60.2 | 28.2 | +3.4 | +8.7 | −3.2 | −7.5 | 17.5 | 11.3 | +0.9 | +1.3 | −2.0 | −3.2 |
| 600–700 | +2.0 | +3.2 | 37.5 | 48.3 | 50.8 | 29.0 | +8.4 | +8.9 | −7.0 | −4.3 | 14.0 | 15.6 | +2.2 | +4.2 | +0.4 | +1.9 |
| B. subtilis | ||||||||||||||||
| 0–100 | +5.4 | +3.1 | 66.8 | 4.3 | 52.7 | 32.2 | +5.5 | +8.6 | +0.9 | −4.5 | 10.1 | 15.8 | +3.4 | +3.2 | −0.2 | +2.1 |
| 100–200 | +4.5 | +2.8 | 59.2 | 15.0 | 65.9 | 22.9 | +7.8 | +6.1 | −8.6 | −1.5 | 9.5 | 9.3 | +3.7 | +3.3 | 0.0 | +2.4 |
| 200–300 | +3.7 | +2.2 | 61.4 | 25.6 | 49.6 | 38.5 | +9.9 | +6.4 | −1.4 | 0.0 | 12.5 | 12.2 | +4.6 | +2.0 | +1.5 | +0.4 |
| 300–400 | +3.3 | +3.3 | 69.2 | 24.1 | 68.1 | 22.6 | +9.9 | +7.2 | +0.2 | −2.2 | 8.4 | 9.5 | +4.5 | +4.4 | +2.1 | −0.8 |
| 400–500 | +2.3 | +3.4 | 64.8 | 23.9 | 65.1 | 19.5 | +3.1 | +2.8 | −1.0 | +0.7 | 9.9 | 14.5 | +2.5 | +3.7 | +3.8 | +2.5 |
| 500–600 | +3.5 | +2.6 | 62.1 | 17.9 | 61.7 | 24.4 | +4.7 | +6.8 | −0.6 | −1.0 | 18.9 | 11.5 | +3.5 | +3.4 | +1.5 | +1.0 |
| 600–700 | +3.5 | +3.9 | 55.1 | 23.4 | 74.0 | 17.0 | +7.6 | +7.5 | −3.5 | −3.6 | 16.6 | 9.1 | +2.9 | +3.7 | −1.0 | −1.3 |
| 700–800 | +4.1 | +2.9 | 81.2 | 11.0 | 68.3 | 21.0 | +9.0 | +3.8 | −7.4 | +0.6 | 7.2 | 10.3 | +5.8 | +3.8 | +2.1 | +2.1 |
| 800–900 | +2.6 | +3.5 | 56.0 | 32.7 | 65.6 | 23.6 | +8.2 | +6.3 | −0.6 | −1.2 | 11.1 | 11.0 | +3.5 | +3.9 | +2.8 | +1.4 |
| 900–1,000 | +3.3 | +2.2 | 64.7 | 17.7 | 58.9 | 30.0 | +4.7 | +7.3 | −0.1 | −0.3 | 11.6 | 11.1 | +1.8 | +3.5 | −0.6 | +1.7 |
| 1,000–1,100 | +3.1 | +3.5 | 60.4 | 32.7 | 61.7 | 20.8 | +11.1 | +5.9 | −2.1 | −1.0 | 10.9 | 9.9 | +2.7 | +3.0 | +2.0 | +0.4 |
| 1,100–1,200 | +2.6 | +3.2 | 53.6 | 37.5 | 67.9 | 21.3 | +9.7 | +8.3 | +0.5 | −2.1 | 11.0 | 10.6 | +3.3 | +3.3 | −0.5 | +2.7 |
| 1,200–1,300 | +3.2 | +3.9 | 60.4 | 21.6 | 78.1 | 11.8 | +6.7 | +2.8 | −1.0 | −2.4 | 15.7 | 9.3 | +3.4 | +5.0 | +1.6 | +2.1 |
| 1,300–1,400 | +3.3 | +4.1 | 68.7 | 18.8 | 79.2 | 9.6 | +5.4 | +5.7 | −0.8 | −3.8 | 11.0 | 11.2 | +3.3 | +4.1 | +2.5 | +2.0 |
| 1,400–1,500 | +3.1 | +2.7 | 61.5 | 24.3 | 53.9 | 26.6 | +5.6 | +5.3 | −1.1 | +1.4 | 14.6 | 18.3 | +3.2 | +3.2 | +2.8 | +0.8 |
| M. genitalium | ||||||||||||||||
| 0–100 | +1.7 | +1.6 | 74.2 | 17.9 | 72.5 | 11.9 | +0.4 | +0.3 | −3.8 | −6.2 | 10.3 | 15.1 | +0.9 | +0.8 | +1.4 | +1.5 |
| 100–200 | +2.2 | +0.8 | 82.4 | 4.1 | 73.8 | 16.5 | −0.5 | −0.1 | −4.5 | +2.8 | 7.9 | 10.3 | +0.7 | +0.6 | +1.6 | −1.6 |
| M. pneumoniae | ||||||||||||||||
| 0–100 | +0.3 | +0.3 | 73.7 | 17.1 | 67.3 | 22.9 | +2.4 | −0.7 | −1.2 | −3.5 | 9.4 | 10.1 | −0.3 | −1.1 | 0.0 | −3.2 |
| 100–200 | +0.7 | +0.8 | 69.8 | 18.3 | 71.3 | 3.7 | +2.1 | 0.0 | −3.3 | −3.6 | 13.4 | 26.7 | +0.3 | +1.6 | +0.5 | +2.4 |
| 200–300 | +0.1 | +0.7 | 69.5 | 15.7 | 91.7 | 1.4 | +0.9 | −0.5 | +0.8 | −0.8 | 13.0 | 5.9 | −0.3 | +0.2 | +1.4 | −2.6 |
| HHV6 | ||||||||||||||||
| 0–50 | +6.6 | +8.2 | 23.9 | 23.6 | 30.9 | 16.2 | +23.6 | +30.2 | −16.8 | −23.7 | 3.2 | 4.1 | +8.9 | +7.8 | −6.5 | −7.1 |
| HHV7 | ||||||||||||||||
| 0–50 | +2.2 | +2.3 | 23.6 | 23.6 | 32.9 | 16.1 | +6.4 | +4.3 | −2.7 | −6.5 | 4.2 | 3.5 | +2.2 | +1.1 | −1.5 | −3.8 |
| HCMV | ||||||||||||||||
| 0–50 | +4.5 | +3.0 | 22.5 | 25.6 | 30.0 | 15.8 | +12.3 | +5.2 | −8.6 | −9.1 | 3.4 | 5.0 | +3.6 | +3.1 | +1.6 | −0.7 |
Overall difference %G − %C in 100-kb regions (50 kb for viral genomes) with respect to the leading strand (not distinguishing coding and noncoding). R and L signify clockwise and counterclockwise from the origin relative to the reported strand in the database, respectively.
Within each 100 kb (50 kb for viral genomes), aggregate genes encoded in the leading strand are labeled + and genes encoded in the lagging strand are labeled −.
The G − C difference in genes from the leading and lagging strands is shown as (fG+ − fC+) − (fG− − fC−) where fG+ (fC+) is the frequency of base G (C) at codon site 3 calculated from the genes of the leading strand and fG− (fC−) are the frequencies for the genes of the lagging strand. Similarly for the A − T difference.
G − C frequencies of combined intergenic sequences in the corresponding 100-kb windows (i.e., excluding coding, rRNA and tRNA genes). G − C frequencies are reported with respect to the leading strand.
Codon site 3 base frequencies were compared in all genes from the leading and lagging strands in the same 100-kb windows. Gene orientation is biased in the E. coli genome near the origin of replication, with more than 70% of genes oriented in the direction of replication. Codon site 3 nucleotide choices show no bias near (<100 kb) the origin of replication. However, there is manifest compositional asymmetry at codon site 3 and in intergenic sequences that are more distant from the origin (>100 kb, see Table 1).
Similar to E. coli, codon site 3 in H. influenzae exhibits a mild asymmetry away from the origin (>300 kb). Gene orientation does not show any consistent trend in H. influenzae but gene orientation bias is pronounced throughout the genome in M. genitalium and in M. pneumoniae with ≈85% of genes encoded from the leading strand. Codon site 3 base composition is congruent for genes from the leading strand and genes from the lagging strands in M. genitalium and in M. pneumoniae (Table 1). These observations suggest that biased gene orientation could be the major cause of the strand compositional asymmetry in these genomes. B. subtilis highlights both biased gene orientation and a strand compositional asymmetry extending from the origin throughout the genome.
A strand compositional asymmetry with respect to A versus T was detected only in M. genitalium and B. subtilis but weaker than that of G versus C. Codon site 3 generally shows T > A, and this tendency is stronger in the genes encoded by the leading strand (Table 1).
Dinucleotide Composition of Leading and Lagging DNA Strand.
In addition to base composition of single nucleotides, we compared dinucleotide compositions of leading and lagging strands. The latter was assessed by dinucleotide relative abundance values. These are defined as ρXY = fXY/fXfY, where fXY denotes frequency of the dinucleotide XY and fX and fY are frequencies of the nucleotides X and Y, respectively (e.g., ref. 19). In a sliding window of 50 kb, no significant differences were observed between pairs of complementary dinucleotide relative abundances across the genome (not shown). The constancy of dinucleotide relative abundances relative to the two strands is consistent with the constancy of the genome signature (defined as the collection ρ*XY = f*XY/f*X f*Y, extending over all dinucleotides XY evaluated for 50-kb contigs where the f*XY are the symmetrized dinucleotide frequencies computed from the contig concatenated with its inverted complementary sequence and similarly for f*X and f*Y; refs. 19 and 20). See Discussion for further interpretations.
Compositional Asymmetry in Large Viral Genomes.
Of the 10 complete herpesvirus genomes, a strand compositional asymmetry is observed only in HHV6, HHV7, and HCMV, all classified as β-herpesviruses. However, another member of the same group, MCMV, does not show strand compositional asymmetry. In the three herpesviruses having strand compositional asymmetry, the preference switches from C > G to G > C at the lytic origin of replication (oriL) located near positions 68 kb in HHV6, 62 kb in HHV7, and 93 kb in HCMV. It is generally believed that replication in herpesviruses commences bidirectionally but later switches to a rolling-circle form (e.g., ref. 21). It is also suspected that a RNA/DNA hybrid is created at the initial stages of herpesvirus replication. Several other long viral genomes (generally >150 kb) including Vaccinia virus, Variola virus, Autographa californica nuclear polyhedrosis virus, Bombyx mori nuclear polyhedrosis virus, Orgyia pseudotsugata nuclear polyhedrosis virus, Molluscum contagiosum virus 1, Lymphocystic disease virus, African swine fever virus, and Paramecium bursaria chlorella virus do not reveal clear strand asymmetry over large portions of the genomes (not shown).
In HHV7 and HCMV, the intergenic regions and codon site 3 of genes in the two strands feature a compositional asymmetry to about the same extent as in B. subtilis (Table 1). Among the herpesviruses, HHV6 shows the sharpest strand compositional asymmetry. In HHV6 there are clear amino acid and codon usage differences between genes transcribed from the leading versus the lagging strand (the definition of leading and lagging strands assumes that replication proceeds bidirectionally from oriL).
DISCUSSION
Possible sources of compositional strand asymmetry are highlighted in Table 2. These include effects of DNA or RNA metabolic asymmetry. For example, in the lagging strand RNA, primase settings are required for initiating replication and producing the Okazaki fragments and at completion for removing the primers. These generally differ from the helicase activity of the leading strand where there are apparently only slight discontinuities in replication processivity. There is currently a scarcity of data to investigate biases in the primer sequences. There may also be different kinds of errors associated with template–primer alignments and insertions. Moreover, architectural asymmetry at the replication forks putatively engenders different types of lesions on the two strands (1). Generally, lagging strand replication is less accurate, and especially deletions of Tn runs are higher in the lagging strand than in the leading strand (22). Efficiency of exonucleolytic proofreading or mismatch repair seems to be influenced by genomic G + C versus A + T richness (1).
Table 2.
Possible sources of strand compositional asymmetry
| Source | Type of asymmetry |
|---|---|
| Possible replication biases | Enzymological asymmetry* |
| Architectural asymmetry at the replication fork | |
| Different mutation rates in leading vs. lagging strand | |
| dNTP pools fluctuate during the cell cycle | |
| Θ-form vs. rolling-circle replication (or bipartite replication) (in large viruses apparently Θ-bidirectional replication initiation but later switches to rolling circle) | |
| Differences in signal or binding sites in the two strands | |
| Possible transcription biases | Expression level—active operons may allow specific biases because transcription-coupled repair is more efficient in active regions |
| Deamination events (C → T mutations in coding sequences) | |
| Biased gene density | Gene density high in the leading strand (applies across E. coli close to oriC and in the entire genomes of M. genitalium, M. pneumoniae, and B. subtilis). Is the asymmetry in gene orientation more pronounced in Gram-positive vs. Gram-negative genomes? |
| Amino acid and codon biases | Dominated by G > C at site 1, A > T at site 2—in conjunction with biased gene density this may cause strand compositional asymmetry |
| Codon site 3 less restricted by protein requirements. | |
| Multiple replication origins | Putatively attenuates strand compositional asymmetry; e.g., phage T4 with multiple transcription primed origins; possibly for many archaeal genomes |
| Selection for regulation and control sites may be strand-biased | |
| Differences in growth rate or in certain gene (or operon) clusters | |
In some bacterial genomes helicase primase bias (primers ≈5% of Okazaki fragments). These may differ about the origin and away from the origin [e.g., in HSV1 the origin-binding protein UL9 (helicase primase) acts at the origin whereas the helicase primase complexes UL5, UL8, UL52 acts away from the origin].
There is considerable variation in dNTP precursor pools and in dNTP metabolizing enzymes during the cell cycle (22). These enzymes and degrees of metabolic efficiency are considered to be linked to the replication machinery. DNA damage can lead to altered dNTP levels and thus to dNTP imbalances, which, in turn, are prone to induce single-point mutations because the probability of correct or incorrect nucleotide incorporation is related to the amounts of the competing substrates. Alterations in dNTP pools can modulate spontaneous and induced genetic changes. dNTP pool disturbances can lead to chromosomal aberrations, mutations, recombination, and repair modifications (22). There are also possibilities of directed mutagenesis like Treffers mutator gene (23).
Compositional strand asymmetry is manifest in several eubacterial genomes including E. coli, B. subtilis, M. genitalium, B. burgdorferi, and marginally in M. pneumoniae, H. pylori, and H. influenzae but insignificantly in the cyanobacterium Synechocystis sp. It is not present in the archaeal genomes of M. jannaschii, M. thermoautotrophicum, and A. fulgidus. Apropos, an origin of replication, has not been identified in the archaea at hand and it has been speculated that many archaeal genomes possess multiple origins of replication (24). Along these lines, strand compositional asymmetry is not apparent in the phage T4 genome nor in the yeast Saccharomyces cerevisiae chromosomes (not shown). The latter are known to possess multiple origins of replication (ARS elements). Transcriptionally primed T4 replicates bidirectionally, with multiple start sites (25). The bacteriophage lambda (λ) genome divides into three parts, each with a single gene orientation, whose boundaries feature a discontinuity in (C − G)/(C + G) and (A − T)/(A + T) counts relative to the two strands. This is consistent with the model in which the strand compositional asymmetry can be attributed to the preferred gene orientation coupled with amino acid and codon usage biases. Lambda replicates as a phage bidirectionally from a single origin near the O gene (26) or as a prophage (when integrated into E. coli) unidirectionally with most genes encoded from the leading strand (27). Genes of bacteriophage T7 are all transcribed in the same orientation, and the DNA strand corresponding to the coding strand features A > T and G > C consistent with amino acid constraints.
Two principal mechanisms have been ventured to explain the extant strand compositional asymmetry. The first (2, 3) emphasizes asymmetric replication and repair mutational tendencies during base incorporation into the leading and lagging strands. This may be related to primer sequence biases of the helicase primase enzyme in initiating Okazaki fragments (see also below). The second hypothesis (4) argues that the principal strand asymmetric mutation biases are generated during transcription and transcription-coupled repair (28) in conjunction with many deamination events imposed on the single-strand coding sequence.
In another perspective, the strand compositional asymmetry may be correlated with selective amino acid and codon constraints (e.g., relatively high acidic residue usages underscoring G at codon site 1 and A at codon site 2). A plethora of genes favoring one strand in conjunction with appropriate residue and codon choices within genes can generate a substantial strand compositional asymmetry. Transcription-coupled repair is known to be more efficient on the template strand than on the coding strand, especially in the removal of cyclobutane dimers (28). On this basis we would expect more rapid repair of adducts on the template strand than on the coding strand. This problem can be somewhat mitigated in coding sequences by selecting against pyrimidine dinucleotides concomitantly increasing the frequencies of A and G. Deamination events in the coding strand during transcription are projected to increase mutations of C → T (4). However, the E. coli genome of average 51% G + C content has C > T at codon site 1 and C ≈ T at codon site 3, contrary to expectations ensuing from a C → T mutational tendency. Only site 2 has T > C, probably because of a preponderance of hydrophobic amino acids encoded by NTN codons (29). Moreover, under an excess of C → T transitions in one strand, the G > C phenomenon should be compensated by T > A and vice versa and the counts of A + G and C + T would stay constant. However, an unequivocal inequality is observed only for G − C and not A − T. Thus, it appears that the observed asymmetry cannot arise only from the above-described biased transition rates. Parenthetically, A⋅T positions putatively mutate faster than G⋅C positions, which may in part account for the diminished asymmetry of A versus T between the two strands (1).
For the bacterial and viral genomes possessing strand compositional asymmetry, nucleotide replication-biased and gene orientation-biased models alone do not adequately explain the strand compositional asymmetry and why and when it exists. Rather, the strand compositional asymmetry arises from a combination of mechanisms related to replication and repair, transcription, and selective constraints affecting amino acid and codon usages. Biased gene orientation does not account for the marginal strand asymmetry in H. influenzae, where gene and replication orientation are uncorrelated. Also, the gene orientation-biased model cannot account for the strand compositional asymmetry in the three human herpesviruses, HHV6, HHV7, and HCMV, where gene orientation is unidirectionally traversing oriL (Table 1), and genes on average contain approximately the same amounts of G and C nucleotides (not shown). In E. coli, distant from the origin, strand compositional asymmetry prevails even when the opposite gene orientation is locally favored (see Fig. 1 and Table 1). In these genomes, an inhomogeneous mutational bias seems to be the predominant cause of the strand compositional asymmetry. However, in spite of a large orientation gene bias there is no significant strand compositional asymmetry at codon site 3 of genes or in intergenic regions in M. pneumoniae and M. genitalium. Near the origin of E. coli the strand compositional asymmetry can be accommodated by a preferred gene direction concordant with replication direction plus the predominance of G > C in coding segments.
It is unclear why the strand compositional asymmetry varies over different regions of the same genome (e.g., E. coli close to the origin versus distant from the origin, see Table 1). Clusters of functionally related genes or genes with similar expression levels may result in specific amino acid or codon choices. For example, a virtual continuum of 26 ribosomal protein genes (“highly expressed”) is present in the leading strand of the E. coli genome about 480 kb left of the origin. However, this region does not seem exceptional in Table 1 or Fig. 1. A similar cluster of 25 ribosomal protein genes is present in H. influenzae about 250 kb right of the origin, but as in E. coli, this region is not distinguished from the rest of the genome. It is known that transcription-coupled repair is more efficient in regions of active genes. On this basis we might expect, especially during the rapid growing phase, that the transcription template strand of highly expressed genes maintains accurate fidelity. Although it is known that the highly expressed genes entail highly biased codon usage (30), these apparently do not correspond to nucleotide biases. Possible differences during replication in mutation rates and mutational biases over different parts of the genome may also contribute to variance in strand compositional asymmetry. Differences in mutation rates along different parts of mammalian genomes may arise from unbalanced nucleotide precursor pool concentrations during replication. Nucleotide precursor pool imbalances can influence replication and repair efficiency in both eukaryotes and prokaryotes (22). However, it is not known if and how the nucleotide precursor pool concentrations vary during replication in different genomes. Along these lines, the G + C content of mammalian genes appears to be uncorrelated with time of replication (31).
Significant extremes in dinucleotide relative abundances are present in most genomes (19). The dinucleotide relative abundance vector is pervasively constant (assessed over 50-kb contigs) throughout the whole genome constituting a genome signature (19, 20). These highly stable normalized DNA doublet frequencies suggest that there may be genome-wide factors such as functions of the replication and repair machinery, context-dependent mutation rates, and base-step conformational tendencies that impose limits on the compositional patterns of a genomic sequence. Intriguingly, dinucleotide relative abundances tend to be symmetric and effectively constant relative to the leading and lagging strand for all dinucleotides (not shown) despite the strand compositional asymmetry. The constancy of dinucleotide relative abundances relative to the two strands is consistent with the constancy of the genome signature. General trinucleotide and higher-order relative abundances are correlated with dinucleotide relative abundances (19). Offhand, mechanisms generating the asymmetry between the two DNA strands seem to operate at the level of individual nucleotides and are not context-dependent. However, mutation is generally regarded as context-dependent (1). Strand asymmetry may exist in the distribution of certain signal sequences probably as a result of biased selection rather then biased mutation tendencies. For example, >75% of Chi-sites GCTGGTGG in the E. coli genome are found in the leading strand (5).
Acknowledgments
We thank Drs. B. E. Blaisdell, C. Burge, A. M. Campbell, and E. Mocarski for valuable comments on the manuscript. This work was supported in part by National Institutes of Health Grants 2R01GM10452-33 and 5R01HG00335-09 and National Science Foundation Grant 9403553-002.
Note Added in Proof
Recently released preliminary sequences of the complete genomes of Chlamydia trachomatis (Stanford University, 1 Mb length) and Mycobacterium tuberculosis (Sanger Centre, 4.4 Mb length) both exhibit an extended compositional asymmetry in C–G counts. C. trachomatis features potently C > G in the region between 200 kb and 700 kb of the preliminary sequence and G > C in the other half of the genome. M. tuberculosis shows G > C in the first half of the genome (0–2.3 Mb) and C > G in the second half of the genome, with few exceptions. A pronounced anomaly is found in the interval 3.90–3.95 Mb, which deviates in both base and dinucleotide compositions from the genome average, suggestive of a recent, laterally transferred region.
References
- 1.Kunkel T A. BioEssays. 1992;14:303–308. doi: 10.1002/bies.950140503. [DOI] [PubMed] [Google Scholar]
- 2.Lobry J R. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 3.Lobry J R. Science. 1996;272:745–746. doi: 10.1126/science.272.5262.745. [DOI] [PubMed] [Google Scholar]
- 4.Francino M P, Ochman H. Trends Genet. 1997;13:240–245. doi: 10.1016/S0168-9525(97)01118-9. [DOI] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Fickett J W, Torney D C, Wolf D R. Genomics. 1992;13:1056–1064. doi: 10.1016/0888-7543(92)90019-o. [DOI] [PubMed] [Google Scholar]
- 7.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R A, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, et al. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 8.Brewer B J. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 9.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 10.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 11.Karlin S, Blaisdell B E, Bucher P. Protein Eng. 1992;5:729–738. doi: 10.1093/protein/5.8.729. [DOI] [PubMed] [Google Scholar]
- 12.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- 15.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 16.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, et al. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 17.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klenk H-P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 19.Karlin S, Burge C. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 20.Karlin S, Mrázek J, Campbell A M. J Bacteriol. 1997;179:3899–3913. doi: 10.1128/jb.179.12.3899-3913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamzeh F M, Lietman P S, Gibson W, Hayward G S. J Virol. 1990;64:6184–6195. doi: 10.1128/jvi.64.12.6184-6195.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas D C, Svoboda D L, Vos J-M H, Kunkel T A. Mol Cell Biol. 1996;16:2537–2544. doi: 10.1128/mcb.16.5.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox E C, Yanofsky C. Proc Natl Acad Sci USA. 1967;58:1895–1902. doi: 10.1073/pnas.58.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen G J, Woese C R. Cell. 1997;89:991–994. doi: 10.1016/s0092-8674(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 25.Mosig G, Colowick N. Methods Enzymol. 1995;262:587–604. doi: 10.1016/0076-6879(95)62046-x. [DOI] [PubMed] [Google Scholar]
- 26.Scherer G. Nucleic Acids Res. 1978;5:3141–3156. doi: 10.1093/nar/5.9.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell A M. Adv Genet. 1962;11:101–146. [Google Scholar]
- 28.Lommel L, Carswell-Crumpton C, Hanawalt P C. Mutat Res. 1995;336:181–192. doi: 10.1016/0921-8777(94)00055-b. [DOI] [PubMed] [Google Scholar]
- 29.Kypr J, Mrázek J. Int J Biol Macromol. 1987;9:49–53. [Google Scholar]
- 30.Sharp P M, Li W-H. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyre-Walker A. Nucleic Acids Res. 1992;20:1497–1501. doi: 10.1093/nar/20.7.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]