Abstract
As the amount of available sequence data increases, it becomes apparent that our understanding of translation initiation is far from comprehensive and that prior conclusions concerning the origin of the process are wrong. Contrary to earlier conclusions, key elements of translation initiation originated at the Universal Ancestor stage, for homologous counterparts exist in all three primary taxa. Herein, we explore the evolutionary relationships among the components of bacterial initiation factor 2 (IF-2) and eukaryotic IF-2 (eIF-2)/eIF-2B, i.e., the initiation factors involved in introducing the initiator tRNA into the translation mechanism and performing the first step in the peptide chain elongation cycle. All Archaea appear to posses a fully functional eIF-2 molecule, but they lack the associated GTP recycling function, eIF-2B (a five-subunit molecule). Yet, the Archaea do posses members of the gene family defined by the (related) eIF-2B subunits α, β, and δ, although these are not specifically related to any of the three eukaryotic subunits. Additional members of this family also occur in some (but by no means all) Bacteria and even in some eukaryotes. The functional significance of the other members of this family is unclear and requires experimental resolution. Similarly, the occurrence of bacterial IF-2-like molecules in all Archaea and in some eukaryotes further complicates the picture of translation initiation. Overall, these data lend further support to the suggestion that the rudiments of translation initiation were present at the Universal Ancestor stage.
Translation initiation is an intriguing problem from two ostensibly diverse perspectives, the biomechanical and the evolutionary: In the former terms, translation initiation becomes a problem of threading an mRNA tape into a ribosomal tape reader, establishing the correct reading frame, and then effecting the first step in the translation elongation cycle; in the latter term, it becomes one of the evolution of translation, particularly the emergence of the initiation process-the rudimentary mechanisms from which it arose, its relationship to translation elongation, the evolutionary stage(s) at which both processes emerged, and what this implies regarding the nature of the universal ancestor (1, 2) and the primary branchings on the phylogenetic tree. A thorough-going comparative approach to translation initiation will obviously reveal a great deal about the evolution of the process. Not so obvious is that such an approach should be an aid to understanding the mechanism of the process as well.
The various components of the translation apparatus tend to be highly conserved over evolutionary time; many are universal in distribution (3, 4). Translation initiation is an exception, however. Although the bacterial (5) and eukaryotic (6) mechanisms resemble one another, their resemblance at the molecular level is slight. It had been generally accepted that bacterial and eukaryotic translation initiation mechanisms, although mechanistically generally similar, were molecularly unrelated and so had evolved independently. The Methanococcus jannaschii genome (7–9), which gave us our first comprehensive look at the componentry of archaeal translation initiation, revealed that archaeal translation initiation showed considerable homology with eukaryotic initiation, which, if anything, reinforced the divide between bacterial initiation and that seen in the other domains. We recently have shown, however, that bacterial translation initiation factor 1 (IF-1), contrary to previously accepted opinion, is related in sequence to its eukaryotic/archaeal (functional) counterpart, eIF-1A, and that several other initiation/translation factors thought to be confined to one or another of the domains have homologs in all domains (10). Thus, it would appear that even translation initiation had its rudiments in a functionality that existed at the universal ancestor stage (10).
In the present communication, we explore the more complex case of the translation initiation factor that shepherds the initiator tRNA into the ribosome. In Bacteria, this is accomplished by IF-2 (5). In eukaryotes, it is accomplished by eIF-2, a protein that comprises three subunits (6). Both factors form the initiation complex by interacting with Met-tRNAiMet, GTP, and the small ribosomal subunit (5, 6). However, the process differs slightly in the two cases, e.g., the stage at which GTP or mRNA participates in the complex (5, 6). It seems that they differ also in evolutionary origin; the γ subunit (only) of eIF-2 is a member of the elongation factor (EF) family, being specifically related therein to bacterial EF-Tu (11). Bacterial IF-2, however, is only a distant sister group of this family (11).
At the end of the initiation step, Eukarya use eIF-5 (whose N-terminal domain is homologous to the C-terminal domain of the eIF-2β; N.C.K., unpublished work) to promote the hydrolysis of GTP bound to the preinitiation complex, the release of eIF-2⋅GDP, and the joining of the 60S ribosomal subunit (13), whereas in Bacteria, this step is performed by IF-2 (14). Continued operation of this initiation cycle also requires regeneration of the spent GTP. In Bacteria, this regeneration seems to occur spontaneously, whereas eukaryotes (in analogy to bacterial EF-Tu) require eIF-2B, a separate factor, comprising five subunits (three of which, α, β, and δ, are related in sequence; Fig. 1; ref. 15) to regenerate the GTP (15, 16).
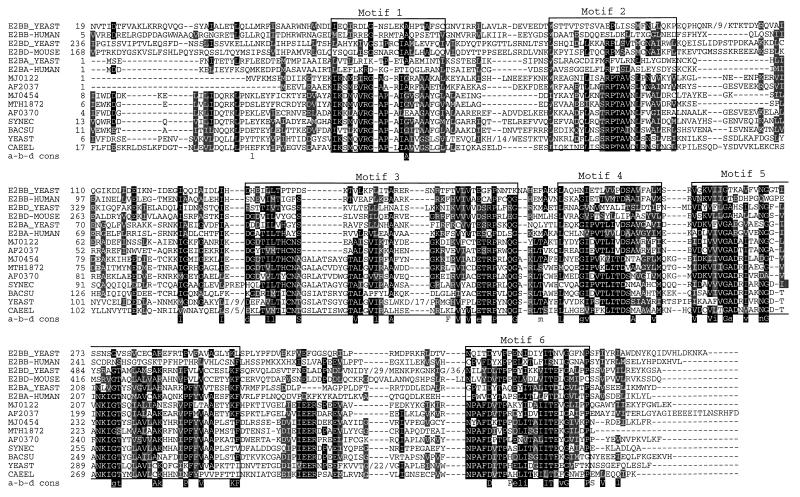
Figure 1.
Multiple sequence alignment of the eIF-2B α-β-δ subunits with their archaeal and bacterial homologs. Identical residues are highlighted black, and conservative substitutions are highlighted gray. Boxed areas mark the motif borders as generated from the meta-MEME motif search tool (29). The archaeal family of aIF-2B subunits appears to have all of the conserved residues shared between the three eukaryotic subunits, together with additional characteristics, absent from any of these three subunits. In particular, motif 1 is absent from all three eukaryotic subunits, and motifs 3, 5, and 6 are well conserved in all three of them. Motif 2 seems to be in part shared only with the δ subunit, whereas motif 4 is shared with the α and β subunits. Finally, motif 5 is less conserved in the β subunits. Protein names and their reference (accession number, when different) are: E2BB YEAST, S. cerevisiae eIF-2B β subunit; E2BB-HUMAN, human eIF-2B β subunit (YS20 HUMAN); E2BD YEAST, S. cerevisiae eIF-2B δ subunit; E2BD-MOUSE, Mus musculus eIF-2B δ subunit (PIR:B55146); E2BA YEAST, S. cerevisiae eIF-2B α subunit; E2BA-HUMAN, human eIF-2B α subunit (EMBL:X95648); MJ0122, M. jannaschii ORF MJ0122 (SW:E2B2 METJA); AF2037, A. fulgidus ORF AF2037 (gi2648498); MJ0454, M. jannaschii ORF MJ0454 (SW:E2B1 METJA); MTH1872, M. thermoautotrophicum ORF MTH1872 (gi2623009); AF0370, A. fulgidus ORF AF0370 (gi2650263); SYNEC, Synechocystis sp. hypothetical protein slr1938 (EMBL:Z99111 27); BACSU, hypothetical gene ykrs (EMBL:D90915 85); YEAST, S. cerevisiae hypothetical protein YPR118w (PIR:S69011); CAEEL, C. elegans hypothetical protein C01G10.9 (EMBL:Z81030 4). The correct annotations of the M. jannaschii functions are available at: http://geta.life.uiuc.edu/~nikos/Methanococcus.html.
Archaea possess a clear homolog of (all three subunits of) eIF-2 (7, 17, 18), whereas the M. jannaschii genome has been reported to have homologs of only the α and δ subunits of eIF-2B. This idea is puzzling in that the γ and ɛ subunits are thought to house the catalytic activity of the proteins (19, 20). More puzzling, the Archaea also possess a homolog of bacterial IF-2, as does yeast (7, 17, 18, 21).
This ostensibly contradictory situation shows that our understanding of translation initiation, not only in the Archaea but in all organisms, is still incomplete. The knowledge of three complete archaeal genomes has revealed that translation initiation in Bacteria and Eukarya is profoundly more interconnected (10) than we previously thought and triggers a more detailed analysis of this process, in the context of a bacterial–archaeal–eukaryotic comparison. Herein, we examine the relationship and phylogenetic distribution of the IF-2/eIF-2/eIF-2B factors.
MATERIALS AND METHODS
Databases.
The nonredundant protein sequence database at the National Center of Biotechnology Information (National Institutes of Health) was used for all of the sequence similarity searches. A query for the mere presence or absence of a sequence from the unpublished genomes of Deinococcus radiodurans and Treponema pallidum was done through the blast server of The Institute for Genomic Research at http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-tigrbl. Database searches were performed with blast (22) and wu-blast 2.0 (23) programs, using the blosum62 substitution matrix and default parameters at http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-newblast?Jform = 1. Access to bibliographical databases was facilitated greatly by using Entrez (24) (at http://www.ncbi.nlm.nih.gov/Entrez/) and Sequence Retrieval System (25) (at http://srs.ebi.ac.uk:5000/).
Sequence Analysis.
Multiple sequence alignments were performed by clustalw (26) and the pileup program of the Genetics Computer Group package, Ver. 8.1 from the University of Wisconsin (27). Visualization of the conserved residues was facilitated by the boxshade (Ver. 3.21) program at http://ulrec3.unil.ch/software/BOX form.html. From the multiple sequence alignment of individual families, sequence profiles (28) were generated and used to search protein sequence databases. Motif identification searches were used with the meta-MEME motif search tool (29) (See also http://www.sdsc.edu/MEME/meme/website/). Unrooted trees were constructed using various phylip package (Ver. 3.55c) programs (30). Trees were visualized by treetool (Ver. 2.0.1. available from the Ribosomal Database Project at the University of Illinois, Urbana-Champaign at http://geta.life.uiuc.edu).
RESULTS AND DISCUSSION
Archaeal Translation IF-2B (aIF-2B).
eIF-2 and eIF-2B function in tandem, the latter being required to recycle the GTP used by the former (6). As previously mentioned, the Archaea possess obvious homologs of all of the eIF-2 subunits but not all of the subunits of eIF-2B (7, 17, 18). Of the five eIF-2B subunits, no homologs of γ and ɛ (which appear to house the catalytic activity of the proteins; refs. 13, 19, and 20) were found; and, of the three remaining subunits, α, δ, and β (which constitute a family; refs. 15 and 16), only the first two were reported as having (specific) homologs in the M. jannaschii genome (7). This initial assessment, however, is not entirely correct. A closer examination shows the two M. jannaschii proteins not to be specifically related to the corresponding two eIF-2B subunits, but both do belong to the same overall gene family, a family that further contains: three additional archaeal genes (two from Archaeoglobus fulgidus and one from Methanobacterium thermoautotrophicum), several bacterial genes, and two uncharacterized eukaryotic ORFs. Although the family does appear universal among the Archaea (to this point), it definitely is not among the Bacteria; the complete genomes of Escherichia coli (31), Haemophilus influenzae (32), Helicobacter pylori (33), both fully sequenced Mycoplasma species (34, 35), Borrelia burgdorferi (36), Treponema pallidum, and Deinococcus radiodurans contain no members.
The more obvious relationships among the various members of this family are the following (see Fig. 2 and Table 1):
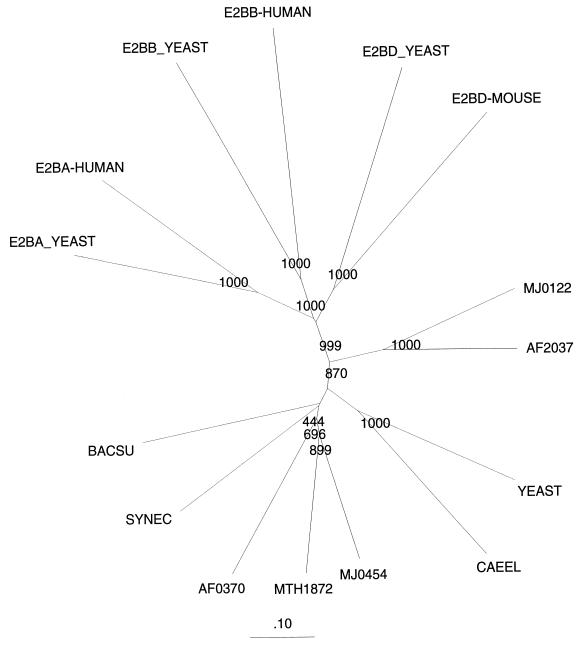
Figure 2.
A neighbor-joining (NJ) tree (41) showing distance relationships as derived from the multiple alignment presented in Fig. 1. The tree was calculated by using clustalw and visualized by treetool. Bootstrap (12) out of 1,000 tests for some important branching patterns is shown. Protein parsimony analysis (protpars) from the phylip package (Ver. 3.55c) (30) gave the same results with the NJ tree. Names and their references are as in Fig. 1.
Table 1.
Pairwise amino acid percentage identity among the IF-2B α-β-δ/I-II subunit family
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. MJ0454 | 60.0 | 54.2 | 48.5 | 53.3 | 44.8 | 41.5 | 39.0 | 36.3 | 30.8 | 26.7 | 26.8 | 23.6 | 22.7 | 21.8 | |
| 2. MTH1871 | 60.0 | 49.2 | 50.1 | 50.8 | 42.3 | 41.3 | 41.6 | 36.5 | 26.5 | 30.1 | 22.9 | 22.8 | 23.5 | 20.0 | |
| 3. SYNEC | 54.2 | 49.2 | 47.9 | 44.8 | 40.2 | 41.0 | 39.0 | 37.8 | 27.5 | 26.6 | 25.3 | 21.3 | 17.0 | 18.7 | |
| 4. BACSU | 48.5 | 50.1 | 47.9 | 42.7 | 36.4 | 35.2 | 42.9 | 38.3 | 23.8 | 23.7 | 22.9 | 23.2 | 22.9 | 22.4 | |
| 5. AF0370 | 53.3 | 50.8 | 44.8 | 42.7 | 41.0 | 35.8 | 40.6 | 37.8 | 27.8 | 26.2 | 25.5 | 24.0 | 21.0 | 21.3 | |
| 6. AF2037 | 44.8 | 42.3 | 40.2 | 36.4 | 41.0 | 58.8 | 34.3 | 34.0 | 30.4 | 31.0 | 26.0 | 27.1 | 26.3 | 23.2 | |
| 7. MJ0122 | 41.5 | 41.3 | 41.0 | 35.2 | 35.8 | 58.8 | 36.9 | 33.5 | 30.8 | 27.8 | 26.5 | 25.4 | 25.0 | 22.6 | |
| 8. YEAST | 39.0 | 41.6 | 39.0 | 42.9 | 40.6 | 34.3 | 36.9 | 46.5 | 24.2 | 23.7 | 24.3 | 19.7 | 22.0 | 23.0 | |
| 9. CAEEL | 36.3 | 36.5 | 37.8 | 38.3 | 37.8 | 34.0 | 33.5 | 46.5 | 24.2 | 27.8 | 24.3 | 22.7 | 18.3 | 17.3 | |
| 10. E2BA-YEAST | 30.8 | 26.5 | 27.5 | 23.8 | 27.8 | 30.4 | 30.8 | 24.2 | 24.2 | 42.5 | 22.8 | 24.4 | 26.7 | 24.5 | |
| 11. E2BA HUMAN | 26.7 | 30.1 | 26.6 | 23.7 | 26.2 | 31.0 | 27.8 | 23.7 | 27.8 | 42.5 | 27.7 | 29.4 | 23.7 | 25.7 | |
| 12. E2BD-YEAST | 26.8 | 22.9 | 25.3 | 22.9 | 25.5 | 26.0 | 26.5 | 24.3 | 24.3 | 22.8 | 27.7 | 35.4 | 23.4 | 21.3 | |
| 13. E2BD MOUSE | 23.6 | 22.8 | 21.3 | 23.2 | 24.0 | 27.1 | 25.4 | 19.7 | 22.7 | 24.4 | 29.4 | 35.4 | 21.5 | 25.0 | |
| 14. E2BB YEAST | 22.7 | 23.5 | 17.0 | 17.0 | 21.0 | 26.3 | 25.0 | 22.0 | 18.3 | 26.7 | 23.7 | 23.4 | 21.5 | 31.5 | |
| 15. E2BB-HUMAN | 21.8 | 20.0 | 18.7 | 18.7 | 21.3 | 23.2 | 22.6 | 23.0 | 17.3 | 24.5 | 25.7 | 21.3 | 25.0 | 31.5 |
Protein names and their reference are according to Fig. 1.
(i) The archaeal members of the family, the bacterial members, and the two uncharacterized eukaryotic ORFs are clearly more similar in sequence to one another than they are to any of the three eIF-2B subunits. This could mean either that the former forms a distinct clade within the family or that the eIF-2B subunits are related more distantly by virtue of being highly derived. We believe the latter to be the case (see below).
(ii) Three obvious clades exist within the family: the first comprises M. jannaschii ORF MJ0454, A. fulgidus AF0370, and the M. thermoautotrophicum MTH1872; the second comprises M. jannaschii ORF MJ0122 and A. fulgidus AF2037; and the third comprises the two hypothetical eukaryotic genes.
Given only two available complete sequences [one from Synechocystis sp. (37) and one from Bacillus subtilis (38)], one yet-to-be released (from Aquifex pyrophilus), and several incomplete sequences (from Thermotoga maritima and Rhodospirillum rubrum), additional information is required to determine whether the bacterial versions form a distinct clade as well.
Less obvious relationships in this overall family are:
(iii) The three eIF-2B subunits form a separate clade not seen in the raw similarity data but indicated by the branching order of Fig. 2 and a profile search based on the three eIF-2B subunit (not shown).
(iv) The subgroup comprising M. jannaschii ORF MJ0122 and A. fulgidus ORF AF2037 bears a specific relationship to the eIF-2B α-β-δ subunit subgroup seen in the tree of Fig. 2, in profile searches, and in cladistic analysis (not shown). Specifically, profiles based on the MJ0122 group vs. the MJ0454 group identify eIF-2Bα, eIF-2Bδ, and eIF2Bβ with Z scores of 24 vs. 13, 18 vs. 6.5, or 13 vs. 4, respectively. Thus, a likely evolutionary scenario for this family is that the ancestral type of the group (which appears most to resemble MJ0454 and its closer relatives) gave rise (probably in the archaeal line of descent) to a MJ0122-type of molecule, which in turn gave rise to the ancestor of the eukaryotic subunits (α, β, and δ). The key questions here are whether the archaeal (as well as the uncharacterized eukaryotic) versions function in translation and, if so, what that function is. In that all of the sequenced archaeal genomes show a complete eIF-2 (all three subunits), it seems likely that their homologs of the α-δ-β subunits of eIF-2B are somehow functionally associated with it, i.e., are involved in translation initiation. Yet, in that the eIF-2B subunits responsible for GTP recycling on eIF-2, namely γ and ɛ (13, 17, 18), are not present in the Archaea, it may be that this GTP recycling mechanism is not needed. An experimental resolution of this problem is needed sorely.
In recognition of their apparent functional uniqueness within this family, we propose renaming the M. jannaschii ORFs MJ0454 and MJ0122 and their specific relatives to aIF-2BI and aIF-2BII, respectively. The absence of aIF-2BII in Methanobacterium is puzzling and suggests that this factor may not be central to archaeal translation initiation.
This is the first identification of a homolog of an eIF-2B subunit in Bacteria. Yet, its very scattered and phylogenetically diverse (39) distribution and the fact that Bacteria show none of the subunits of the companion factor eIF-2 make the functional role of this eIF-2B-related protein in bacteria problematic. One also wonders why it is found in some eukaryotes, in addition to the normal eIF-2B subunits.
The IF-2 Question.
IF-2 and eIF-2 perform essentially the same function in bacterial and eukaryotic translation initiation, respectively, i.e., the introduction of the initiator Met-tRNAiMet into the ribosome. Yet the two proteins appear unrelated specifically: IF-2 function resides in a single polypeptide chain, whereas eIF-2 comprises three subunits and requires an additional multi-subunit factor, eIF-2B, for recycling GTP. It is puzzling to find both eIF-2 and an IF-2-like molecule in all sequenced archaeal genomes (7, 17, 18) and in at least one eukaryotic lineage as well (21).
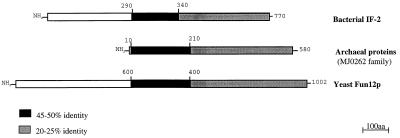
An alignment of IF-2 sequences (not presented) shows that all of them contain a highly conserved “G-domain” (40–50% sequence identity among Archaea, Eukarya, and Bacteria) and a much less conserved, but still homologous, C-terminal domain (20–25% sequence identity between Bacteria and Archaea). Whereas the G-domains seem to be truly orthologous throughout, this is not so for the C-terminal domain, where homology tends to be spotty in distribution (see Fig. 2). The N-terminal domain of bacterial IF-2 (Fig. 3) does not have homologs/counterparts in the archaeal and eukaryotic examples, but this may not be significant, for even among the Bacteria, sequence in this section of the molecule is exceedingly variable (5, 14). The question becomes the function of IF-2 in Archaea (and eukaryotes). Because Archaea lack an homolog of eukaryotic eIF-5 (which hydrolyzes GTP during translation initiation), it is conceivable that the archaeal homolog of bacterial IF-2 performs this role (as it does in Bacteria).
Figure 3.
A schematic representation of the sequence relationship among the bacterial IF-2 protein family (average sequence is displayed), with the four currently available Archaeal proteins (average sequence from M. jannaschii ORF MJ0262; M. thermoautotrophicum ORF MTH259; A. fulgidus ORF AF0768; and S. acidocaldarius gene infB is displayed) and the yeast Fun120p (SW:YAD5 YEAST) gene product.
CONCLUSIONS
The purpose of the present communication is to focus attention on our incomplete understanding of archaeal translation initiation (indeed translation initiation in general) and to bring some definition to the problem, but the issues we raise cannot be resolved without appropriate experimentation. Existing data do not support archaeal translation initiation as being chimeric (a blend of bacterial and eukaryotic features), as some would favor (40). Rather, they support a now familiar theme seen over and over in the information processing-associated systems of the cell; namely, that the archaeal version of a process resembles most the eukaryotic version, but a simplified, if not the ancestral, counterpart thereof.
Archaeal factor IF-1/eIF-1A, although universal, shows more features in common with its eukaryotic than its bacterial counterpart (10). eIF-2, found in Archaea and Eukarya, shows only analogy, not homology, with its bacterial counterpart. Although the presence of IF-2 in Archaea might suggest some relationship to bacterial initiation, the molecule also occurs in (some) eukaryotes and suggests more our incomplete understanding of the process than a specific bacterial/archaeal relationship. The same might be said for the occurrence of the uncharacterized members of the eIF-2B α-β-δ subunit family, found in Archaea, Bacteria, and Eukarya. As mentioned above, archaeal aIF-2B appears to lack the eukaryotic type of in situ guanine recycling function (performed by the eIF-2B γ and ɛ subunits). This suggests that Archaea might have no need for such a factor, as is the case in bacterial translation initiation. The Archaea, like their eukaryotic counterparts, seem to have an eIF-5A function, which also does not occur in Bacteria. Archaea do lack the equivalent of eIF-4 (which is mainly concerned with recognition of the eukaryotic-specific mRNA cap structure) and eIF-5; and, none of the many eIF-3 subunits are found in the Archaea, except for a (yeast) SUI1 homolog, implying that SUI1 might function as a separate unit (as is the case for mammalian eIF-1) (10).
In total, it seems that the rudiments of translation initiation were somehow present in the universal ancestor state but that much of the process then evolved separately on the bacterial and combined archaeal/eukaryotic lines of descent, with significant further refinement occurring on the eukaryotic line once it separated from the archaeal.
Acknowledgments
We thank R. V. Swanson for providing unpublished information for the genome of Aquifex pyrophilus. We also thank C. Ouzounis (EBI/UK) for critical reading of the manuscript. This work was supported by grants of National Aeronautics and Space Administration (NAG 54500) and Department of Energy (DEFG C02-95ER61963) to C.R.W.
ABBREVIATIONS
- IF
initiation factor
- eIF
eukaryotic IF
- aIF
Archaeal IF
References
- 1.Woese C R. In: Organization and Control in Prokaryotic and Eukaryotic Cells. Charles H P, Knight B C J G, editors. Cambridge, U.K.: Cambridge Univ. Press; 1970. pp. 39–53. [Google Scholar]
- 2.Woese C R, Fox G F. J Mol Evol. 1977;10:1–6. doi: 10.1007/BF01796132. [DOI] [PubMed] [Google Scholar]
- 3.Ouzounis C, Kyrpides N. FEBS Lett. 1996;390:119–123. doi: 10.1016/0014-5793(96)00631-x. [DOI] [PubMed] [Google Scholar]
- 4.Kyrpides, N., Overbeek, R. & Ouzounis, C. (1998) J. Mol. Evol., in press. [DOI] [PubMed]
- 5.Gualerzi C O, Pon C L. Biochemistry. 1990;29:5881–5888. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- 6.Pain V M. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 7.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 8.Kyrpides N, Olsen G, Klenk H-P, White O, Woese C. Microb Compar Genomics. 1996;1:329–338. [PubMed] [Google Scholar]
- 9.Andrade M, Casari G, de Daruvar A, Sander C, Schneider R, Tamares J, Valencia A, Ouzounis C. CABIOS. 1997;13:481–483. doi: 10.1093/bioinformatics/13.4.481. [DOI] [PubMed] [Google Scholar]
- 10.Kyrpides N C, Woese C R. Proc Natl Acad Sci USA. 1998;95:224–228. doi: 10.1073/pnas.95.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keeling P J, Doolittle W F. Proc Natl Acad Sci USA. 1995;92:5761–5764. doi: 10.1073/pnas.92.13.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 13.Das K, Chevesich J, Maitra U. Proc Natl Acad Sci USA. 1993;90:3058–3062. doi: 10.1073/pnas.90.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laalami S, Sacerdot C, Vachon G, Mortensen K, Sperling-Petersen H U, Cenatiempo Y, Grunberg-Manago M. Biochimie. 1991;73:1557–1566. doi: 10.1016/0300-9084(91)90191-3. [DOI] [PubMed] [Google Scholar]
- 15.Pavitt G D, Yang W, Hinnebusch A G. Mol Cell Biol. 1997;17:1298–1313. doi: 10.1128/mcb.17.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price N T, Mellor H, Craddock B L, Flowers K M, Kimball S R, Wilmer T, Jefferson L S, Proud C G. Biochem J. 1996;318:637–643. doi: 10.1042/bj3180637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 19.Koonin E V. Protein Sci. 1995;4:1608–1617. doi: 10.1002/pro.5560040819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price N T, Kimball S R, Jefferson L S, Proud C G. Biochem J. 1996;318:631–636. doi: 10.1042/bj3180631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keeling P J, Baldauf S L, Doolittle W F, Zillig W, Klenk H-P. Syst Appl Microbiol. 1996;19:312–321. [Google Scholar]
- 22.Altchul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Altchul S F, Gish W. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- 24.Schuler G D, Epstein J A, Ohkawa H, Kans J A. Methods Enzymol. 1996;266:141–162. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- 25.Etzold T, Argos P. Comput Appl Biosci. 1993;9:49–57. doi: 10.1093/bioinformatics/9.1.49. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devereux J, Haeberli P, Smithies N O. Nucleic Acids Res. 1984;12:387–396. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gribskov M, McLachlan A D, Eisenberg D. Proc Natl Acad Sci USA. 1987;89:10915–10919. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey T L, Elkan C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 30.Felsenstein J. Phylogenetic Inference Program (phylip) Manual, Ver. 3.5. Seattle: Univ. of Washington; 1990. [Google Scholar]
- 31.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 32.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 33.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, et al. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 34.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 35.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R A. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 37.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 38.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 39.Woese C R. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koonin E V, Mushegian A R, Galperin M Y, Walker D R. Mol Microbiol. 1997;25:619–637. doi: 10.1046/j.1365-2958.1997.4821861.x. [DOI] [PubMed] [Google Scholar]
- 41.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]