Abstract
The molecular mechanisms of head development are a central question in vertebrate and invertebrate developmental biology. The anteriorly expressed homeobox gene otd in Drosophila and its homolog Otx in mouse are required for the early development of the most anterior part of the body, suggesting that a fundamental genetic program of cephalic development might be conserved between vertebrates and invertebrates. We have examined this hypothesis by introducing the human Otx genes into flies. By inducing expression of the human Otx homologs with a heat shock promoter, we found that both Otx1 and Otx2 functionally complement the cephalic defects of a fly otd mutant through specific activation and inactivation of downstream genes. Combined with previous morphological studies, these results are consistent with the view that a common molecular ground plan of cephalization was invented before the diversification of the protostome and the deuterostome in the course of metazoan evolution.
The developmental mechanisms and the evolution of the vertebrate and invertebrate cephalic structures have been debated extensively since the last century (1, 2). Based on anatomical and paleontological observations, it traditionally has been accepted that vertebrate and invertebrate head and brain were invented independently. Although this notion still is contested intensively, recent molecular and developmental studies have revealed a common mechanism of axial patterning for the trunk regions of protostomes and deuterostomes. A striking example is seen in the conserved colinearity between the expression patterns along the antero-posterior axis and the chromosomal locations of the Drosophila homeotic (HOM) complex genes and the vertebrate Hox complex genes (3–5). It also has been shown that the molecular mechanisms underlying dorsoventral patterning are conserved between flies and vertebrates (6).
The basic organizations of the insect and the vertebrate heads have been studied intensively in the past years. The developing vertebrate brains are proposed to be organized in distinct subregions that reflect a basic metameric regionalization (7, 8). Similarly, morphological and developmental studies on insect brains reveal an underlying metameric organization (9–11). Among the regulatory genes that control cephalic development, the anteriorly expressed homeobox-containing gene orthodenticle (otd) in Drosophila (12, 13) and its homolog Otx in mice (14–19) play important roles in early patterning of the most anterior parts of the body, suggesting that fundamental genetic programs of cephalic development might be conserved (20, 21, 22). Here, we show that, despite the evolutionary distance and sequence diversity, the human Otx genes rescue developmental cephalic defects of a Drosophila otd mutant, indicating that the human Otx genes are functional homologs of the Drosophila gene. Our findings suggest that the basic regulatory interactions controlling the cephalic development may be conserved between the flies and mammals and are consistent with the view that a common molecular ground plan of cephalic development might have been established before the diversification of the protostome and the deuterostome.
MATERIALS AND METHODS
Transformant Flies.
The human Otx genes (Fig. 1; ref. 15) were subcloned downstream of a heat shock protein (hsp) 70 promoter in pNHT4 heat shock P-element vector (23). The recombinant plasmid was introduced into ry506 flies by germ line transformation (24).
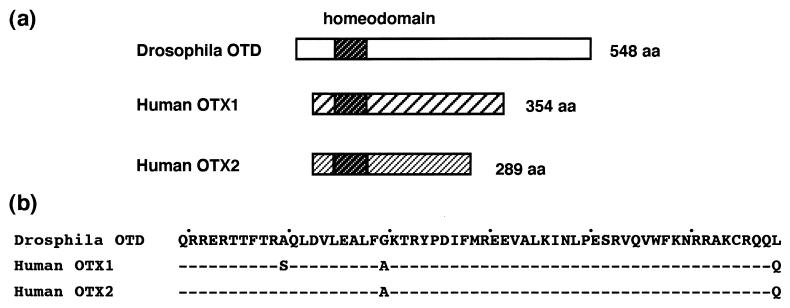
Figure 1.
(a) Structure comparison of the Drosophila OTD and the human OTX proteins. The Drosophila OTD protein is 548 aa long and the human OTX1 and OTX2 proteins are 354- and 289 aa long, respectively. All three proteins have the conserved homeodomain near the amino terminus and carry a long carboxyl terminus. In addition to the homeodomain conservation, the three OTD/OTX proteins share a short Tyr-Pro—–Arg-Lys stretch immediately upstream of the homeodomain as well as a tripeptide, Phe/Tyr-Leu-Lys, at the amino terminus. Little homology exist between the nonhomeodomain regions of the Drosophila OTD and the human OTX proteins. The two human OTX proteins show strong similarity to each other in the amino terminus and weak similarity in the carboxyl-terminus. (b) Amino acid sequences of the OTD/OTX homeodomains. Dashes indicate identical residues.
Heat Shock Protocols.
Transformant males were crossed with ocelliless (oc)1 mutant females, and the expression of the hsp-otd/Otx gene was induced with multiple 1-h heat pulses at 37°C (four times at 2-h intervals in the indicated time window) in the third instar larval stage. The treated larvae were raised at 25°C, and the heads of the adult flies were mounted in glycerol.
Signal Detection.
An mAb, 4D9 (25), was used to detect EN protein. hedgehog (hh; ref. 26) and wingless (wg; ref. 27) expression was monitored with complementary RNA probes. After heat pulses, larvae were kept at 18°C for 36 h and the discs were dissected for signal detection by using standard protocols (28, 29).
RESULTS
The Vertex of the Drosophila Head.
The dorsal region, or vertex, of the head is considered one of the most anterior structures of the adult fly (10). The medial subdomain of the vertex contains the light sensing organs, ocelli, and a set of characteristic sensory bristles [the large ocellar bristles (ocb), the postvertical bristles (pvb), and the small interocellar bristles; see Fig. 2a]. During pupal development, the medial vertex is formed by the fusion of the dorsomedial domains of the eye–antennal discs (30). The development of this region is controlled by the otd gene and is affected severely in oc1 mutant flies, in which otd expression in the primordium of the head vertex specifically is eliminated (12, 30). Flies homozygous or hemizygous for oc1 mutations lack the ocelli and most of the associated bristles (Fig. 2b, arrowhead indicates a remaining pvb).
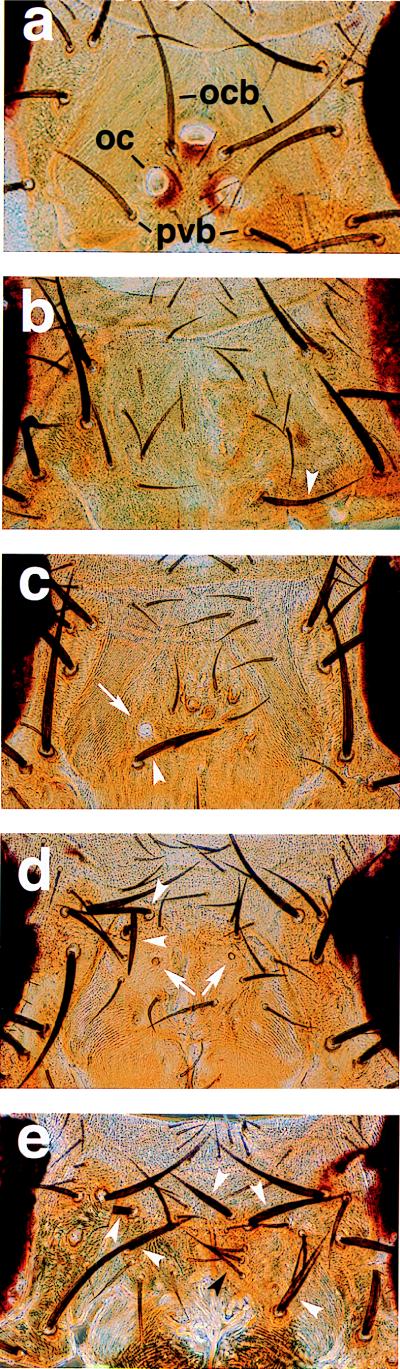
Figure 2.
Developmental rescue of the vertex structures with the human Otx genes. (a) Wild-type (Oregon R) vertex. Positions of ocelli (oc), ocellar bristles (ocb), and postvertical bristles (pvb) are indicated. Three to four pairs of smaller interocellar bristles also are located in the medial region. (b) oc1 vertex. (c) hsp-otd (line 5A) vertex with oc1 background. Arrowheads in b and c indicate postvertical-like bristles. (d) hsp-Otx1 (line 7.5.4) vertex with oc1 background. Arrows in c and d indicate rescued ocelli lenses. (e) hsp-Otx2 (line 12.12.8) vertex with oc1 background. White arrowheads in d and e indicate rescued large bristles. Black arrowhead in e indicates rescued interocellar-like bristles.
Rescue of the Medial Vertex Structures with Multiple Heat Pulses.
It has been shown that the Drosophila oc1 defects can be rescued with a hsp promoter-driven otd gene (hsp-otd) by a single 45-min heat pulse at 94 h after egg laying in the late third instar stage (30). To improve reproducibility, we applied four consecutive heat pulses of the hsp-otd gene at 37°C during the third instar stage. This multi-pulse protocol resulted in partial morphological rescue of vertex structures in a significant fraction of eclosed flies (Fig. 2C; Table 1). Heat pulses were most effective when applied in the temporal window of either 72–85 h after egg laying or 84–97 h after egg laying (Table 1). The efficiency of this protocol drops in later stages, and it becomes virtually ineffective at the end of the third larval instar. This complementation effect was specific to the otd gene because no rescue was obtained with a hsp-driven empty spiracles (ems) gene, which is another anteriorly expressed homeobox gene (31, 32).
Table 1.
Rescue of the medial vertex structures with multiple heat pulses
| Flies | Heat induction* | Rescue of ocelli structures, %
|
Heads examined, n | ||
|---|---|---|---|---|---|
| Total | With lens | With pigment | |||
| oc1/Y; hsp-otd/+ | 72–85 | 44 | 25 | 29 | 72 |
| 84–97 | 37 | 29 | 7 | 68 | |
| 96–109 | 4 | 4 | 0 | 55 | |
| 108–121 | 4 | 4 | 0 | 27 | |
| oc1/Y; hsp-ems/+ | 72–85 | 0 | 0 | 0 | 52 |
| 84–97 | 0 | 0 | 0 | 50 | |
| oc1/Y; +/+ | 72–85 | 0 | 0 | 0 | 51 |
| 84–97 | 0 | 0 | 0 | 50 | |
Hours after egg laying.
Morphological Rescue of the Drosophila Cephalic Defects by the Human Otx Homologs.
The human Otx homologs Otx1 and Otx2 are highly diverged in amino acid sequence from the fly otd gene with little homology outside the homeodomain (Fig. 1; ref. 15). The two genes are expressed in overlapping patterns in the developing forebrain and midbrain of the mouse (14). Experiments with Otx knockout mice have shown that the murine Otx1 and Otx2 genes, which are almost identical to the human homologs, are required for cephalic development, including formation of the rostral part of the brain (16–19). Similarly, the Drosophila otd gene activity is required for cephalic and brain development (12, 13, 33), suggesting that the fundamental regulatory functions of the otd/Otx homologs are conserved between flies and mammals.
To test the functional conservation of the otd/Otx homologs, we constructed transformant hsp-Otx1 and hsp-Otx2 flies and induced expression of these human homologs by heat pulses in oc1 background. Both human Otx1 and Otx2 homologs complemented the oc1 defect, generating either ocellar lenses (arrows in Fig. 2d) or associated ocellar pigments (Table 2). In some cases, both lenses and pigments were formed. Formation of the vertex bristles also was enhanced by the human Otx genes (arrowheads in Fig. 2 d and e). Whereas human Otx homologs tended to produce more bristles on the median vertex than the otd gene (Table 2), the fly gene was more potent in making postvertical-like bristles at the right position (arrowhead in Fig. 2c). In wild-type flies, the postvertical bristles are located at the posterior edge of the medial vertex.
Table 2.
Developmental rescue of the medial vertex structures with the human Otx genes
| Flies | Ocelli rescue*
|
Macrochetae rescue†
|
|||
|---|---|---|---|---|---|
| Lens | Pigment | Heads examined, n | Vertex macrochetae, n | Heads examined, n | |
| oc1/Y; hsp-Otx1/+ | |||||
| line 3.19.1 | 30 | 0 | 46 | 2.5 ± 1.7 | 53 |
| line 7.5.4 | 25 | 12 | 52 | 1.2 ± 1.1 | 51 |
| oc1/Y; hsp-Otx2/+ | |||||
| line 12.12.8 | 5 | 4 | 55 | 3.4 ± 1.5 | 53 |
| line 8.13.6 | 12 | 0 | 52 | 2.3 ± 1.5 | 44 |
| line 7.7.3 | 8 | 2 | 52 | 1.9 ± 1.3 | 54 |
| oc1/Y; hsp-otd/+ | |||||
| line 5A | 18 | 2 | 50 | 1.5 ± 0.9 | 48 |
| oc1/Y; hsp-ems/+ | |||||
| line 13-1 | 0 | 0 | 52 | 0.9 ± 0.9 | 50 |
| oc1/Y; +/+‡ | 0 | 0 | 51 | 1.0 ± 0.9 | 50 |
Heat pulses were applied between 72 and 85 h after egg laying.
Heat pulses were applied between 84 and 97 h after egg laying. Large bristles, which are similar in size to ocellar bristles and postvertical bristles, were counted. The wild-type vertex has four such bristles.
oc1 larvae were heat treated without crossing to transformants. Similar results were obtained with untreated oc1 larvae.
Molecular Genetic Interactions in the Vertex Primordium.
The primordium of the vertex is situated near the dorsomedial edge of the eye–antennal disc (30). A network of cross-regulatory segment polarity gene interactions is involved in the development of the vertex primordium: en and hh are expressed and wg is suppressed in a medial patch of cells in the late third instar stage (Fig. 3 a, f, and k; ref. 34). These genetic interactions are unique to the vertex primordium and are very different from those in trunk development, where wg acts to maintain en and hh, and en and hh act to maintain wg (35). The oc1 mutation causes specific loss of expression of en and hh in this region whereas expression of wg is maintained in a continuous crescent-like pattern at the dorsomedial region of the eye-antennal disc. To determine whether the morphological complementation by the human Otx genes is a result of similar genetic interactions, we examined the expression of en, hh, and wg in the vertex primordium cells after heat induction in oc1 background.
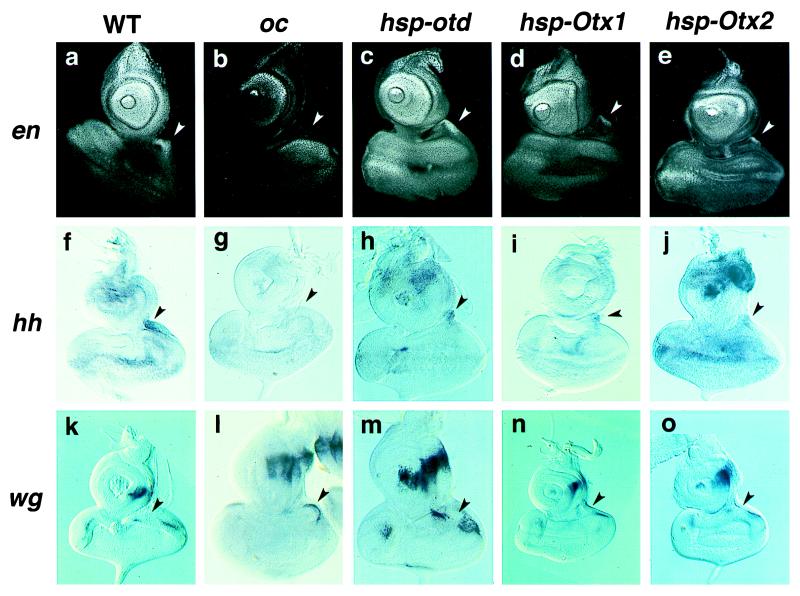
Figure 3.
Activation and repression of the segment polarity genes by the human Otx homologs. (a–e) EN protein expression. (f–j) hh RNA expression. (k–o) wg RNA expression. (a, f, and k) Wild type. (b, g, and l) oc1. (c, h, and m) hsp-otd line 5A with oc1 background. (d, i, and n) hsp-Otx1 line 3.19.1 with oc1 background. (e, j, and o) hsp-Otx2 line 8.13.6 with oc1 background. Arrowheads indicate the vertex primordia. An mAb (4D9) was used to detect EN protein. hh and wg expressions were monitored with specific RNA probes. Heat pulses were applied four times in 72–85 h after egg laying. Larvae were kept at 18°C for 36 h after the last heat pulse, and the discs were dissected for signal detection.
Induction of the fly otd gene stimulated en and hh and repressed wg in the vertex primordium, confirming previous observations (Fig. 3 c, h, m; refs. 30 and 34). Despite the ubiquitous induction of the otd gene by the hsp promoter over the eye–antennal disc, the regulatory effects on these downstream genes were restricted to the vertex primordium, suggesting that specific cofactor(s) in this region are present.
Consistent with the morphological results described above, induction of the human Otx1 (Fig. 3 d, i, n) and Otx2 (Fig. 3 e, j, o) genes resulted in activation of en and hh and repression of wg in the vertex primordium. The specific regulatory effects on these downstream genes were somewhat more variable than those induced by the fly otd gene (Table 3). However, similar cell type specificity was observed despite the ubiquitous induction of the human Otx homologs, suggesting that the induced human OTX proteins might be able to interact with the vertex cofactor(s). Heat induction of the ems gene failed to affect expression of these segmentation genes, indicating that the downstream controls are specific to the otd/Otx homologs. Hybridization with a probe specific to the Drosophila otd sequence showed that the expression of the endogenous otd gene remained at the mutant level (Table 3), indicating that the specific downstream regulations by the human Otx genes are not mediated by the activation of the endogenous Drosophila homolog.
Table 3.
Activation and repression of the vertex regulatory genes
| Flies | engrailed, % (n) | hedgehog, % (n) | wingless, % (n) | orthodenticle,* % (n) |
|---|---|---|---|---|
| Wild type | 100 (9) | 100 (15) | 100 (6) | 100 (11) |
| oc1/Y; +/+ | 0 (28) | 0 (27) | 17 (23) | 0 (16) |
| oc1/Y; hsp-otd/+ | ||||
| line 5A | 73 (19) | 71 (28) | 100 (19) | 13 (16) |
| oc1/Y; hsp-Otx1/+ | ||||
| line 3.19.1 | 27 (15) | 52 (23) | 70 (23) | 0 (18) |
| oc1/Y; hsp-Otx2/+ | ||||
| line 8.13.6 | 56 (27) | 19 (27) | 65 (20) | 0 (12) |
| oc1/Y; hsp-ems/+ | ||||
| line 13-1 | 0 (19) | 0 (12) | 8 (12) | 0 (17) |
Numbers of examined discs are shown in parentheses. Discs with positive induction in the vertex primordium were counted for engrailed and hedgehog. Discs showing suppression in the vertex primordium were counted for wingless. Heat pulses were applied four times between 72 and 85 h after egg laying.
Signals were detected with a specific RNA probe for the fly otd sequence. Residual otd expression was present in oc1 discs. Transformant discs with similar basal expression were not counted. Broad staining over the eye–antennal discs was observed for hsp-otd discs because of the ubiquitous induction of the otd gene by the heat shock promoter.
DISCUSSION
By introducing the human Otx homologs into Drosophila, we have shown that both Otx1 and Otx2 genes are able to rescue a developmental vertex defect caused by loss of otd expression in the vertex primordium. In addition to the morphological results, molecular genetic data have revealed that the rescue potential of the human Otx genes are based on their conserved regulatory activities that result in specific activation of en and hh and repression of wg in the developing vertex head primordium. It has been shown that homologous vertebrate Hox genes functionally substitute the Drosophila HOM complex genes (36–39) in the development of the trunk segments. Our work extends the concept of this evolutionary conservation of the regional specification mechanisms to the cephalic development.
The human OTX1 and OTX2 proteins significantly differ from the Drosophila OTD protein in both size and sequence (Fig. 1). The OTD protein shares little amino acid sequence similarity with the human OTX proteins except for 60 amino acid residues of the homeodomain of the entire 548 residues. The homeodomains of the human OTX1 and OTX2 proteins differ from that of the Drosophila OTD protein at three and two positions, respectively. Limited additional similarity is present outside of the homeodomain (15): Both of the human OTX proteins share with the Drosophila OTD protein a short Tyr-Pro—–Arg-Lys stretch immediately upstream of the homeodomain. The three OTD/OTX proteins also share a similar tripeptide, Phe/Tyr-Leu-Lys, at the amino terminus. It thus seems likely that the majority of the conserved functional specificity of the otd/Otx homologs is mediated by the evolutionary conserved homeodomain. Alternatively, it is also conceivable that the additional weak similarities between these proteins in the nonhomeodomain regions also might be responsible for the conserved regulatory specificity. Furthermore, simple amino acid stretches of alanine, serine, or histidine are scattered in the nonhomeodomain regions of these three proteins. It is intriguing to note that similar simple amino acid stretches have been shown to be important to the functions of homeobox-containing regulatory proteins HOXD13 (40), EN(41), and EVEN-SKIPPED (42).
Our data also suggest that the conserved functional specificity of the OTD/OTX proteins might be mediated by the interaction with specific cofactor(s) in the vertex primordium cells. Similar mechanisms of cofactor and homeodomain protein interactions have been demonstrated for the determination of the downstream target specificities of the HOM/HOX proteins (43). It is intriguing that extradenticle, one of such cofactors for the HOM/HOX proteins, is expressed beyond the trunk regions in the head vertex primordium and the developing brain (refs. 44 and 45 and T.N. unpublished work).
Although our results indicate that both Otx1 and Otx2 are mostly as potent as the Drosophila otd gene in the vertex development, it is important to know whether similar functional conservation can be demonstrated for other developmental processes that require otd/Otx gene activity. Concerning this point, it is noteworthy that the human Otx transgenes used in this study are able to rescue the embryonic brain defects of a lethal otd allele (S.L., unpublished work). Furthermore, recent reciprocal experiments have shown that the Drosophila otd gene introduced via homologous recombination into the murine Otx1 locus also restores most of the developmental abnormalities of the Otx1 mutant brain (D.A. and A.S., unpublished work).
Based on the conservation of the mechanisms of dorsoventral specification and other processes, it has been proposed that protostomes and deuterostomes are derived from a common ancestor, Urbilateria, which may have had a longitudinal central nervous system along the antero-posterior body axis (6). Our data imply that at least a certain fraction of genetic interactions controlling the cephalic development may be conserved between the protostome and the deuterostome, suggesting that the origin of the molecular interactions that underlie cephalic development also could be traced back to this primitive ancestor. It is interesting to note in this regard that, in addition to the molecular conservation, similar morphological ground plans have been proposed for the vertebrate and invertebrate brains (22, 46, 47). It is also noteworthy that recent studies demonstrate that the otd/Otx genes are instrumental in the formation of the cephalic structures (refs. 48 and 49 and S.L., unpublished work). Based on these findings, we suggest that, despite tremendous anatomical diversity, the cephalic regions, including the brains of the vertebrate and the invertebrate, might be homologous at a deep level (50) as the structures formed most anteriorly along a common axial design.
Acknowledgments
We thank Dr. Uwe Walldorf for hsp-ems flies and Drs. Sigeo Hayashi and Tetsuya Tabata for gene probes. mAb (4D9) was obtained from the Developmental Studies Hybridoma Bank. We also thank the members of the fly laboratories at the Institute of Biological Sciences for their kind help in stock maintenance. This work was supported by the Swiss National Science Foundation (K.F.T. and H.R.), the Italian Telethon program and the Italian Association for Cancer Research (A.S.), the Roche Research Foundation, the Yamada Science Foundation, Grant-in-Aid for Scientific Research on Priority Areas from MESSC, Japan, and the University of Tsukuba (K.F.T.).
References
- 1.Gee H. Before the Backbone. London: Chapman & Hall; 1996. [Google Scholar]
- 2.Jefferies R P S. The Ancestry of the Vertebrates. London: British Museum; 1986. [Google Scholar]
- 3.Gehring W J, Qian Y Q, Billeter M, Furukubo-Tokunaga K, Schier A F, Resendez-Perez D, Affolter M, Otting G, Wüthrich K. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 4.Duboule D. Guidebook to the Homeobox Genes. Oxford: Oxford Univ. Press; 1994. [Google Scholar]
- 5.Sharkey M, Graba Y, Scott M P. Trends Genet. 1997;13:145–151. doi: 10.1016/s0168-9525(97)01096-2. [DOI] [PubMed] [Google Scholar]
- 6.De Robertis E M, Sasai Y. Nature (London) 1996;380:37–40. doi: 10.1038/380037a0. [DOI] [PubMed] [Google Scholar]
- 7.Lumsden A, Krumlauf R. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- 8.Puelles L, Rubenstein J L R. Trends Neurosci. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 9.Cohen S M, Jürgens G. Trends Genet. 1991;7:267–272. doi: 10.1016/0168-9525(91)90327-M. [DOI] [PubMed] [Google Scholar]
- 10.Jürgens G, Hartenstein V. In: The Development of Drosophila. Bate M, Martinez-Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 687–746. [Google Scholar]
- 11.Reichert H, Boyan G. Trends Neurosci. 1997;20:258–263. doi: 10.1016/s0166-2236(96)01034-x. [DOI] [PubMed] [Google Scholar]
- 12.Finkelstein R, Smouse D, Capaci T M, Spradling A C, Perrimon N. Genes Dev. 1990;4:1516–1527. doi: 10.1101/gad.4.9.1516. [DOI] [PubMed] [Google Scholar]
- 13.Hirth F, Therianos S, Loop T, Gehring W J, Reichert H, Furukubo-Tokunaga K. Neuron. 1995;15:769–778. doi: 10.1016/0896-6273(95)90169-8. [DOI] [PubMed] [Google Scholar]
- 14.Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nature (London) 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- 15.Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D’Apice M R, Nigro V, Boncinelli E. EMBO J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- 17.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Genes Dev. 1995;9:2346–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 18.Ang S-L, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. Development. 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- 19.Acampora D, Mazan S, Avantaggiato V, Barone P, Tuorto F, Lallemand Y, Brulet P, Simeone A. Nat Genet. 1996;14:218–222. doi: 10.1038/ng1096-218. [DOI] [PubMed] [Google Scholar]
- 20.Finkelstein R, Boncinelli E. Trends Genet. 1994;10:310–315. doi: 10.1016/0168-9525(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 21.Thor S. Neuron. 1995;15:975–977. doi: 10.1016/0896-6273(95)90084-5. [DOI] [PubMed] [Google Scholar]
- 22.Arendt D, Nübler-Jung K. BioEssays. 1996;18:255–259. doi: 10.1002/bies.950180314. [DOI] [PubMed] [Google Scholar]
- 23.Gibson G, Gehring W J. Development. 1988;102:657–675. [Google Scholar]
- 24.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 25.Patel N H, Martin-Blanco E, Coleman K G, Poole S F, Ellis M C, Kornberg T B, Goodman C S. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- 26.Tabata T, Kornberg T B. Cell. 1994;76:89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 27.DiNardo S, Kuner J M, Theis J, O’Farrell P H. Cell. 1985;43:59–69. doi: 10.1016/0092-8674(85)90012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel N H. In: Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. Goldstein L S B, Fyrberg E, editors. New York: Academic; 1994. pp. 445–487. [Google Scholar]
- 29.Lehmann R, Tautz D. In: Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. Goldstein L S B, Fyrberg E, editors. New York: Academic; 1994. pp. 445–487. [Google Scholar]
- 30.Royet J, Finkelstein R. Development. 1995;121:3561–3572. doi: 10.1242/dev.121.11.3561. [DOI] [PubMed] [Google Scholar]
- 31.Dalton D, Chadwick R, McGinnis W. Genes Dev. 1989;3:1940–1956. doi: 10.1101/gad.3.12a.1940. [DOI] [PubMed] [Google Scholar]
- 32.Walldorf U, Gehring W J. EMBO J. 1992;11:2247–2259. doi: 10.1002/j.1460-2075.1992.tb05284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen S M, Jürgens G. Nature (London) 1990;346:482–485. doi: 10.1038/346482a0. [DOI] [PubMed] [Google Scholar]
- 34.Royet J, Finkelstein R. Development. 1996;122:1849–1858. doi: 10.1242/dev.122.6.1849. [DOI] [PubMed] [Google Scholar]
- 35.Hammerschmidt M, Brook A, McMahon A P. Trends Genet. 1997;13:14–21. doi: 10.1016/s0168-9525(96)10051-2. [DOI] [PubMed] [Google Scholar]
- 36.Malicki J, Schughart K, McGinnis W. Cell. 1990;63:961–967. doi: 10.1016/0092-8674(90)90499-5. [DOI] [PubMed] [Google Scholar]
- 37.McGinnis N, Kuziora M A, McGinnis W. Cell. 1990;63:969–976. doi: 10.1016/0092-8674(90)90500-e. [DOI] [PubMed] [Google Scholar]
- 38.Zhao J J, Lazzarini R A, Pick L. Genes Dev. 1993;7:343–345. doi: 10.1101/gad.7.3.343. [DOI] [PubMed] [Google Scholar]
- 39.Lutz B, Lu H-C, Eichele G, Miller D, Kaufman T C. Genes Dev. 1996;10:176–184. doi: 10.1101/gad.10.2.176. [DOI] [PubMed] [Google Scholar]
- 40.Muragaki Y, Mundlos S, Upton J, Olsen B R. Science. 1996;272:548–551. doi: 10.1126/science.272.5261.548. [DOI] [PubMed] [Google Scholar]
- 41.Han K, Manley J L. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han K, Manley J L. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- 43.Mann R S, Chan S K. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez-Crespo S, Morata G. Development. 1995;121:2117–2125. doi: 10.1242/dev.121.7.2117. [DOI] [PubMed] [Google Scholar]
- 45.Aspland S E, White R A. Development. 1997;124:741–747. doi: 10.1242/dev.124.3.741. [DOI] [PubMed] [Google Scholar]
- 46.Williams N A, Holland P W H. Nature (London) 1996;383:490. [Google Scholar]
- 47.Lacalli T. Philos Trans R Soc Lond B. 1996;351:243–263. [Google Scholar]
- 48.Andreazzoli M, Pannese M, Boncinelli E. Development. 1997;124:1733–1743. doi: 10.1242/dev.124.9.1733. [DOI] [PubMed] [Google Scholar]
- 49.Gammill L S, Sive H. Development. 1997;124:471–481. doi: 10.1242/dev.124.2.471. [DOI] [PubMed] [Google Scholar]
- 50.Bolker J A, Raff R A. BioEssay. 1996;18:489–494. doi: 10.1002/bies.950180611. [DOI] [PubMed] [Google Scholar]