Abstract
The mosquito Aedes aegypti is the world’s most important vector of yellow fever and dengue viruses. Work is currently in progress to control the transmission of these viruses by genetically altering the capacity of wild Ae. aegypti populations to support virus replication. The germ-line transformation system reported here constitutes a major advance toward the implementation of this control strategy. A modified Hermes transposon carrying a 4.7-kb fragment of genomic DNA that includes a wild-type allele of the Drosophila melanogaster cinnabar (cn) gene was used to transform a white-eyed recipient strain of Ae. aegypti. Microinjection of preblastoderm mosquito embryos with this construct resulted in 50% of the emergent G0 adults showing some color in their eyes. Three transformed families were recovered, each resulting from an independent insertion event of the cn+-carrying transposon. The cn+ gene functioned as a semidominant transgene and segregated in Mendelian ratios. Hermes shows great promise as a vector for efficient, heritable, and stable transformation of this important mosquito vector species.
The recent application of molecular biological tools to the analyses of insect vectors of disease and vector–parasite interactions has required the development of efficient methods for germ-line transformation (1, 2). Although there have been preliminary reports of transformation (3–5), and a number of methods have been used for transient expression of gene products (6–8), a method for efficient, heritable, and stable integration of exogenous DNA into a mosquito genome has yet to be developed. Such a tool would enable study of the effects of various synthetic genes on parasite development in the insect and could contribute to the development of novel methods for the control of diseases caused by mosquito-borne pathogens (1).
Recently, it was shown that the class-II transposon Hermes, a member of the hAT family of elements isolated from the housefly, Musca domestica, is capable of plasmid-to-plasmid mobility in the embryonic soma of the yellow fever mosquito, Aedes aegypti (9). Here we report the results of experiments performed to determine if the observed mobility would translate into a method for the insertion of exogenous genes into the chromosomes of the whole organism. A critical feature of these experiments was the use of a marker gene that allowed the rapid and unequivocal detection of potentially transformed mosquitoes by screening for complementation of a mutant gene with an eye-color phenotype. Ommochrome pigments produce the deep purple color of the adult mosquito eye. Ommochromes are synthesized in a well-characterized series of enzymatically catalyzed steps from the precursor amino acid, tryptophan (10). Null mutations in two of the enzymes in this pathway, tryptophan oxygenase and kynurenine hydroxylase, result in eye color mutant phenotypes in the adults of many insects (10, 11). An Ae. aegypti mutation, white-eye (designated w), was originally described by Bhalla (11) as a defect in the kynurenine hydroxylase that converts kynurenine to 3-hydroxykynurenine. The corresponding gene identified by this mutation was postulated to be homologous to the wild-type cinnabar (cn) gene described in Drosophila melanogaster. The product of the reaction, 3-hydroxykynurenine, is non-cell-autonomous and can diffuse from one cell to another. Therefore, we were able to show with a transient assay that the D. melanogaster cn+ gene can complement the white-eye phenotype of the Bhalla Ae. aegypti w mutant strain (12). Microinjection of white-eye Ae. aegypti embryos with a 4.7-kb fragment of D. melanogaster genomic DNA that includes a wild-type copy of the cn gene (13) resulted in some adult mosquitoes with partial restoration of eye color. We have adopted the designation kynurenine hydroxylase-white (khw) for the Ae. aegypti mutation described by Bhalla to distinguish this gene from others which when mutated may also cause a white-eye phenotype. Here we report the successful development of a germ-line transformation system for Ae. aegypti with the Hermes transposon in conjunction with the cn+ gene.
MATERIALS AND METHODS
Plasmid Construction.
The transformation vector pH[cn], which includes the cn+ gene fragment from D. melanogaster (13) flanked by the terminal inverted repeats of the Hermes transposon, was developed as follows. All Hermes fragments have been described by Warren et al. (14). A 1.6-kb EcoRI/BamHI fragment containing ≈0.8 kb of the left arm of Hermes and 0.8 kb of flanking Musca domestica genomic DNA (derived from the E1 fragment) was ligated into EcoRI/BamHI-digested pUC19. The unique EcoRI site was eliminated by EcoRI digestion and filled by using the Klenow DNA polymerase. This plasmid was amplified by using a long reverse oligonucleotide primer (5′-CACAGGAAACAGCTATGACCATG-3′) and a second primer complementary to the region at the 5′ end of the LacZ gene of pUC19 (5′-GCGGTACCTGGCCAACGCGCGGGGAGAGGC-3′), which added a unique KpnI site. This product and a 0.8-kb fragment containing 0.5 kb of the right half of Hermes and ≈0.3 kb of flanking genomic DNA (derived from the B6 fragment) were digested with SalI/KpnI and ligated. The resulting plasmid contained unique BamHI and SalI sites between the two arms into which the LacZ alpha peptide of pBluescript SK (Stratagene) was inserted as a BglII/XhoI-digested fragment produced by PCR by using the primers, 5′-GCAGATCTATCAGGGCGATGGCC-3′ and 5′-GCCTCGAGCTGGCACGACAGGTTTCCCG-3′. This plasmid, pH[LacZ], was modified further by cutting with XbaI and SacII, and inserting a 4.7-kb XbaI/SacII fragment of D. melanogaster genomic DNA containing a wild-type copy of the cn gene (13). The resulting plasmid pH[cn] and the transposase encoding helper plasmid pHSHH1.9 (15) were purified through a cesium chloride gradient by ultracentrifugation, washed five times with isopropanol saturated with NaCl, and precipitated in three volumes of 70% ethanol. The dried pellet was resuspended in sterile, distilled H2O.
Embryo Microinjection.
Embryos homozygous for the khw mutation were collected and prepared for microinjection in the manner described by Morris (16). Embryos were injected with a solution of the plasmids, pH[cn] and pHSHH1.9, with a final concentration of 0.5 mg/ml and 0.3 mg/ml, respectively, in 5 mM KCl and 0.1 mM NaH2PO4 (pH 6.8). Microinjected G0 embryos were exposed to a heat shock of 39–41°C for 1 h to induce the expression of the Hermes transposase, which is under the control of the D. melanogaster hsp70 gene promoter. Embryos were then placed at 27°C, 80% relative humidity, and estivated for 5 days before hatching.
Mosquito Rearing.
General aspects of mosquito rearing are described by Munstermann (17). The khw strain was obtained from the University of Notre Dame Ae. aegypti stock center in 1992 by the senior author. In the course of validating that the khw strain contained the same mutation described by Bhalla (11), several thousand individuals were examined without detecting any segregation of colored eyes. Individual G0 males with colored eyes were mated with 15 khw strain virgin females. Single G0 females with colored eyes were mated with 3–5 khw strain males. Males and females with white eyes were mated together in several pools. Females were allowed to feed on blood and the G1 eggs collected. The adult mosquito eye starts differentiation during the later larval instars and pigment is readily visible by the late fourth instar. Phenotypic differences in eye color among the khw strain, wild-type mosquitoes, and putative transformants may be scored at the late fourth instar; however, routine screening of eye color was done on adults.
Southern Blot Analysis.
Southern blot analyses were done as described in Sambrook et al. (18). The various probes that were used are illustrated in Fig. 2A. The Hermes left-hand probe (LH) was made by gene amplification of a 0.76-kb DNA fragment with the following primers: 5′-GATACTTATGCGCCACTTGC-3′ and 5′-CCGACAATCTCGTACCACCC-3′, with pH[cn] as the template. The cn probe (cn+) was the complete 4.7-kb XbaI/SacII D. melanogaster fragment purified from pH[cn]. Probes to the M. domestica genomic DNA flanking the Hermes element in pH[cn] (MD) were made by gene amplification of the left-hand sequence (E1) with the following primers, E1REV, 5′-ACAGACTGCATTGCAGTTGG-3′, and E1FOR, 5′-AAAGCTGGTACCAGATCTGC-3′, and the right-hand sequence (B6) with the primers B6REV, 5′-ATCTCCGAACTCGATCTAAG-3′, and B6FOR, 5′-GGTCAAAACGTGCCTAGTCC-3′. By using these primers and pH[cn] as template DNA, probes of 0.9 and 0.21 kb in length were produced for the left- and right-hand sequences, respectively.
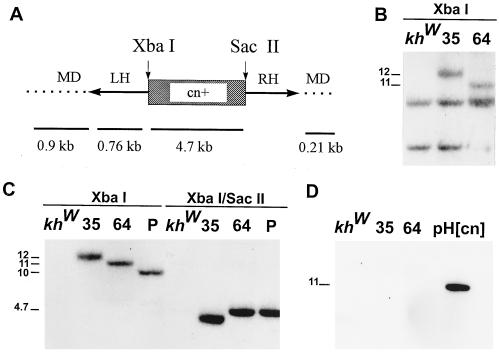
Figure 2.
(A) A representational map of the partial structure of the Hermes construct, pH[cn]. The Hermes left (LH) and right (RH) inverted terminal repeats (arrows) flank the 4.7-kb genomic fragment of D. melanogaster DNA carrying a wild-type copy of the cinnabar gene (cn+) (boxed region). This potentially mobile portion of the pH[cn] construct is flanked by M. domestica genomic DNA (MD, dotted lines). The positions of two restriction endonuclease-cleavage sites (SacII and XbaI) are indicated. The relative extents and sizes of the DNA fragments used as probes in the Southern analyses are shown below the map (numbers and solid lines). (B) Southern analysis of genomic DNA isolated from G1 progeny of transformed families 35 and 64, and the control recipient strain, khw, digested with XbaI, which cuts once in the integrated fragment, hybridized with the 0.76-kb LH-labeled fragment. Unique hybridization signals associated with insertions in families 35 and 64 are seen at 12 and 11 kb, respectively. Lower molecular weight signals corresponding to cross-hybridizing fragments are seen in the transformed families and recipient strain. (C) Southern blot analysis of genomic DNA isolated from G2 mosquitoes of families 35, 64, and the pool (P), and the recipient strain digested with either XbaI or XbaI/SacII, and hybridized with the 4.7-kb cn+ labeled fragment. High molecular weight signals corresponding to fragment lengths of 12, 11, and 10 kb are seen in the XbaI digests of DNA from families 35, 64, and the pool, respectively. The expected 4.7-kb fragment size is evident in the XbaI/SacII digest of DNA from family 64 and the pool. However, a truncated signal at 4.5 kb is seen in family 35 DNA, indicating loss of some DNA. (D) Southern blot analysis of genomic DNA isolated from G2 mosquitoes of families 35, 64, and the recipient strain, khw digested with XbaI, and hybridized with the MD-labeled fragments. The positive control is XbaI-digested pH[cn].
RESULTS
Germ-Line Transformation and Genetic Analysis.
A total of 925 preblastoderm embryos were microinjected with pH[cn] and the helper plasmid, pHSHH1.9, and subsequently heat-shocked for 1 h. Hermes transposase produced from pHSHH1.9 can act in trans to mobilize the Hermes construct in pH[cn]. A total of 120 adults (G0 generation) resulted from the microinjected embryos, and of these, 60 (31 males, 29 females) showed color in their eyes. These mosquitoes were used as single founders of families by mating them with homozygous khw individuals. The 60 white-eyed G0 adults were mated together in pools. Only 24 (40%) of the single founder families produced G1 progeny. Poor fertility and fecundity were also observed in the pooled white-eyed G0 families. Either late fourth instar larvae or adult G1 progeny were screened visually for changes in eye color that might result from integration of the cn+ transgene into the germ line of a G0 mosquito. Four of the founder families, 32, 35, 36, and 64, and one of the pools produced G1 progeny with colored eyes (Table 1). The eye colors were all different, varying from light to dark red. Two families, 32 and 36, showed mosaicism in their eye color. Families 35 and 64 produced multiple G1 progeny with colored eyes, a “cluster” phenomenon observed in other transposon-mediated transformation experiments with D. melanogaster and the Medfly, Ceratitis capitata (19, 20). A total of 63 G1 progeny with colored eyes were found in ≈6,800 mosquitoes screened.
Table 1.
G0 families showning colored eyes in the G1 progeny
| G0 family | No. of G1 with colored eyes | No. of G1 with mosaic eyes | Total G1 screened |
|---|---|---|---|
| 32 | 0 (0%) | 1 (0.2%) | 357 |
| 35 | 27 (56%) | 0 (0%) | 48 |
| 36 | 0 (0%) | 1 (0.8%) | 117 |
| 64 | 31 (19%) | 0 (0%) | 163 |
| Pool* | 3 (11%) | 0 (0%) | 26 |
The three G1 progeny with colored eyes were mated separately to khw strain individuals and lines were established. Subsequent Southern analysis showed that all three G1 progeny resulted from a single event, and G2 offspring from these lines were combined.
Test crosses were set up with colored-eyed G1 mosquitoes by crossing them to homozygous khw individuals. In the case of families 35 and 64, where multiple colored eye G1 animals were available, intercrosses also were made between colored-eyed siblings. In the test crosses, families 32 and 36 failed to produce G2 progeny with colored eyes. However, families 35 and 64, and the three colored-eye G1 mosquitoes from the pooled G0 animals produced G2 progeny distributed in phenotypic classes with numbers consistent with the conclusion that each contains a single insertion capable of segregation (Table 2).
Table 2.
Results from test crosses and intercrosses of G1 colored-eye progeny of families 35 and 64, and the pool
| Experiment | Family | Phenotypic class | No. of animals | Total | χ2 |
|---|---|---|---|---|---|
| Test cross* | 35 | Colored-eye | 142 | 253 | 3.78† |
| White-eye | 111 | ||||
| 64 | Colored-eye | 39 | 85 | 0.57† | |
| White-eye | 46 | ||||
| Pool | Colored-eye | 121 | 282 | 5.67† | |
| White-eye | 161 | ||||
| Intercross‡ | 35 | Colored-eye | 133 | 169 | 1.23§ |
| White-eye | 36 | ||||
| 64 | Dark red-eye | 28 | 139 | 5.24¶ | |
| Red-eye | 65 | ||||
| White-eye | 48 |
*G1 colored-eye mosquito crossed with homozygous recessive khw.
Phenotypic distributions are consistent with 1:1 segregation of a single integrated marker gene as evaluated by χ2, df = 1.
G1 colored eye mosquitoes crossed with one another.
All colored-eye phenotypes were treated as equal for family 35, and the phenotypic distributions are consistent with 3:1 segregation of a single integrated marker gene as evaluated by χ2, df = 1.
Phenotypic distributions are consistent with the 1:2:1 ratio of genotypes as evaluated by χ2, df = 2.
Family 64 produced colored-eye progeny that could be interpreted to result from complementation by a gene with a semidominant effect. Three distinct eye-color phenotypes, dark red, medium red, and white, could be seen in the progeny of the intercross, and these were interpreted to result from the homozygous dominant (two copies of the transgene), heterozygous (one copy of the transgene), and homozygous recessive (no copies of the transgene), progeny respectively (Fig. 1, Table 2).
Figure 1.
Eye color phenotypes resulting from the G1 intercross of family 64. Animals were photographed with incident light under a dissecting microscope. (Upper Left) khw recipient strain white eye phenotype. (Upper Right) Wild-type eye color. Transformed members of family 64 carrying two copies (Lower Right) and one copy (Lower Left) of an insertion.
The analysis of the G2 progeny of family 35 was made more complex by the presence of at least four distinct eye color phenotypes. The lighter colored eyes were remarkable in that they showed mosaic patches of darker facets among a lighter pink background eye color. However, dividing the animals into two phenotypic classes, colored eyes and white eyes, resulted in numbers consistent with a 3:1 ratio also indicative of a single integration event (Table 2).
Southern Blot Analysis.
Southern blot analyses were performed on genomic DNA isolated from families 35, 64, and the pool to determine if the presence of DNA fragments corresponding to the transgene was correlated with complementation of the phenotype and if these fragments had the structure characteristic of a transposition event. Three types of DNA fragments were used as probes in the Southern blot analysis (Fig. 2A). A 0.76-kb fragment, LH, complementary to the left-hand end of Hermes was used to verify the presence of the inverted repeat region of the transformation construct. The 4.7-kb fragment, cn+, of D. melanogaster DNA containing the wild-type copy of the cn gene was used to verify the presence of the marker gene. Two probes, MD, were made by gene amplification of the M. domestica DNA flanking the Hermes element in pH[cn]. These fragments were used to verify that the integrations into the genome did not include the flanking plasmid DNA, as would be expected of precise excision and transposition.
Genomic DNA was digested with XbaI, which cuts once in the putative transposed fragment, or with XbaI and SacII, which results in the precise excision of the 4.7-kb fragment containing the D. melanogaster cn+ gene (Fig. 2A). Families 35, 64, and the pool all have unique hybridizing fragments of ≈12, 11 and 10 kb, respectively, in the XbaI-digested DNA hybridized with the LH or cn+ fragments (Fig. 2 B and C). Additional strong hybridizing fragments were seen in both experimental and control DNAs when using the LH fragment (Fig. 2B). Family 64 and the pool have the expected XbaI/SacII signal at 4.7 kb representing the internal cn+ fragment. However, family 35 shows an anomalously smaller signal suggesting loss of DNA within this internal fragment (Fig. 2C). Diagnostic digests with StyI and hybridization with the 4.7-kb fragment show loss of DNA in the portion of the D. melanogaster genomic fragment that is outside the coding region and to the 5′ end of the cn gene (data not shown). Hybridization to XbaI-digested genomic DNAs with the MD probes failed to produce a signal (Fig. 2D).
DISCUSSION
These data indicate that transformation constructs based on the Hermes transposable element can integrate into the genome of the mosquito Ae. aegypti in an efficient and heritable manner. Three stable independent insertion events were recovered, two of which were from single founder families, resulting in a transformation efficiency of ≈8% (2/24) of single fertile G0 founders or 0.3% (3/925) when expressed in terms of events per injected embryo. The transgenic lines have been stable through more than 10 generations at the time of submission of this report (data not shown).
In addition, we have confirmed that a wild-type allele of the cn gene from D. melanogaster functions as a semidominant transgene when introduced into the khw strain of Ae. aegypti. The transgenic phenotype is readily observed, and it is possible in family 64 to discriminate between homozygous and heterozygous animals. It is quite probable that the D. melanogaster cn+ gene also can be used as a transformation marker in other Diptera, and possibly other orders that contain species with kynurenine hydroxylase mutants. Of particular note, both colored-eyed G0 founders (represented by the single families) and white-eyed G0 founders (represented by the pool) produced transformed families. Therefore the presence or absence of color in a G0 animal is not an accurate indication that it has been genetically transformed.
Genetic analysis of G1 families showing complementation demonstrated that the changes in eye color were heritable and could segregate as Mendelian factors, suggesting that the transformation events were caused by insertions in chromosomes. The results of test crosses revealed that the three families produced G2 progeny in the 1:1 phenotypic ratio expected for a single chromosomal insertion of the transgene. However the insertion observed in the pool family appears to have occurred in a chromosomal region that results in a reduction in the number of colored-eyed individuals observed.
The results of intercrosses performed for families 35 and 64 also were consistent with a single chromosomal insertion of the transgene. Family 35 has an interesting insertion that results in variable expression of the phenotype, reminiscent of either position-effect variegation or somatic excision of the transposon (21). Due to this effect it was not possible to determine which animals were homozygous or heterozygous for the transgene; therefore the results were evaluated based on a 3:1 ratio of colored eye/white eye phenotypic segregation. For family 64 the distribution of phenotypes produced the expected dark red eye/medium red eye/white eye ratio (1:2:1) for a semidominant transgene.
Southern blot analyses of the transformed lines confirmed the presence of the transgene and provided further evidence for integration into mosquito chromosomes. The results observed with the LH and cn+ probes indicate that hybridizing fragments containing both the Hermes element and the cn+ marker gene were integrated. The size and number of the observed fragments were consistent with a single insertion of the transformation construct into the genome. The different sizes of the XbaI fragments from each of the lines demonstrates that they are independent insertions. The single unique hybridizing fragment in each case demonstrates that only a single copy of the transgene was integrated. The lack of hybridization to MD probes is a result consistent with transposase-mediated insertion as only sequences delimited by the Hermes inverted terminal repeats would be expected to have transposed into the genome.
The nature of the DNA producing the common hybridizing signal with the LH probe has not been determined, but it may represent cross-hybridization of the probe with an endogenous hAT element. If such an endogenous hAT element does exist, activity of its transposase may have been responsible for a number of the results observed, including the small deletion in family 35, the absence of transformants in the G2 generation of families 32 and 36, as well as the observed mosaicism. Although the cn+ gene product and its enzymatic substrate and product are thought to be cell nonautonomous, it appears that they can display “local autonomy,” presumably as a result of the product of the enzyme reaction, 3′-hydroxy kynurenine, being immediately available for the next step in the pigment biosynthetic pathway and thus does not diffuse out of the cell.
We have presented both genetic and molecular evidence that we interpret to demonstrate that the observed phenotypic complementation is associated with single insertions of the transgene into mosquito chromosomes of three independent lines. Given the previous demonstration of the ability of the Hermes element to accurately transpose within the embryonic soma of Ae. aegypti (9), it is probable that the integration events observed in these experiments were also caused by a Hermes-mediated transposition event. However, Hermes integration events that included flanking plasmid sequences have been observed (A.A.J., unpublished data). Various protocols including inverse and universal PCR were performed to determine the nature of the junction between the integrated transgene and the mosquito genomic DNA. These attempts were unsuccessful and the characterization of these junction fragments is the basis of ongoing research.
The Hermes transposon coupled with the cn+ marker gene and khw recipient strain, along with the similar development of mariner-based transformation in Ae. aegypti (22), provide powerful tools for the molecular analysis of this major vector mosquito species. They also make it possible to develop stable lines of virus-resistant mosquitoes (8) that can be used in genetic control strategies to interfere with the transmission of pathogenic agents.
Acknowledgments
We acknowledge the pioneering efforts of Drs. Alison C. Morris and Kurt Yardley in the development of mosquito trangenesis, Ms. L. Miyashiro and Ms. P. Nguyen for mosquito care, Dr. A. J. Howell for the gift of the D. melanogaster cn+ gene, Drs. D. O’Brochta and P. Atkinson for the Hermes plasmids, Dr. D. Prager for continuous support of the project, and L. M. Olson for help in preparing the manuscript. This research was supported by grants from the National Institutes of Health (AI29746 and AI32730 to A.A.J.), the United Nations Development Programme/World Bank/World Health Organization Special Programme for Research and Training in Tropical Diseases (to F.H.C. and M.Q.B.), and the John D. and Catherine T. MacArthur Foundation (to F.H.C. and A.A.J.).
ABBREVIATIONS
- cn
cinnabar
- khw
kynurenine hydroxylase-white
- LH
left hand probe
Footnotes
A commentary on this article begins on page 3349.
References
- 1.Collins F H, James A A. Sci Med. 1996;3:52–61. [Google Scholar]
- 2.Meredith S E O, James A A. Ann Parasitol Humaine Comp. 1990;65:S1:113–118. doi: 10.1051/parasite/1990651113. [DOI] [PubMed] [Google Scholar]
- 3.Miller L H, Sakai R K, Romans P, Gwadz R W, Kantoff P, Coon H G. Science. 1987;237:779–781. doi: 10.1126/science.3039658. [DOI] [PubMed] [Google Scholar]
- 4.McGrane V, Carlson J O, Miller B R, Beaty B J. Am J Trop Med Hyg. 1988;39:502–510. doi: 10.4269/ajtmh.1988.39.502. [DOI] [PubMed] [Google Scholar]
- 5.Morris A C, Eggleston P, Crampton J M. Med Vet Entomol. 1989;3:1–7. doi: 10.1111/j.1365-2915.1989.tb00467.x. [DOI] [PubMed] [Google Scholar]
- 6.Morris A C, Pott G B, Chen J, James A A. Am J Trop Med Hyg. 1995;52:456–460. doi: 10.4269/ajtmh.1995.52.456. [DOI] [PubMed] [Google Scholar]
- 7.Shotkoski F, Morris A C, James A A, ffrench-Constant R H. Gene. 1996;168:127–133. doi: 10.1016/0378-1119(95)00756-3. [DOI] [PubMed] [Google Scholar]
- 8.Olson K E, Higgs S, Gaines P J, Powers A M, Davis B S, Kamrud K I, Carlson J O, Blair C D, Beaty B J. Science. 1996;272:884–886. doi: 10.1126/science.272.5263.884. [DOI] [PubMed] [Google Scholar]
- 9.Sarkar A, Yardley K, Atkinson P W, James A A, O’Brochta D A. Insect Biochem Mol Biol. 1997;27:359–363. doi: 10.1016/s0965-1748(97)00018-0. [DOI] [PubMed] [Google Scholar]
- 10.Beard C B, Benedict M Q, Primus A P, Finnerty V, Collins F H. J Heredity. 1995;86:375–380. doi: 10.1093/oxfordjournals.jhered.a111606. [DOI] [PubMed] [Google Scholar]
- 11.Bhalla S C. Mosq News. 1968;28:381–385. [Google Scholar]
- 12.Cornel A J, Benedict M Q, Salazar Rafferty C, Howells A J, Collins F H. Insect Biochem Mol Biol. 1997;27:993–997. doi: 10.1016/s0965-1748(97)00084-2. [DOI] [PubMed] [Google Scholar]
- 13.Warren W D, Palmer S, Howells A J. Genetica. 1996–1997;98:249–262. doi: 10.1007/BF00057589. [DOI] [PubMed] [Google Scholar]
- 14.Warren W D, Atkinson P W, O’Brochta D A. Genet Res. 1994;64:87–97. doi: 10.1017/s0016672300032699. [DOI] [PubMed] [Google Scholar]
- 15.O’Brochta D A, Warren W D, Saville K J, Atkinson P W. Genetics. 1996;142:907–914. doi: 10.1093/genetics/142.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris A. In: The Molecular Biology of Insect Vectors of Disease. Crampton J M, Beard C B, Louis C, editors. London: Chapman & Hall; 1997. pp. 423–429. [Google Scholar]
- 17.Munstermann L E. In: The Molecular Biology of Insect Vectors of Disease. Crampton J M, Beard C B, Louis C, editors. London: Chapman & Hall; 1997. pp. 13–20. [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Lidholm D A, Lohe A R, Hartl D L. Genetics. 1993;134:859–868. doi: 10.1093/genetics/134.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loukeris T G, Livadaras I, Arca B, Zabalou S, Savakis C. Science. 1995;270:2002–2005. doi: 10.1126/science.270.5244.2002. [DOI] [PubMed] [Google Scholar]
- 21.Henikoff S. BioEssays. 1996;18:401–409. doi: 10.1002/bies.950180510. [DOI] [PubMed] [Google Scholar]
- 22.Coates C J, Jasinskiene N, Miyashiro L, James A A. Proc Natl Acad Sci USA. 1998;95:3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]