Abstract
The mariner transposable element is capable of interplasmid transposition in the embryonic soma of the yellow fever mosquito, Aedes aegypti. To determine if this demonstrated mobility could be utilized to genetically transform the mosquito, a modified mariner element marked with a wild-type allele of the Drosophila melanogaster cinnabar gene was microinjected into embryos of a kynurenine hydroxylase-deficient, white-eyed recipient strain. Three of 69 fertile male founders resulting from the microinjected embryos produced families with colored-eyed progeny individuals, a transformation rate of 4%. The transgene-mediated complementation of eye color was observed to segregate in a Mendelian manner, although one insertion segregates with the recessive allele (female-determining) of the sex-determining locus, and a separate insertion is homozygous lethal. Molecular analysis of selected transformed families demonstrated that a single complete copy of the construct had integrated independently in each case and that it had done so in a transposase-mediated manner. The availability of a mariner transformation system greatly enhances our ability to study and manipulate this important vector species.
The incidence of vector-borne diseases is on the rise. As part of a multifaceted effort to control the transmission of diseases, we are developing tools for the molecular genetic manipulation of mosquitoes (1). We intend to use the tools and techniques of modern molecular biology to generate strains of mosquitoes that are incapable of transmitting a specific pathogen. These strains will be used selectively in release programs to reduce disease transmission. One of the key requirements for this effort is a method for introducing genes into mosquitoes. Recently, the Hermes transposable element from the housefly, Musca domestica, was shown to integrate into the germ line of the yellow fever mosquito, Aedes aegypti (2). The identification of Hermes as a viable candidate for mosquito transgenesis resulted from a strategy that first evaluated the ability of the element to mobilize (excise and insert) in the embryonic soma of the mosquito (3), followed by demonstration that it would integrate into the germ line (2). By using this approach, we show that a modified mariner transposable element efficiently and stably integrates into the germ line of Ae. aegypti.
Transposition assays based on the mobilization of a marked transposon from a donor to a target plasmid (4) were used to show that the mariner element, Mos1, from Drosophila mauritiana (5), was capable of mobility in embryos of Ae. aegypti. Subsequently, a genetic transformation experiment showed that Mos1 could integrate into the germ line of the mosquito. This experiment exploited the recently demonstrated ability of a wild-type copy of the Drosophila melanogaster cinnabar (cn+) gene to complement the white-eye phenotype of the kynurenine hydroxylase-white (khw) strain of Ae. aegypti (6–8). We report the successful generation of transgenic Ae. aegypti lines that contain a stable integrated copy of a Mos1-cn+ transgene. These results demonstrate the robustness of the strategy for identifying potential transformation vectors in mosquitoes, and provide the basis for the development of an additional, independent transformation system for this vector mosquito.
METHODS
Plasmid Construction.
The transposition assay plasmids, pKhsp82MOS, pBSMOSoriKan, and pMOS5′+3′oriKan, have been described (4). The transformation vector, pM[cn], was constructed by inserting a 4.7-kb SacII/XbaI cn+ D. melanogaster genomic fragment (ref. 6; gift of A. J. Howells, GenBank accession no. U56245) into the unique SalI site of pBSMOS (5).
Embryo Microinjection and Mosquito Rearing.
The interplasmid transposition assays were conducted in the Rockefeller strain of Ae. aegypti (obtained from the University of Notre Dame), essentially as described (4), with the heat shock being performed at 39°C for 1 h. For the transformation experiment, embryos homozygous for the khw mutation were collected and prepared for microinjection as described (9). Embryos were injected with a solution of pM[cn] and pKhsp82MOS, each at a final concentration of 0.5 mg/ml in 5 mM KCl and 0.1 mM NaH2PO4 (pH 6.8). Approximately 16 h postinjection, the embryos were exposed to a heat shock at 39°C for 1 h after which they were placed at 27°C, 80% relative humidity and estivated for 5 days. The embryos were hatched and the resulting larvae were allowed to develop to adults (G0 generation). General aspects of mosquito rearing were performed as described (10). Individual G0 males were mated with 10 khw virgin females. Pools of 2–10 G0 females were mated with 3 khw males. Progeny from these crosses (G1 generation) were screened for the presence of eye color. Those G1 individuals showing complementation of their eye color were back-crossed to the parental genotype, khw/khw. Colored-eyed progeny from this cross (G2 generation) were intercrossed to create functionally homozygous individuals (G3 generation) containing two copies of the transgene.
Southern Blot Analysis.
Southern blot analyses were done as described (11). Genomic DNA isolated from G2 individuals was digested with a restriction endonuclease, SacI, which cuts twice within the transformation construct, prepared for Southern blot analysis, and hybridized with a radiolabeled cn+ gene fragment (Fig. 1).
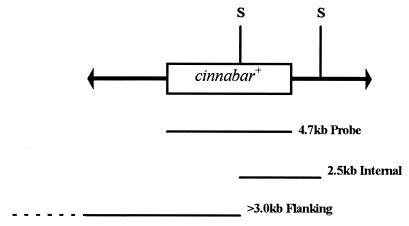
Figure 1.
Schematic diagram of the mobile portion of the pM[cn] construct. The mariner inverted terminal repeats are represented as arrows flanking the 4.7-kb genomic DNA fragment from D. melanogaster that includes a copy of the cn+ gene. The relative positions of the SacI restriction endonuclease-cleavage sites are shown (S). Listed below are the relative extents and sizes of the fragment hybridized with the genomic DNA and the expected hybridizing genomic fragments from the transformed families.
Inverse PCR Analysis.
Genomic DNA was digested to completion with Sau3AI and ligated under conditions of dilute DNA concentration with excess T4 DNA ligase. Gene amplification was performed with the following oligonucleotide primers: MLF1, 5′-TTGTTTACTCTCAGTGCAGTCAACATGTCG-3′ (148–177); MLR1, 5′-TTCGACAGTCAAGGTTGACACTTCACAAGG-3′ (114–85); MRF1, 5′-AAGACGATGAGTTCTACTGGCGTGGAATCC-3′ (1121–1150); MRR1, 5′-CTTGCCGTATGTGATGGAGCGTTGTCATGG-3′ (941–912).
The numbers in parentheses indicate the nucleotide positions in the Mos1 sequence (GenBank accession no. X78906). The reactions were performed under the following amplification conditions: 1 cycle of 95°C for 5 min, 40 cycles of 95°C for 30 sec, 65°C for 30 sec, 72°C for 1 min, and 1 cycle of 72°C for 5 min. Amplification products were cloned into pGEM-T (Promega), and the DNA sequence was determined by using the MLR1 and MRF1 primers.
RESULTS
Mariner Transposition Assays.
These assays are designed to detect the transposition of a marked mariner element from a donor plasmid to a target plasmid and were performed in the presence and absence of a helper plasmid supplying the transposase protein. In the presence of the helper plasmid, 12 independent transposition events (0.0009% of donor plasmids) were observed (Table 1). In all cases the modified mariner element inserted at the 3′ end of a TA dinucleotide. This insertion was accompanied by a duplication of the TA residues at the 3′ end of the modified element (data not shown). No transposition events were observed in the absence of the helper plasmid.
Table 1.
Data from transposition assays in Ae. aegypti embryos
| Helper plasmid present | No. of experiments | No. of donor plasmids recovered | No. of transposition events observed (%) |
|---|---|---|---|
| − | 3 | 550,000 | 0 |
| + | 5 | 1,396,000 | 12 (0.0009) |
Germ-Line Transformation and Genetic Analysis.
A total of 1,625 khw embryos (G0 generation) were micro-injected with pM[cn] and the helper plasmid pKhsp82MOS. Of the injected embryos, 231 (14.2%) survived to become adults, and 86 of these (37%, 46 males and 40 females) had colored eyes. All 121 G0 males (both colored- and white-eyed) were used as single founders of families, and all 110 females were mated in one of 13 pools. A total of 69 (57%) of the single male founders were fertile and all of the female pools produced G1 progeny. G1 progeny were screened visually as adults for changes in eye color. Three of the single male founders, families 11, 90 and 128, and two of the female pools, 122 and 137, produced G1 progeny with colored eyes (Table 2). The observed eye color varied between families, ranging from a light orange to a dark purple/black that was close to wild type. The eye color within each family was constant, except for family 137 in which a few individuals had lighter colored-eyes than the majority. A total of 38 colored-eyed G1 progeny were observed from ≈20,700 mosquitoes screened and the transformation efficiency was approximately 4% (calculated as the percentage of fertile male founder families with G1 progeny showing complementation). The three transformed G0 male founder families produced relatively few progeny in total and the colored-eyed progeny were among the last adults to emerge from multiple cycles of blood feeding and egg laying (this was observed also for the two female pools). The G1 progeny of family 128 were unusual in that the distribution of colored-eyed individuals was biased in favor of the females, suggesting that the insertion may be sex-linked.
Table 2.
G0 families producing G1 progeny with colored eyes
| G0 family | Total G1 progeny screened | No. of G1 progeny with colored eyes (%) |
|---|---|---|
| 11 | 79 | 1 (1.3) |
| 90 | 189 | 3 (1.6) |
| 122 (female pool) | 548 | 3* |
| 128 | 29 | 11† (37.9) |
| 137 (female pool) | 459 | 20* |
The percentage of progeny was not determined for the female pools as it was not known how many individuals each transformed female contributed to the total.
A total of 10/11 females had colored eyes and only 1/18 males had colored eyes.
Test crosses were set up with colored-eyed G1 mosquitoes by mating them to homozygous khw individuals. The individuals from family 90 were sterile and did not produce G2 progeny. The remainder of the families produced both colored-eyed and white-eyed G2 progeny in approximately 1:1 numerical ratios consistent with the insertion and subsequent Mendelian segregation of a transgene on a single chromosome, although colored-eyed individuals were underrepresented in families 128 and 137 (Table 3). Family 137 continued to show variable expression of the eye color phenotype in the G2. Intercrosses were set up with colored-eyed G2 mosquitoes within each family (Table 4). Two colored-eye phenotypic classes were observed in families 11 and 122, one class being the same color as the previous generation and the other class having a darker color. We interpreted the novel phenotype, darker color in the eyes, to result from the presence of two transgenes in each animal, a genetic condition functionally equivalent to homozygosity. By using this interpretation, these results are consistent with a 1:2:1 ratio of genotypes and phenotypes anticipated from a cross between two animals heterozygous for a single insertion of a transgene with a semidominant effect. There were no white-eyed females recovered in family 128, a result consistent with the interpretation of transgene linkage to the recessive sex-determining locus. In addition, the eye color of family 128 is so dark that it is not possible to distinguish animals carrying one or two copies of the transgene. As with the previous generations of family 137, a few individuals were observed that had a lighter eye color than the majority. In addition, no phenotype was observed that was consistent with a homozygous genotype. This result was interpreted to indicate that homozygosity of the insertion in family 137 results in a lethal condition.
Table 3.
Results from test crosses of G1 colored-eyed progeny with homozygous recessive khw
| Family | Phenotypic class | No. of animals | Total | χ2 |
|---|---|---|---|---|
| 11 | Colored-eye | 288 | 569 | 0.08* |
| White-eye | 281 | |||
| 122 | Colored-eye | 69 | 143 | 0.16* |
| White-eye | 74 | |||
| 128 | Colored-eye | 147 | 346 | 7.6* |
| White-eye | 199 | |||
| 137 | Colored-eye | 183 | 464 | 20.68 |
| White-eye | 281 |
Phenotypic distributions are consistent with 1:1 segregation of a single marker gene as evaluated by χ2, df = 1.
Table 4.
Results from self-cross of G2 colored-eye progeny
| Family | Phenotypic class | No. of animals | Total | χ2 |
|---|---|---|---|---|
| 11 | White-eye | 86 | 273 | 7.32* |
| Heterozygous colored-eye | 132 | |||
| Homozygous colored-eye | 55 | |||
| 122 | White-eye | 99 | 422 | 0.80* |
| Heterozygous colored-eye | 211 | |||
| Homozygous colored-eye | 112 | |||
| 128 | White-eye males | 93 | 320 | 11.34† |
| Colored-eyed males | 97 | |||
| White-eyed females | 0 | |||
| Colored-eyed females | 130 | |||
| 137 | White-eye | 75 | 235 | 0.23‡ |
| Heterozygous colored-eye | 160 | |||
| Homozygous colored-eye | 0 |
Phenotypic distributions are consistent with a 1:2:1 ratio as evaluated by χ2, df = 2.
Phenotypic distributions are consistent with segregation of a sex-linked gene as evaluated by χ2, df = 3.
The mariner insertion in this family results in a homozygous lethal phenotype and a 2:1 ratio of colored-eye/white-eye surviving animals. The observed phenotypic distributions are consistent with this interpretation as evaluated by χ2, df = 2.
Southern Blot and Inverse PCR Analyses.
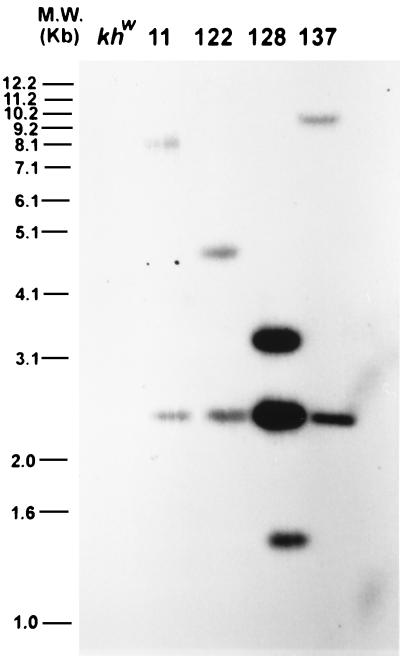
All families except for family 90 were subjected to a molecular analysis to determine whether the observed phenotypic complementation was correlated with the insertion of the mariner construct into the mosquito genome. The 4.7-kb probe corresponding to the D. melanogaster genomic DNA fragment should hybridize to two fragments in SacI-digested genomic DNA isolated from a family. An internal 2.5-kb fragment should be visible in all families along with a single fragment whose size, >3.0 kb, is unique to each family (Fig. 1). The 2.5-kb internal SacI fragment was present in all four families, and each family has the predicted additional fragment (Fig. 2). These results are consistent with unique insertions into the mosquito genome. Family 128 also had a 1.4-kb hybridizing fragment, indicating that this family may contain an additional, partial copy of the mariner construct. It is unknown why the observed hybridizing signal was higher for family 128 as the amount of DNA loaded on the agarose gel was approximately the same as for the other families.
Figure 2.
Southern blot analysis of G2 genomic DNA from the transformed families, digested with SacI and hybridized with the 4.7-kb cn+ gene fragment. The predicted 2.5-kb internal fragment is observed in each case, as is a flanking fragment of greater than 3.0 kb, representing a single complete insertion event. Family 128 contains an additional 1.4-kb hybridizing fragment.
An inverse gene amplification protocol was performed on G2 genomic DNA isolated from families 11 and 137 to determine the primary structure of the junction between the mariner inverted terminal repeats and the mosquito DNA. Both insertion site junctions have putative TA duplications immediately adjacent to the mariner inverted terminal repeats (Fig. 3). GenBank analyses of the flanking DNA revealed no significant matches to any known sequences.
Figure 3.
Primary DNA sequence of the junctions between the mariner inverted terminal repeats and the Ae. aegypti genomic DNA. The mariner sequence is shown in lowercase letters, the genomic Sau3AI sites are shown in bold (the complete left hand sequence from family 137 is not shown because the PCR product extends for several hundred base pairs), and the flanking TA residues are underlined.
DISCUSSION
We have shown that a modified mariner transposable element is capable of mediating germ-line transformation of Ae. aegypti. This important vector species joins the relatively small list of insects for which a stable germ-line transformation system is available (12, 13). Furthermore, we have provided additional support for the approach of using the interplasmid transposition assays as predictors for the ability of a transposable element to insert into an insect genome (2, 3). Recently, mariner has been shown to be capable of transforming the protozoan parasite, Leishmania (14), and with the success of these efforts in Ae. aegypti, it is likely that mariner will function in a wide variety of animal species. Interestingly, searches for mariner-like elements in the Ae. aegypti genome so far have been unsuccessful (15). In our transposition assay experiments, the lack of transposition events in the absence of supplied transposase supports the hypothesis that the genome is empty of mariner-like elements. This finding will be advantageous as there will be no endogenous homologous elements to re-mobilize integrated transgenes.
Our analysis of mariner has shown that it functions as a heritable, stable, and efficient mediator of gene insertion into Ae. aegypti. It is important to note that families founded by both colored-eyed G0 founders (11 and 90) and white-eyed G0 founders (128) produced G1 transformed progeny, and thus the presence or absence of eye color in a G0 individual is not indicative of a transgene insertion. Eye-color complementation in G0 individuals most likely results from a combination of cn+ expression from lingering plasmid DNA and somatic transposition events.
Genetic analyses of G1 progeny from the putatively transformed lines showed that the events associated with complementation could segregate as Mendelian factors, and this is additional evidence for mariner insertion into mosquito chromosomes. In test crosses, families 11 and 122 produced G2 progeny in approximately 1:1 phenotypic ratios that would be expected if each line contained a single chromosomal insertion of mariner. The numbers of G2 progeny in test crosses of families 128 and 137 also could be interpreted to result from single insertion events, but in addition, each insertion was into a region of DNA that when interrupted results in a reduction in the number of colored-eyed individuals recovered.
The results of intercrosses performed between respective G2 siblings of families 11 and 122 provided evidence that these individuals are heterozygous for a single transgene inserted into an autosome. The distribution of the phenotypes produced colored eye/white eye phenotypic ratios (3:1), and dark color eye/light color eye/white eye phenotypic ratios (1:2:1). The distribution of phenotypes for family 128 confirmed that the insertion is sex-linked, with all of the females having colored-eyes and the males segregating in an approximately 1:1 ratio. The insertion associated with family 137 appears to be homozygous lethal as no homozygous individuals were observed on the basis of phenotype.
There was a consistent appearance of lighter colored-eyed individuals in family 137, even though these individuals were not used as progenitors for each following generation. The molecular basis for this effect is unknown but it may be the result of position effect variegation resulting in a lower level of cn+ expression in those individuals. Position effect variegation of white gene expression in D. melanogaster is usually associated with a mosaic expression pattern in the eyes (16); however, the cn+ gene product is cell nonautonomous and thus may explain the lack of this phenotype.
Southern blot analyses of DNA isolated from transformed mosquitoes confirmed the presence of input plasmid DNA and provided evidence that this DNA was integrated into the mosquito genome. Transposase-mediated excision from the donor plasmid of a fragment of DNA containing the mariner inverted terminal repeats flanking the cn+ gene, and their proper integration into the genome should produce a characteristic number of DNA fragments upon digestion with an appropriate restriction endonuclease. These fragments can be revealed by hybridization with a selected DNA probe. In this case, hybridization of the 4.7-kb SacII/XbaI cn+ D. melanogaster genomic fragment to SacI-digested genomic DNA of each transformed family, reveals an internal 2.5-kb fragment, and a fragment whose size is greater than 3.0 kb and includes adjacent mosquito genomic DNA. These fragments were present in all four families, providing evidence of insertion into the mosquito genome. The extra fragment of DNA seen in family 128 cannot be explained by a normal mariner transposition event. It is possible that there has been a rearrangement of the marker gene; further molecular analysis should resolve this. The putative TA duplications flanking the insertions into families 11 and 137 are characteristic of precise mariner transposition and insertion as was observed in the interplasmid transposition assays. Only sequences delimited by the mariner inverted terminal repeats were inserted and the flanking DNA was unique.
The frequency, 0.0009%, of plasmid-to-plasmid mobility observed for mariner is lower than the frequency, 0.003%, observed for Hermes in similar experiments (2). The differences in actual transformation efficiencies, 4% and 8%, for mariner and Hermes (2), respectively, reflect these numbers; however, the predictive value of using mobility frequencies to estimate actual transformation efficiencies has to be rigorously tested taking into account that none of these systems have been optimized. However, we conclude that an effective approach to establishing transformation systems in other important insect species is to determine empirically the frequency of mobility of a number of elements and then demonstrate germ-line integration with the element showing the highest frequency of mobility. With this positive baseline, it then becomes possible to evaluate other elements and different parameters for individual elements. Because cross-mobilization between hAT-like (Hermes) and Tc1-like (mariner) transposons is not expected, the availability of these two independent transformation systems in Ae. aegypti should enhance our ability to study and manipulate this disease vector by allowing insertions of different transgenes within a single genome.
Acknowledgments
We thank Drs. Alison Morris and Kurt Yardley for their preliminary efforts in mosquito transgenesis, Ms. P. Nguyen for mosquito care, Drs. F. Collins and A. Howells for the khw strain and D. melanogaster wild-type cn gene, Dr. D. L. Hartl for the gift of the pBSMOS plasmid, Dr. D. Prager for continuous support of the project, and L. Olson for help in preparing the manuscript. This work was supported by grants from the National Institutes of Health (NIAID 29746 and 32730) and the John D. and Catherine T. MacArthur Foundation.
ABBREVIATIONS
- cn, cinnabar
khw, kynurenine hydroxylase-white
Footnotes
References
- 1.Collins F H, James A A. Sci Med. 1996;3:52–61. [Google Scholar]
- 2.Jasinskiene, N., Coates, C. J., Benedict, M. Q., Cornel, A. J., Salazar Rafferty, C., James A. A. & Collins, F. H. (1998) Proc. Natl. Acad. Sci. USA, 3743–3747. [DOI] [PMC free article] [PubMed]
- 3.Sarkar A, Yardley K, Atkinson P W, James A A, O’Brochta D A. Insect Biochem Mol Biol. 1997;27:359–363. doi: 10.1016/s0965-1748(97)00018-0. [DOI] [PubMed] [Google Scholar]
- 4.Coates C J, Turney C L, Frommer M, O’Brochta D A, Atkinson P W. Mol Gen Genet. 1997;253:728–733. doi: 10.1007/s004380050377. [DOI] [PubMed] [Google Scholar]
- 5.Medhora M, Maruyama K, Hartl D L. Genetics. 1991;128:311–318. doi: 10.1093/genetics/128.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren W D, Palmer S, Howells A J. Genetica. 1996–1997;98:249–262. doi: 10.1007/BF00057589. [DOI] [PubMed] [Google Scholar]
- 7.Cornel A J, Benedict M Q, Salazar Rafferty C, Howells A J, Collins F H. Insect Biochem Mol Biol. 1997;27:993–997. doi: 10.1016/s0965-1748(97)00084-2. [DOI] [PubMed] [Google Scholar]
- 8.Bhalla S C. Mosq News. 1968;28:381–385. [Google Scholar]
- 9.Morris A C. In: The Molecular Biology of Insect Vectors of Disease. Crampton J M, Beard C B, Louis C, editors. London: Chapman & Hall; 1997. pp. 423–429. [Google Scholar]
- 10.Munstermann L E. In: The Molecular Biology of Insect Vectors of Disease. Crampton J M, Beard C B, Louis C, editors. London: Chapman & Hall; 1997. pp. 13–20. [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Spradling A C, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 13.Loukeris T G, Livadaras I, Arca B, Zabalou S, Savakis C. Science. 1995;270:2002–2005. doi: 10.1126/science.270.5244.2002. [DOI] [PubMed] [Google Scholar]
- 14.Gueiros-Filho F J, Beverley S M. Science. 1997;276:1716–1719. doi: 10.1126/science.276.5319.1716. [DOI] [PubMed] [Google Scholar]
- 15.Robertson H M, MacLeod E G. Insect Mol Biol. 1993;2:125–39. doi: 10.1111/j.1365-2583.1993.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S. BioEssays. 1996;18:401–409. doi: 10.1002/bies.950180510. [DOI] [PubMed] [Google Scholar]