Abstract
Thousands of genes have recently been sequenced in organisms ranging from Escherichia coli to human. For the majority of these genes, however, available sequence does not define a biological role. Efficient functional characterization of these genes requires strategies for scaling genetic analyses to the whole genome level. Plasmid-based library selections are an established approach to the functional analysis of uncharacterized genes and can help elucidate biological function by identifying, for example, physical interactors for a gene and genetic enhancers and suppressors of mutant phenotypes. The application of these selections to every gene in a eukaryotic genome, however, is generally limited by the need to manipulate and sequence hundreds of DNA plasmids. We present an alternative approach in which identification of nucleic acids is accomplished by direct hybridization to high-density oligonucleotide arrays. Based on the complete sequence of Saccharomyces cerevisiae, high-density arrays containing oligonucleotides complementary to every gene in the yeast genome have been designed and synthesized. Two-hybrid protein–protein interaction screens were carried out for S. cerevisiae genes implicated in mRNA splicing and microtubule assembly. Hybridization of labeled DNA derived from positive clones is sufficient to characterize the results of a screen in a single experiment, allowing rapid determination of both established and previously unknown biological interactions. These results demonstrate the use of oligonucleotide arrays for the analysis of two-hybrid screens. This approach should be generally applicable to the analysis of a range of genetic selections.
The sequencing of the Saccharomyces cerevisiae genome has identified an estimated 6,000 genes, fewer than half of which have a known biological function (1). Understanding how these genes function is a major challenge for researchers in the postgenome era (2). Protein and mRNA gene expression patterns, disruption phenotypes, and protein–protein interactions need to be determined for every gene in a genome (3). However, the application of traditional methods of functional analysis to every gene in a genome may be limited by the need to manipulate and sequence numerous DNA clones in plasmid-based genetic screens such as the two-hybrid screen. We demonstrate here the use of DNA arrays containing oligonucleotides complementary to nearly every gene in the S. cerevisiae genome to analyze the results from plasmid-based genetic screens.
MATERIALS AND METHODS
Plasmids and Strains.
For the Ymr117c screen, the yeast strains used for two-hybrid screening were CG1945 and Y187 (CLONTECH). The FRYL library was constructed by cloning yeast genomic DNA into a pACTII-derived vector. The pAS2ΔΔ bait vector was constructed from the pAS2 plasmid (CLONTECH) by deletion of the CYH2 gene and the HA epitope. The bait plasmid was constructed by PCR amplification of YMR117c from genomic DNA and cloning into pASΔΔ as a BamHI-PstI fragment. The bait plasmid was verified by sequencing after cloning. For the Ymr138w screen, the yeast strains used were the Y190 and Y187 cyh2R marked derivatives of Y159 and Y153, respectively. The library was a yeast cDNA library fused to the transcriptional activation domain of GAL4 (gift of S. Elledge, Baylor College of Medicine, Houston). The bait vector pTS434 was constructed by cloning CIN4 into pAS1-CYH2 (CLONTECH) as a NcoI-BamHI fragment.
Two-Hybrid Screens.
For the Ymr117c screen, CG1945 yeast cells were transformed with the bait vector and used in a mating strategy (4). Y187 cells were first transformed with DNA from the FRYL two-hybrid library, transformants were pooled, and aliquots of the cell suspension were frozen. The two strains were mixed, concentrated onto filters, and incubated on rich medium for 4.5 h at 30°C. The cells were collected, and a 10−3 dilution was spread on −L, −LW, and −W plates to score the number of parental cells and the number of diploids. The rest of the cell suspension was spread on −LWH plates and incubated for 3 days at 30°C. Diploids (8.5 × 107) were screened, and 5,800 His+ colonies were selected. Ten milliliters of a 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) mixture (0.5% agar/0.1% SDS/6% dimethylformamide/0.04% X-Gal) was poured on the plates, and the plates were incubated at 30°C. Blue clones were checked after a 30 min to 18 h incubation and streaked on −LWH selective plates. One hundred and eight total clones were identified as positive by the X-Gal assay and processed as described below. For the YMR138w screen, Y190 containing pTS434 was transformed with cDNA library by using a lithium acetate-based protocol. Transformants (5 × 106) were screened by plating on −Ade-selective media, and 114 colonies Ade+ were selected. All 114 colonies were patched onto Ade+ plates and lifted onto BA85 nitrocellulose filters (Schleicher & Schuell) and immersed in liquid nitrogen for 10 s. The filters were then soaked with 3 ml of Z buffer (60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgSO4/50 mM 2-mercaptoethanol, pH 7.0) containing 0.05% X-Gal. Filters were incubated at 30°C for 6 h and scored for the development of blue color. Eighty-six clones were positive by a lacZ filter assay. All 86 clones passed testing for solo activation by streaking strain Y190 carrying the library isolate and pTS434 on −L plates plus 5 μg/ml cycloheximide. The strains were confirmed to have lost the TRP-containing plasmid by failure to grow on −W media. Eighty-one clones passed testing for specificity by mating strain Y190 carrying library plasmids with Y187 carrying the negative controls pAS-CDK2, pAS10-lamin, pAS1-p53, and pAS1-rev (a gift of D. Amberg, State University of New York, Health Science Center at Syracuse). Library plasmid inserts from both screens were amplified by PCR and the insert junctions with the Gal4 domain were sequenced and precisely identified in the yeast genome by using the blast program, the Saccharomyces Genome Database (http://genome-www.stanford.edu), and the Yeast Protein Database (http://www.proteome.com). In parallel, clones were used to inoculate 200 μl cultures. Saturated cultures were collected, pooled, and processed as described below.
PCR Amplification and Labeling of DNA from Pooled Clones.
Approximately 1 × 107 cells of each positive two-hybrid clone were pooled (Fig. 1) and DNA was isolated as previously described (5). Using the vector-based primers T7FOR (5′-GAATTGTAATACGACTCACTATAGGGAGGTGATGAAGATACCCCACC-3′) and T3REV (AGATGCAATTAACCCTCACTAAAGGGAGACGGGGTTTTTCAGTATCTACGATTC-3′), all library inserts were PCR-amplified in a single reaction. The 50-μl PCR contained: 2.5 units of Taq DNA polymerase, 10 mM Tris (pH 8.5), 50 mM KCl, 1.5 mM MgCl2, 0.2 μM each primer, and 250 μM each dNTP. Conditions used for amplification were as follows: 30 cycles at 96°C for 30 s, 62°C for 30 s, 72°C for 2 min. Reaction products were purified in a Qiaquick spin column (Qiagen). One microgram total PCR product was fragmented with 0.1 unit DNase I (amplification grade, GIBCO/BRL) for 2 min in 35 μl containing: 10 mM Tris-acetate (pH 7.5), 10 mM magnesium acetate, 50 mM potassium acetate, and 15 mM CoCl. The DNase I reaction was then boiled for 15 min, chilled on ice, and incubated with 1 μmol biotin-ddATP (NEN) and 25 units terminal transferase (Boehringer Mannheim) for 1 h at 37°C. SSPE-T hybridization buffer (0.9 M NaCl/60 mM NaH2PO4/6 mM EDTA/0.005% Triton X-100) was added to a final volume of 200 μl.
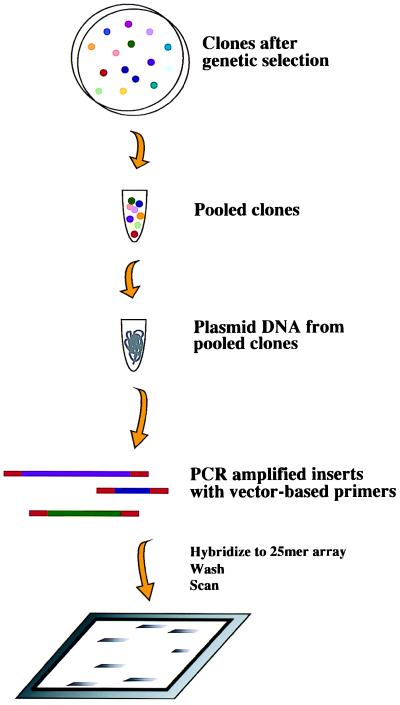
Figure 1.
Strategy for identifying sequences after a genetic selection. Rather than individual purification and dideoxy sequencing, all clones are pooled from plates and plasmid DNA is isolated in a single purification. PCR amplification using primers with 3′ sequence corresponding to vector sequence is used to selectively enrich for insert DNA from the plasmid pool. Amplified insert DNA is fragmented with DNase I, labeled with biotin-ddATP, and hybridized to an array containing oligonucleotide probes for every gene in the yeast genome.
Generation of cDNA Product from PCR Product.
RNA was transcribed from 240 ng of purified PCR product by using T7 polymerase (Ambion). The reaction was incubated an additional hour with 20 units DNase I. RNA was purified by using an RNA spin column (Qiagen). Two micrograms of RNA was used for first-strand cDNA synthesis (Promega). Reaction products were purified in a Qiaquick spin column (Qiagen), and 1 μg total PCR product was digested and prepared for hybridization.
Hybridization of DNA to the High-Density Oligonucleotide Array.
Arrays were prewashed with hybridization buffer 5 min prior to sample hybridization. Following a 5-min incubation at 99°C, the sample was chilled on ice, allowed to return to room temperature, and applied to the array. After a 12-h hybridization at 42°C, the array was washed 10 times with 6× SSPE-T, washed with 0.5× SSPE-T for 15 min, and stained with a streptavidin-phycoerythrin conjugate (Molecular Probes) for 10 min, all at 42°C. The staining buffer contained 6× SSPET, 0.5 mg/ml BSA, and 1 mg/ml streptavidin-phycoerythrin. The array was washed five times with 6× SSPE-T prior to scanning. Hybridization patterns were detected by using an argon ion laser to excite phycoerythrin; the resulting emission was detected by using a photomultiplier tube through a 560-nm bandpass filter (Molecular Dynamics). The entire array was read at a resolution of 7.5 μm in less than 20 min, generating quantitative signal for each probe element. The collected data were analyzed with image and data analysis software (Affymetrix, Santa Clara, CA).
Criteria for Gene Detection.
On chips A, B, C, and D, which contain an average of 20 oligonucleotide probes per gene, the presence of a gene fragment was determined by visual and quantitative detection of three contiguous positive probes. On the E chip, which contains probes for 5′ sequence from genes that are longer than 1 kb, detection of two contiguous positive probes was considered sufficient to detect a gene fragment.
RESULTS
Oligonucleotide arrays containing more than 65,000 DNA synthesis features were prepared by using light-directed, solid-phase combinatorial chemistry as previously described (6, 7). Each 50 × 50 μm synthesis feature is composed of more than 107 copies of a specific 25-mer oligonucleotide that is complementary to a portion of a yeast gene. The full set of oligonucleotides includes an average of 40 synthesis features for each of the 6,321 genes identified from the Saccharomyces cerevisiae genome. These arrays were originally designed and used for the analysis of mRNA gene expression levels (8).
Oligonucleotide arrays were first tested for the ability to identify specific gene fragments. A fluorescence image of an array after hybridization of 11 labeled PCR products reveals intense signals at discrete positions, with minimal background (Fig. 2a). Because the probes for a given gene are synthesized in adjacent positions, hybridization of PCR products is detected as horizontal rows of high intensity (Fig. 2b). Signal corresponding to all 11 genes was detected in the correct locations. No significant signal was detected for any other genes in the genome. Each experiment was performed in duplicate, and hybridization results were found to be reproducible (data not shown).
Figure 2.
Fluorescence images of a high-density oligonucleotide array containing 25-mer probes for nearly every gene on Saccharomyces cerevisiae chromosomes 5–10. (a) Fluorescence pattern obtained after hybridization of 11 control genes: YEL002c, YEL003w, YEL005c, YEL006w, YEL018w, YEL019c, YEL021w, YEL024w, YHL014c, YHL045w, and YHL044c. Dark areas correspond to probes for genes not present in the control pool. (b) A close-up view of gene YHL014c shows the exact probe features that hybridize to the insert. Red grid highlights all probe features for YHL014c. Top row of probe elements contains oligonucleotides perfectly complementary to gene sequence, whereas bottom rows contain a mismatch in the central position of the oligonucleotide. Approximate locations of complementary oligonucleotide probes along the YHL014c ORF are also shown.
After a biological selection, library elements in high abundance can be identified by dideoxy sequencing. However, detection of rare elements might require the sequencing of thousands of clones. To determine the ability to detect very rare elements using array hybridization, the control PCR products were remade without the 600-bp YEL006c gene fragment, and known amounts of this sequence were added to the pool. Concentrations of spiked YEL006c DNA as low as 5 pM were detectable by hybridization. Therefore, array hybridization is sensitive to library elements that comprise less than 1:10,000 of the total pool. This is consistent with previous gene expression experiments in which rare mRNAs present at frequencies below 1:100,000 were detected quantitatively (7).
Whole genome yeast arrays were then used to analyze results from two-hybrid screens for protein–protein interactions. Identification of proteins that physically interact within the cell can suggest how a gene product participates in cellular processes (9–12). In the two-hybrid screen, two proteins are expressed in yeast as fusions to either the DNA-binding domain or the activation domain of a transcription factor. Physical interaction of the two proteins reconstitutes transcriptional activity, turning on a gene essential for survival under selective conditions (9). In screening for novel protein–protein interactions, yeast cells are first transformed with a plasmid encoding a specific DNA-binding fusion protein. A plasmid library of activation domain fusions derived from genomic DNA is then introduced into these cells. Transcriptional activation fusions found in cells that survive selective conditions are considered to encode peptide domains that may interact with the DNA-binding domain fusion protein.
To demonstrate the analysis of a genetic selection using oligonucleotide arrays, a two-hybrid screen was conducted for the S. cerevisiae gene YMR117c. YMR117c is a previously uncharacterized ORF recently found by two-hybrid analysis to interact with the U2 snRNP-associated splicing factor, Prp11p (4).
A total of 108 clones were isolated from the YMR117c two-hybrid screen and mixed into a single pool. Plasmid DNA was purified from the pooled clones, and primers containing vector sequence at the 3′ end were used to PCR amplify gene inserts from the plasmid mixture. DNA products generated from the library plasmid pool were partially DNase I digested, biotinylated, and hybridized to whole genome arrays (Fig. 3). Orientation of genes was determined by hybridization of forward-strand cDNA products. All genes identified by array hybridization are listed in Table 1.
Figure 3.
Fluorescence image of a portion of a high-density oligonucleotide array containing 25-mer probes to nearly every gene on Saccharomyces cerevisiae chromosomes 5–10 after hybridization of YMR117c two-hybrid sample. The three lighted strips correspond to probes covering nucleotides 156–654 of ORF YER018c, nucleotides 1860–2484 of YER032w, and nucleotides 4092–4452 of YGL197w. Terminal probes are described as the most 5′ nucleotide of the most 5′ probe and the most 3′ nucleotide of the most 3′ probes that gave a positive signal. Dark areas correspond to probes for genes not present after genetic selection.
Table 1.
Yeast ORFs identified by array analysis of two-hybrid screens
| Clone type | YMR117c | YMR138w (CIN4) |
|---|---|---|
| Genes | YBR020w | YDL117w |
| YCL032W (STE50) | YDR087c | |
| YCR073c (SSK22) | YGL172w (NUP49) | |
| YDR104c | YHR141c (MAK18) | |
| YER018c | YLR109w | |
| YER032w (FIR1) | YNR050c (LYS9) | |
| YFR046c | YPL241c (CIN2) | |
| YGL197w | ||
| YIL144w | ||
| YLR319c (BUD6) | ||
| YLR419w | ||
| YML049c | ||
| YMR224c (MRE11) | ||
| YOL18c | ||
| YOL34w | ||
| YOR206w | ||
| YPR010c (RPA135) | ||
| YPR145w (ASN1) | ||
| DNA not encoding protein | 18S and 25S rRNA | |
| Reverse orientation | YNL291c | YBR189w |
| YDR381w | ||
| YNL301c (RP28B) | ||
| YNR035c | ||
| YOL056w (GPM3) |
ORF loci and names are listed for genes detected by array hybridization of PCR products derived from end products of a two-hybrid screen. Because inserts in the noncoding orientation comprise a significant proportion of false positives in the two-hybrid screen, RNA was transcribed from the upstream T7 promoter and used to generate exclusively antisense cDNA strands with reverse transcriptase. cDNA products were then biotinylated, fragmented, and hybridized as described. Genes detected by a double-stranded DNA hybridization but absent in cDNA hybridization are considered to be in reverse orientation. Control experiments were performed to confirm that this method is orientation-specific (data not shown).
For the YMR117c screen, hybridization results were compared with results obtained by dideoxy sequencing of all 108 DNA clones. Nineteen of 22 independent loci were identified by hybridization, with no false positives. Based on analysis of the hybridizing array elements, we were also able to identify the region of the gene present in each insert (Table 2).
Table 2.
Comparison of sequencing and hybridization for clone 5′ ends
| ORF name | ORF size, nt | 5′ end by sequencing | 5′ end array probe |
|---|---|---|---|
| YBR020w | 1,584 | 1,151 | 1,164 |
| YCL032w | 1,038 | 131 | 168 |
| YDR104c | 3,735 | 3,230 | 3,234 |
| YER032w | 2,775 | 1,808 | 1,860 |
| YFR046c | 1,083 | 4 | 114 |
| YGL197w | 4,461 | 3,974 | 4,092 |
| YML049c | 4,083 | 2,597 | 2,616 |
| YMR224c | 2,076 | 531 | 566 |
| YOL018c | 1,191 | 257 | 324 |
| YOL034w | 3,279 | 620 | 669 |
ORF name, ORF size, and the 5′ ends of identified genes, determined either by sequencing or array hybridization, for 10 clones from the YMR117c screen. For genes sequenced multiple times as different inserts, the end of the most 5′ clone is listed. The 5′ end as detected by array hybridization indicates the most 5′ nucleotide of the most 5′ probe detected as positive. Small disparities between sequencing and hybridization are the result of insert 5′ ends falling in between probes on the array. Although array hybridization does not confirm the inserts are in-frame with respect to the start codon, previous work has shown that frameshifting events generally lead to production of protein regardless of the precise fusion junction between gene insert and transcriptional activation domain (4).
The three loci that were not detected by array hybridization were either not represented on the array or were resistant to PCR amplification. One of the undetected inserts, YLR276c, was difficult to amplify by PCR and could only be sequenced after plasmid rescue. The other two undetected inserts start within 200 bases upstream of the 3′ end of the gene, in regions covered by one or no probes. Therefore, the signal for these genes was not recognized as significant because there was not a consistent pattern of hybridization extending across multiple probes.
The relative abundance of a gene in the output of a plasmid library selection can provide information in addition to gene identity and sequence. For example, in the two-hybrid screen, the identification of independent clones containing different, overlapping inserts from the same gene is an important parameter in evaluating the heuristic value of an interaction (4). In principle, it is possible to selectively label the ends of PCR-amplified inserts before hybridization to arrays, resulting in the identification of the 5′ and 3′ ends of each insert. The presence of numerous independent inserts from the same gene could be detected by hybridization patterns identifying multiple 5′ and 3′ ends. However, this approach requires higher probe density and sequence resolution than is provided by the arrays used here.
To further demonstrate this method, a two-hybrid screen for the gene YMR138w was also carried out and analyzed by array hybridization. YMR138w (CIN4) is a gene in which mutations cause supersensitivity to the anti-microtubule drug benomyl, as well as increased rates of chromosome loss (13). Ymr138w is homologous to the ARF1-class of small GTP-binding proteins, but a distinct role in microtubule function is not yet known. The complete results for this screen are listed in Table 1.
Both two-hybrid screens identified interactors consistent with known results for each gene. The previously detected interaction of Ymr117c with Prp11p splicing factor has suggested that Ymr117c could have a functional connection with the U2snRNP (4). Several of the interactors found in this screen also have known associations with the U2snRNP. For example, Yml049c has previously been found to interact with the Prp9p splicing factor (4). Like CIN4, YPL241c (CIN2) was first isolated as a mutation displaying supersensitivity to anti-microtubule agents (13). Mutations in both CIN2 and CIN4 have already been shown to be epistatic to mutations in CIN1, a gene implicated in the postchaperonin folding of yeast tubulin (14). However, these results are evidence for a physical interaction between CIN2 and CIN4 and suggest that they may act as a complex to regulate specific protein-folding pathways. Further investigations are needed to establish the biological significance of interactions from both screens.
The discovery of thousands of uncharacterized genes by genome sequencing projects has increased the need for methods of large-scale functional analysis (15). Several approaches have been initiated to identify genes that when disrupted or removed lead to selective growth disadvantages (15–17). A promising, complementary approach is the application of established genetic screens to every gene in an organism, in an attempt to assign a biological function to every ORF. Genome-wide analyses based on two-hybrid screens, enhanced synthetic lethal screens, and screens for signal peptide sequences have been proposed (18–20). However, because repetitive dideoxy sequencing is required to exhaustively identify the results of a screen, application of these methods to tens of thousands of genes may be limited by time, labor, and expense.
Two-hybrid screens for protein–protein interactions provide a genetic tool that can be applied, in principle, to every gene in a genome. The Escherichia coli bacteriophage T7 genome has already been characterized with exhaustive two-hybrid screening and sequencing for each known gene. Even with the use of novel strategies for highly efficient two-hybrid screening, however, an analysis of all genes encoded in the human genome would require sequencing of approximately 1 × 106 sequence fragments. As an alternative, genes may be individually cloned into two-hybrid vectors and tested in a pairwise manner. One disadvantage of this approach is that testing only the full-length form of a gene might fail to identify those interactions that occur only with isolated domains of a protein (21). Functional selections that need to be performed in mammalian cells would also benefit from more highly parallel analysis. For example, it is conceivable to select for human genes that yield phenotypes such as increased drug or pathogen resistance when overexpressed in cell lines. The use of array hybridization to analyze results from these screens would eliminate the need to maintain large numbers of individual clones in tissue culture until they can be sequenced.
Oligonucleotide arrays can be synthesized for any organism for which complete or partial sequence information is available. These arrays permit highly parallel identification of the sequence and orientation of nucleic acid elements in a pool. The time to analyze the results of a genetic selection can be drastically reduced, making it feasible to apply conventional screens to very large numbers of genes in a mammalian genome. Analysis of screens by array hybridization is adaptable to any genome-wide functional selection or experiment where the output is a set of nucleic acid sequences.
Acknowledgments
We thank D. Shoemaker, M. Campbell, E. Winzeler, J. Pinto, N. Thayer, and D. Amberg for helpful discussions. This work was supported by a National Institutes of Health Institutional Training Grant in Genome Science, the European Union (Biotech 95-0009, the TAPIR network), and the French Ministere de la Recherche (ACC-SV1).
References
- 1.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 2.Oliver S G. Nature (London) 1996;379:597–600. doi: 10.1038/379597a0. [DOI] [PubMed] [Google Scholar]
- 3.Fields S. Nat Genet. 1997;15:325–327. doi: 10.1038/ng0497-325. [DOI] [PubMed] [Google Scholar]
- 4.Fromont-Racine M, Rain J C, Legrain P. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman C S, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 6.Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 7.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittman M, Wang C, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 8.Wodicka L, Dong H, Mittman M, Lockhart D J. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 9.Fields S, Sternglanz R. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 10.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendelsohn A R, Brent R. Curr Opin Biotech. 1994;5:482–486. doi: 10.1016/0958-1669(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 12.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 13.Stearns T, Hoyt M A, Botstein D. Genetics. 1990;124:251–262. doi: 10.1093/genetics/124.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stearns T. Ph.D. thesis. Massachusetts Institute of Technology; 1988. [Google Scholar]
- 15.Lander E S. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 16.Shoemaker D D, Lashkari D A, Morris D, Mittmann M, Davis R W. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 17.Smith V, Chou K N, Lashkari D, Botstein D, Brown P O. Science. 1996;274:2069–2074. doi: 10.1126/science.274.5295.2069. [DOI] [PubMed] [Google Scholar]
- 18.Klein R D, Gu Q, Goddard A, Rosenthal A. Proc Natl Acad Sci USA. 1996;93:7108–7113. doi: 10.1073/pnas.93.14.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroll E S, Hyland K M, Hieter P, Li J J. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel P L, Roecklein J A, SenGupta D, Fields S. Nat Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- 21.Amberg D C, Basart E, Botstein D. Nat Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]