Abstract
HeT-A was the first transposable element shown to have a bona fide role in chromosome structure, maintenance of telomeres in Drosophila melanogaster. HeT-A has hallmarks of non-long-terminal-repeat (non-LTR) retrotransposable elements but also has several unique features. We have now isolated HeT-A elements from Drosophila yakuba, showing that the retrotransposon mechanism of telomere maintenance predates the separation of D. melanogaster and D. yakuba (5–15 million years ago). HeT-A elements from the two species show significant sequence divergence, yet unusual features seen in HeT-Amel are conserved in HeT-Ayak. In both species, HeT-A elements are found in head-to-tail tandem arrays in telomeric heterochromatin. In both species, nearly half of the HeT-A sequence is noncoding and shows a distinctive imperfect repeat pattern of A-rich segments. Neither element encodes reverse transcriptase. The HeT-Amel promoter appears to be intermediate between the promoters of non-LTR and of LTR retrotransposons. The HeT-Ayak promoter shows similar features. HeT-Amel has a frameshift within the coding region. HeT-Ayak does not require a frameshift but shows conservation of the polypeptide sequence of the frameshifted product of D. melanogaster.
Drosophila telomeres appear to be very different from those of other organisms. (Telomeres are ends of chromosomes and have important roles in chromosome organization.) In Drosophila melanogaster the telomeres are not maintained by telomerase, which maintains telomeres in most animals, plants, and single-celled eukaryotes (1). Instead, Drosophila telomeres are elongated by transposition of two unusual retrotransposons, HeT-A and TART, onto chromosome ends (2). This unusual telomere mechanism offers insights into the requirements for telomere function. It also raises the possibility that transposable elements evolved from normal cellular elements, such as telomeres (3).
Retrotransposon-type telomeres have been reported only for D. melanogaster and the closely related Drosophila simulans. It is of interest to know how many other species share this mechanism of telomere maintenance. This information will be helpful in estimating the antiquity of the mechanism and understanding how it has evolved. Furthermore, comparison of telomere transposon sequences from different species can give insight into the characteristics of these elements that are crucial for their role in telomeres.
Analyses of several HeT-A sequences isolated from D. melanogaster have shown that intact and potentially functional elements can differ markedly in both coding and noncoding regions (4–7). Because HeT-A variation increases rapidly with evolutionary distance, it is difficult to use sequence homology to search for HeT-A elements in other species. We have used the very low level of cross-hybridization between HeT-A from D. melanogaster (HeT-Amel) to DNA from Drosophila yakuba [separated from D. melanogaster by 5–15 million years (My) (8)] to clone HeT-A elements from D. yakuba (HeT-Ayak). The sequence of HeT-Ayak differs significantly from that of HeT-Amel over its entire length. In spite of this extensive sequence divergence, the unusual features of HeT-Amel are also found in HeT-Ayak. The conservation of these features argues that they are important for the role of HeT-A elements in telomeres (see Fig. 1 for diagrams of HeT-A elements).
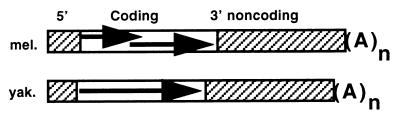
Figure 1.
Diagrams of HeT-Amel and HeT-Ayak element, shown as the sense strand of their RNA transposition intermediates. 5′ and 3′ noncoding regions are striped. Coding regions are marked with arrows, The −1 frameshift in HeT-Amel is indicated by overlapping arrows. (A)n indicates the poly(A) tail on the RNA.
MATERIALS AND METHODS
Drosophila Stocks.
The following stocks were maintained in the laboratory: D. melanogaster, D. simulans, D. mauritiana, D. teissieri, D. yakuba, D. miranda, D. pseudoobscura, D. hydei, and D. virilis. All have been used in this study although not all data are presented.
In Situ Hybridization.
In situ hybridization was performed as described by Pardue and Dawid (9). Hybridization was overnight at 68°C in 2× TNS (TNS = 0.15 M NaCl/0.01 M Tris⋅HCl, pH 6.8).
Southern Blot Hybridization.
DNA was fractionated and analyzed as previously described (10). Low-stringency hybridization was done at 55°C in buffer containing 1 M NaCl, 100 mM Na2HPO4, 7.5 mM EDTA, 1% sarcosyl, 0.1% sodium pyrophosphate, 0.1% polyvinyl pyrrolidone, 0.1% Ficoll, and salmon sperm DNA (50 μg/ml) (D. Nurminsky, personal communication). Stringency was varied by changing conditions of the final wash of the blots. Conditions varied from 55°C in 1× SSC/0.5% SDS to 65°C in 0.1% SSC/0.5% SDS (SSC = 0.15 M NaCl/0.015 M sodium citrate, pH 7.0).
DNA Cloning and Sequencing.
Minilibraries were made from gel-fractionated restriction fragments of D. yakuba DNA cloned in Bluescript SKII (Stratagene). Clones selected from these libraries were subcloned and sequenced by Applied Biosystems Prism Systems (MIT Biopolymers Laboratory). PCR amplification and cloning of products were as described by Pardue et al. (6).
DNA Sequence Analysis.
Analyses were made with the wingenesys programs (Team Associates, Westerville, OH) and programs from the University of Wisconsin Genetics Computer Group (11). Dot plots were made with wingenesys programs using a window of 20, a criterion of 7, and the Unitary cost matrix. Pairwise alignments were made using the Unitary cost matrix. Multiple sequence alignments were made with the multalin program, Version 3.0 (12), using the default parameters.
RESULTS
HeT-Amel Shows Significant Cross-Hybridization Only With DNA From D. melanogaster Sibling Species.
Our earlier studies showed that HeT-Amel cross-hybridized very strongly with DNA from the D. melanogaster sibling species, D. simulans and D. mauritiana (separated by 2–3 My). We have not cloned DNA from either of these two sibling species but we have analyzed PCR-amplified fragments from D. simulans (6). These fragments are as similar to HeT-Amel elements as different HeT-Amel elements are to each other. The sequence analyses (6) and Southern blot hybridization results (Fig. 2) show that, in this group of sibling species (D. melanogaster, D. simulans, and D. mauritiana), the amount of variation between species is nearly equal to the amount of variation within species.
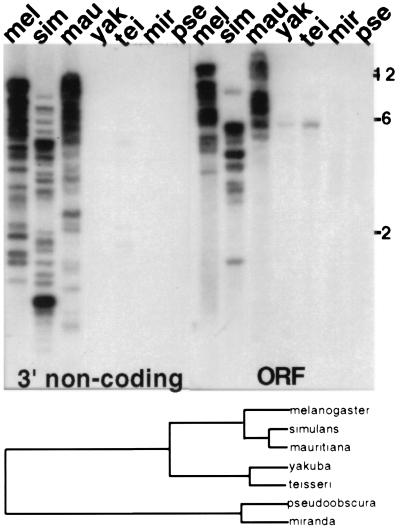
Figure 2.
(Upper) Southern blot showing that HeT-Amel DNA cross-hybridization decreases sharply with evolutionary distance. 32P-labeled sequence from the 3′ noncoding region shows strong hybrdization to DNA from D. melanogaster (Oregon R stock), D. mauritiana, and D. simulans (separation 2–3 My). No hybridization is detected with more distantly related species. Sequence from the coding region (ORF) shows almost the same hybridization as the noncoding region but also reveals faint bands of hybridization to D. yakuba and D. teissieri (separation from D. melanogaster, 5–16 My). DNA was cut with EcoRI. The final wash of blot was 2× SSC at 65°C. Size markers (in kb) are from a DNA ladder. (Lower) Diagram of the evolutionary relationships of species used in this work (largely from ref. 13).
In Southern blot hybridization HeT-Amel cross-hybridization drops dramatically when DNA from more distant species is probed (Fig. 2). Signal from 3′ noncoding region probes is detected only within D. melanogaster sibling species [separation 2–3 My (13)]. HeT-Amel coding sequence probes show a very weak band of hybridization to DNA from either D. yakuba or D. teissieri, two species separated from D. melanogaster by 5–15 My (8, 13). No hybridization is seen with either probe when DNA from D. pseudoobscura or D. miranda is tested. These last two species are separated from D. melanogaster by approximately 46 My (14)).
D. yakuba Has HeT-A Elements With ≈55% Nucleotide Sequence Identity to HeT-Amel.
We began the search for HeT-A elements in D. yakuba by making a minilibrary of D. yakuba DNA from the region of an agarose gel containing fragments hybridizing with the HeT-Amel coding region on Southern blots. That library yielded a cloned 2.2-kb HindIII DNA fragment with significant similarity to HeT-Amel over its entire length (see Fig. 3 for diagrams of cloned sequences). We used this cloned sequence to probe EcoRI-digested D. yakuba DNA. The heaviest hybridization was to fragments of ≈5 kb and a minilibrary of clones was prepared from this region of the gel. A cloned fragment of 5.5 kb isolated from this library completely overlapped the 2.2 kb of sequence in the original clone. The DNA sequence resembled HeT-Amel over the entire fragment, with an overall similarily of ≈55%. When sequence from this D. yakuba clone was used to probe DNA from different Drosophila species, strong hybridization was seen to DNA from D. yakuba and the closely related D. teissieri and very weak hybridization was seen to DNA from the D. melanogaster sibling group (Fig. 4).
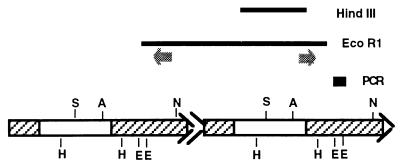
Figure 3.
Diagram of cloned D. yakuba DNA fragments (bars HindIII, EcoRI, and PCR) showing their relation to the deduced organization of the HeT-A elements from which they were derived. The deduced head-to-tail elements are diagrammed below the three cloned fragments. 5′ and 3′ noncoding regions are striped and coding regions are open. Arrowheads at 3′ ends represent oligo(A) sequences. The double arrowhead between the two elements indicates the 25-bp 3′ end at the junction. The two dark arrows below the EcoRI clone indicate the location and orientation of PCR primers used to amplify the segment between the two EcoRI sites. The bar labeled PCR indicates only the new sequence in the PCR fragment and not the sequence that overlaps the two ends of the EcoRI clone. The PCR bar is arbitrarily placed below the right end of the EcoRI clone although it could have come from either end. S, SmaI; A, AlfIII; N, NdeI; H, HindIII; E, EcoRI.
Figure 4.
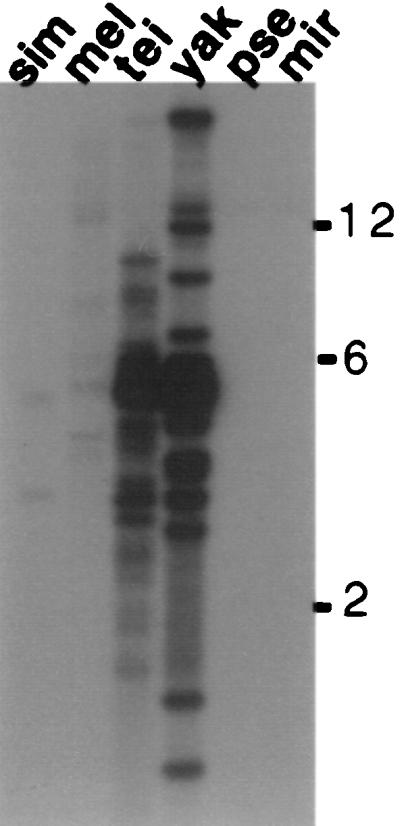
Southern blot showing that HeT-Ayak probes give strong hybridization to DNA from D. yakuba and D. teissieri but very little to DNA from more distantly related species. EcoRI-digested DNA from the species shown in Fig. 2 was probed with 32P-labeled coding sequence from HeT-Ayak. A long exposure is shown because faint hybridization to DNA from D. melanogaster and D. simulans can be detected. The final wash was 1× SSC at 65°C.
HeT-Ayak Elements Are Found in Head-to-Tail Repeats.
The cloned 5.5-kb D. yakuba DNA fragment was circularly permuted so that the 3′ most sequence of HeT-A was at the 5′ end of the fragment (Fig. 3). (For simplicity, the ends of the DNA element are denoted 5′ and 3′ as for HeT-A RNA.) The permuted sequence indicated that the fragment had been cut from a head-to-tail pair of elements with EcoRI sites in their 3′ noncoding regions. EcoRI cleavage had left the 3′ end of the upstream element linked to the 5′ end of the adjacent element, presumably because the fragment had been cut from a tandem array. On Southern blots, EcoRI fragments of about 5.5 kb hybridized heavily with the HeT-Ayak probe, suggesting that these fragments represented monomer subunits cut from a tandem array of several complete elements. If this suggestion is correct, other enzymes cutting once per monomer should also generate hybridizing fragments of 5.5 kb, each enzyme generating monomers of the same size but with permuted sequence. To test this, D. yakuba DNA was cut with AlfIII, NdeI, and SmaI, enzymes that have only one site in the HeT-Ayak sequence. D. yakuba DNA cut with each of these enzymes had a major band of hybridization to HeT-Ayak probes at 5.7 kb, rather than the 5.5 kb seen for EcoRI-cut DNA.
The finding that several single-cutting enzymes produce hybridizing fragments of 5.7 kb is evidence that the cloned fragment is from a chain of tandem repeats of nearly identical HeT-Ayak elements. Because the EcoRI fragment is only 5.5 kb, rather than the 5.7 kb produced by other single cutters, there must be at least one additional EcoRI site in HeT-Ayak. The second site should be about 200 bp from the other site. This second site would produce a 0.2-kb fragment that would have been lost on the Southern blot used. To find the putative small fragment, we used PCR amplification from D. yakuba DNA. The primers (Fig. 3, dark arrows) used were complementary to sequence about 100 bp from each of the ends on the cloned EcoRI fragment so that the amplified product would have about 100 bp of known sequence at either end to verify that amplification had been from the correct DNA. Between the known sequences on the ends we expected to find about 200 bp, representing the postulated second EcoRI fragment. As expected, a fragment of ≈450 bp was obtained from PCR. This fragment was cloned and sequenced. The sequence obtained confirmed that HeT-Ayak has two EcoRI sites separated by 220 bp (Fig. 3, bar labeled PCR).
Because the cloned fragment is defined by EcoRI sites in the 3′ noncoding region, it consists of parts of two different HeT-Ayak elements. The junction between the two elements in the cloned D. yakuba fragment displays a second feature typical of complete HeT-Amel elements. That feature is an extremely short 3′ end (25 bp) of an element lying between what appear to be two full-size elements (Fig. 3, indicated by double arrowheads in tandem elements). We have shown that transcription of HeT-Amel RNA begins at one of two sites (−62 or −31 bp) within the 3′ noncoding region of the element immediately upstream of the transcribed element (15). As a result, a small fragment of sequence from the upstream element is included at the 5′ end of each HeT-Amel RNA molecule. This fragment is added to the chromosome when HeT-A RNA is reverse transcribed, although it may undergo some erosion in length. Thus the remnant of 3′ sequence becomes part of the junction when the next HeT-A element is added to the chromosome. The 25-bp junction fragment in the HeT-Ayak clone argues that transcription of HeT-Ayak RNA resembles that of HeT-Amel, with the promoter located in the 3′ end of the upstream element.
HeT-Ayak Is Localized to Telomere Regions of Polytene Chromosomes.
Polytene chomosomes permit us to map DNA sequences with great precision. The sensitivity of the technique is such that as few as 40 bp of homologous sequence can be detected in euchromatin. In spite of this, no HeT-A hybridization is seen in euchromatic regions. In the D. yakuba stock we have studied, HeT-Ayak probes hybridize strongly to telomeres. (As in D. melanogaster, the amount of hybrid tends to be telomere-specific.) There is also strong hybridization to one part of the chromocenter (Fig. 5). In some nuclei this chromocentral region has been pulled out with chromosome 4. Thus the hybridization appears to be on the telomere of the short arm of this chromosome. This chromocentral hybridization may also include the telomere of the short arm of the X chromosome, the other telomere that is located in the chromocentral heterochromatin. This chromocentral hybridization is not seen in D. melanogaster, suggesting that these species differ in the telomeres of chromosomes X and 4 or in their polytenization in salivary gland nuclei.
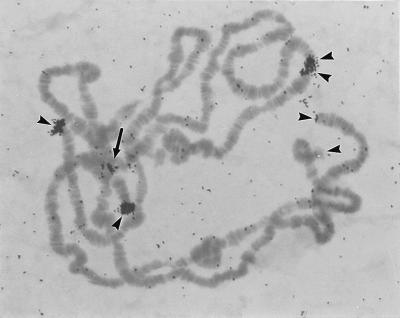
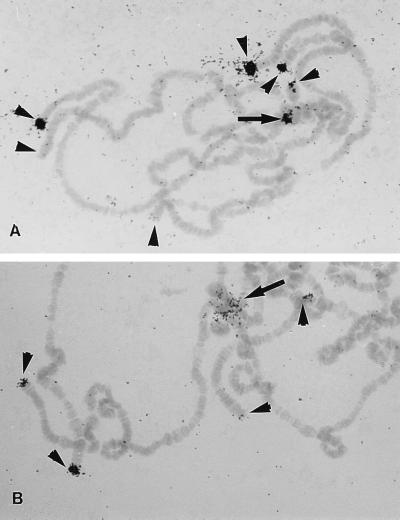
Figure 5.
Autoradiograph of 3H-labeled HeT-Ayak 3′ noncoding probes hybridized with D. yakuba polytene chromosomes. Hybridization is seen on telomeres (arrowheads). The double arrowheads mark ectopically paired telomeres. The hybridization within the chromocenter (arrow) appears to represent the telomere of the short arm of chromosome 4 and perhaps also the short arm of the X chromosome. Probes for other regions of HeT-Ayak sequence show identical patterns of hybridization.
Chromocentral hybridization, similar to that seem in D. yakuba, is also seen in D. simulans. We have analyzed HeT-A localization in D. simulans by using separate probes for the HeT-Amel coding and noncoding regions. In D. simulans both probes hybridize to telomeres and, in addition, bind to DNA within the pericentric heterochromatin that makes up the chromocenter (Fig. 6); however, the two probes do not have exactly the same distribution in the chromocenter. Coding sequence probes hybridize to a discrete cluster of DNA within the chromocentral region in a pattern similar to that seen in D. yakuba (Fig. 6A, arrow). In D. simulans the 3′ noncoding sequence of HeT-Amel also binds within the chromocentral heterochromatin but hybridizes much more generally over this region than does the coding sequence (Fig. 6B, arrow). The difference in the hybridization patterns indicates that the HeT-A noncoding sequence can be found independently of the coding region in some regions of the chromocenter. These regions with only noncoding region hybridization probably are from pericentric heterochromatin. Although HeT-A-related sequences are not seen in pericentric heterochromatin in D. melanogaster, sequences related to the noncoding region of HeT-A are found in the heterochromatic Y chromosome (10). (The Y chromosome is not amplified in polytene nuclei and these Y chromosome sequences are not seen in polytene nuclei.)
Figure 6.
Autoradiographs of 3H-labeled HeT-Amel probes hybridized to D. simulans polytene chromosomes. (A) Coding region probes hybridize to telomeres (arrowheads) and to one spot in the pericentric heterochromatin (arrow). (As with D. melanogaster, there are different levels of hybrid over different chromosome ends. Amounts of hybridizing material tend to be chromosome-specific within a given stock.) The pericentric spot may represent telomeres of the short arms of chromosomes 4 and X. (B) Probes from the 3′ noncoding region hybridize to telomeres and generally over the pericentric heterochromatin, showing that in pericentric regions some 3′ noncoding sequence may exist free of the coding regions.
The HeT-Ayak Coding Region Does Not Require an Internal Frameshift.
One of the distinctive features of HeT-Amel is the −1 frameshift that is required for translation of the entire coding region (4–6). Such frameshifts are typically found between the gag and pol genes of retroelements (16) but HeT-Amel is unusual in that frameshifting has been found within the gag coding region. The overlap gives the coding region the potential of producing two proteins, a nonframeshifted polypeptide of ≈57 kDa and a frameshifted one of ≈110 kDa. [Sizes are only approximate because elements show size polymorphisms (6).] Retroviral gag proteins are polyproteins that must be proteolytically cleaved into several polypeptides to make an infective virus (17). The HeT-Amel frameshift may be an alternative mechanism for deriving two proteins from a single coding region.
The HeT-Ayak coding region does not require a frameshift, yet it produces a polypeptide that has strong similarities to segments of HeT-Amel both before and after its frameshift (i.e., segments in both the 57- and the 110-kDa products). We have sequenced a second cloned fragment of HeT-Ayak. This second element also does not require a frameshift for complete translation.
One of the cloned HeT-Amel elements, 9D4, has a single nucleotide insertion at position 1,144 in the coding region (5). This insertion moves translation into the frame that other HeT-Amel elements achieve by frameshifting. It also eliminates the potential for 9D4 to produce the shorter polypeptide that other HeT-Amel elements can produce. The insertion makes a significant difference in the 9D4 translation product. The 60 amino acids between the 9D4 insertion and the place where other elements must move into the new frame show no significant conservation, only 10% identity with the sequence translated from other HeT-Amel elements and 8% identity with this region of the HeT-Ayak protein. In contrast, frameshifting HeT-Amel elements have 80% amino acid identities in this region and also have 47–55% identities with the HeT-Ayak protein. This sequence conservation is additional evidence that frameshifting takes place in HeT-Amel elements and suggests that 9D4 is an aberrant element.
The HeT-Ayak Coding Region Shares 64% Nucleotide Sequence Identity with the HeT-Amel Coding Region.
The low level of cross-hybridization between the HeT-Amel coding region and DNA from D. yakuba indicates that HeT-A sequences in the two species are significantly diverged. The divergence is confirmed by comparing the sequence of HeT-Ayak with the three HeT-Amel coding regions available. The coding region of HeT-Ayak shows 64–65% nucleotide identity (with 22–26 gaps) to HeT-Amel, depending on the HeT-Amel element used for comparison. It should be noted that the HeT-Amel elements differ among themselves by as much as 16%, yet they show almost identical divergence from the HeT-Ayak elements. Regions of identity are distributed throughout the coding region evenly enough so that a dot matrix comparison at moderate stringency shows a nearly continuous line over the entire coding region (Fig. 7A).
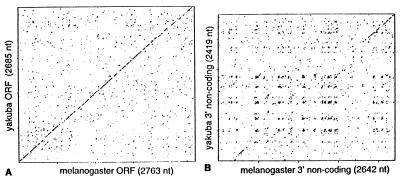
Figure 7.
Dot matrix comparisons of HeT-Ayak with HeT-Amel nucleotide sequences. (A) The coding regions have 64% identity. Identical nucleotides are spread relatively evenly and thus give a nearly continuous diagonal line over the entire region. (B) The 3′ noncoding regions have only 48% identity. This identity is most pronounced in the most 3′ sequences (upper right) and a diagonal line is detected at this location. In most of the comparison, there is not enough sequence identity to yield a diagonal line; however, off-diagonal clusters indicate a pattern of sequence repeats that is conserved.
The amino acid sequence of the HeT-Ayak protein shows no more conservation than does its nucleotide sequence. HeT-Ayak has 57% amino acid sequence identity with each of the frameshifting HeT-Amel elements and 54% identity with the nonframeshifting element 9D4. Nevertheless, the HeT-Ayak protein shows conservation of the motifs that we have noted in HeT-Amel (6). The most distinctive motif in retroelement gag proteins is the zinc knuckle (CCHC box) (18, 19). HeT-Amel belongs to a subgroup of non-long-terminal-repeat (non-LTR) elements with three zinc knuckles of the form (i) CX2CX4HX4C, (ii) CX2CX3HX4C, and (iii) CX2CX3HX6C (6). This region is strongly conserved in HeT-Ayak. There are no deviations in spacing within the knuckles or in the spacing between them. There are few amino acid changes and almost all are conservative. The other motif that has been detected for retroviral gag proteins is the Major Homology Region (20, 21). The motif is found slightly N-terminal of the zinc knuckles and the consensus sequence is QX2EX7R. A highly conserved sequence with this same consensus is found in the appropriate location in the HeT-A proteins. A third conserved region in the HeT-A proteins is a proline-rich region just after the frameshift (8 prolines in 33 amino acids).
The 3′ Noncoding Region of HeT-Ayak Has <50% Nucleotide Sequence Identity With the Same Region of HeT-Amel but Shows a Similar Pattern of Sequence Repeats.
Both HeT-Ayak and HeT-Amel, have large segments of noncoding DNA at the 3′ end. There is less than 50% nucleotide sequence identity in this region, giving an explanation for our failure to detect cross-hybridization with HeT-Amel noncoding probes. Sequence divergence is not distributed evenly along the region. Dot matrix analyses show a diagonal line of similarity over approximately the 3′ most 500 bp of the element (Fig. 7B). This 500 bp contains the promoter for HeT-Amel and possibly also sequences necessary for initiation of reverse transcription. In other parts of the 3′ region the linear identity is not high enough to produce a diagonal line on the plot. It is striking that, despite the obvious sequence divergence, HeT-Ayak has conserved the same pattern of irregular A-rich repeats that distinguishes the 3′ region of HeT-Amel. These repeats can be seen in the pattern of off-diagonal clusters in the dot matrix plot.
DISCUSSION
Insect Telomeres.
In nearly all eukaryotes, chromosome ends are maintained by telomerase, an enzyme that copies an RNA template to add DNA repeats to the end of the chromosome. The known exceptions to this scheme for telomere maintenance have been found among the insects. A limited number of insects have been studied; some appear to have telomerase and some do not. The best studied insect with telomerase is Bombyx mori, shown by Okazaki et al. (22) to have telomere repeats differing by only 1 nucleotide from the repeat on human chromosomes. The B. mori telomere sequence cross-hybridized with DNA from a number of species of Arthropods, although several species in the study showed no cross-hybridization. Species that had no cross-hybridization included the five Diptera and three of the eight Coleoptera studied. Lack of cross-hybridization does not prove that a species lacks telomerase; however, at least two Diptera, D. melanogaster and Chironomous pallidivittatus, are known to have chromosome end sequences that differ significantly from the simple repeats generated by telomerase (2, 23, 24). Thus these species have either lost telomerase or, as we have argued (3, 25), their telomerase has undergone significant evolution. In either case, these species must have shared ancestors with species that now have typical telomerase repeats. Furthermore, comparison of the Drosophila telomeres with those of Chironomous, as well as the phylogenetic distribution of species that did not cross-hybridize to B. mori telomere DNA (22) suggest that modification of the mechanism for telomere maintenance has occurred more than once in insect evolution.
The telomere sequences of Drosophila and Chironomous appear to be quite different from each other. Telomeres in D. melanogaster are composed of the retrotranposons, HeT-A and TART, whereas telomeres in C. pallidivittatus and Chironomous thummi are composed of large complex repeats (23, 24). The mechanism by which the Chironomous repeats are generated is not known. Possibilities suggested by Edstrom and coworkers (24) are gene conversion, unequal recombination of chromosome ends, and RNA-templated DNA synthesis. A study of one telomere of Anopheles gambiae suggests that this species may completely lack telomere-specific repeats (26); however, this conclusion must be accepted with caution because the telomere studied is an atypical one. The chromosome end was produced by accidental insertion of a heterologous P-element that is now maintained in the population by continuous selection for a drug-resistance gene.
Studies of additional insect telomeres are necessary to help us understand the evolution of chromosome ends. The characterization of a D. yakuba telomere-specific element illustrates one of the difficulties encountered in searching for such elements in distantly related species. Sequence divergence between D. melanogaster and D. yakuba allows only minimal cross-hybridization between HeT-A elements in the two species. It seems likely that our failure to detect cross-hybridization with more distantly related species is explained by even greater sequence divergence with increased genetic distance. Finding telomere-specific elements in these species will require greater efforts.
Comparisons of HeT-Ayak and HeT-Amel.
The high level of sequence divergence between HeT-Amel and HeT-Ayak impeded the search for HeT-Ayak; however, in compensation, the divergence strengthens conclusions from analysis of the two elements. Features that are conserved in spite of much sequence change are more likely to be of importance for the element. HeT-Amel differs from typical non-LTR retrotransposable elements in several ways. These features are conserved in HeT-Ayak, bolstering the idea that they are important to HeT-A’s role at the telomere.
HeT-Amel is unusual because it does not encode its own reverse transcriptase. Although this coding sequence could have been lost, failure to detect it in any of the D. melanogaster elements, even those that had transposed very shortly before they were sequenced (5), argues that HeT-Amel acquires reverse transcriptase activity in trans. HeT-Ayak does not encode this enzyme either, showing that the lack of reverse transcriptase coding predates the separation of these two species. We have suggested (3, 25) that HeT-A has evolved from telomerase. Perhaps a gene encoding one of the noncatalytic polypeptide components of telomerase became linked to the gene for telomerase RNA to produce a transcript capable of encoding a protein and also serving as a template for the telomere. If something of this sort produced HeT-A, the element may be reverse-transcribed by the cellular gene for the catalytic subunit of telomerase. The recently published sequences of this subunit from yeast and humans are consistent with this suggestion (27–29) The catalytic subunit of telomerase is related to reverse transcriptase of non-LTR retrotransposons, the class that includes HeT-A.
A second unusual feature of HeT-Amel is the large 3′ noncoding region. HeT-Ayak has a similar noncoding region. Although sequence divergence is even more marked in this region than in the coding regions, both HeT-Amel and HeT-Ayak have distinctive irregular sequence repeats in their 3′ regions. Again, conservation of this feature argues that it is significant. The repeat nature suggests that the sequence has a role in directing chromatin structure, perhaps by specific protein binding. Such a role would be consistent with the chromosomal localizations of HeT-A and HeT-A-related sequences. Intact HeT-A elements are found only in telomere regions, regions identified as heterochromatic by Muller (30). The association of HeT-A with heterochromatin is further supported by a second unusual finding about this element; long segments of the 3′ noncoding region (without associated coding regions and 5′ ends) have been incorporated into families of tandem repeats (HeT-A-related repeats) found at several loci along the length of the heterochromatic Y chromosome (10). Thus the noncoding sequence of HeT-Amel is well represented in two different heterochromatic environments, telomeres and the Y chromosome. This sequence is completely absent in euchromatin.
HeT-Amel has a most unusual promoter. It is located in the 3′ end of the element and directs transcription of its neighbor element immediately downstream (15). Because the 3′ end of the downstream element is a direct repeat of the 3′ end of the element transcribed, it is structurally and functionally reminiscent of the promoters of LTR retroelements. Transcription starts within this promoter and thus adds a few nucleotides of 3′ sequence to the 5′ end of the RNA transcript. The 25 nt of 3′ sequence at the 5′ end of the cloned HeT-Ayak are evidence that transcription of HeT-Ayak also starts in the 3′ end of the upstream element. We predict that the HeT-Ayak promoter will also be in the upstream element.
HeT-Amel is unusual in having a frameshift within the gag coding region rather than between the gag and pol coding regions. Surprisingly, HeT-Ayak has no frameshift; nevertheless its amino acid sequence is similar to the frameshifted product of HeT-Amel. The frameshift gives HeT-Amel the ability to product two polypeptides, one of ∼57 kDa and the other of ∼110 kDa (31). Our preliminary antibody studies suggest that both polypeptides are made; an antibody raised against the N-terminal end of the two polypeptides (the shared amino acids) recognizes Drosophila proteins of sizes appropriate for both the frameshifted and the nonframeshifted products (O.N.D. and M.-L.P., unpublished results). HeT-Ayak should give only the larger polypeptide as a primary translation product. However, the HeT-Ayak translation product might be proteolytically cleaved to yield smaller polypeptides. Such cleavage might be analogous to the cleavage of gag proteins during retroviral maturation (17). It is interesting that one of the proteins associated with telomerase, the protein encoded by the Saccharomyces cerevisiae est-3 gene, requires a frameshift for proper translation. This frameshift appears to be dispensable; a coding region engineered to give the same polypeptide can substitute for the original gene (32). The involvement of frameshifting in HeT-A and est-3 translation is especially intriguing because both are involved in telomere extension. The precise roles of the two proteins are not yet defined but they may well be similar.
This study has demonstrated that the low level of hybridization of HeT-Amel to D. yakuba DNA is explained by sequence divergence between HeT-Amel and HeT-Ayak rather than by lack of HeT-A elements in D. yakuba DNA. The extensive sequence divergence appears to be completely compatible with retention of features distinguishing HeT-A from other transposable elements. Extrapolation from these results suggests that more distantly related species also have HeT-A elements but that sequence divergence has made those elements even more diffficult to detect by cross-hybridization. Comparison of HeT-Ayak and HeT-Amel also suggests an approach to increase the power of searches for HeT-A in more distantly related species. The coding region of HeT-Ayak is equally diverged from each of the HeT-Amel coding regions studied, despite the fact that these HeT-Amel sequences differ from each other by as much as 16%. Because cross-hybridizing sequences in different HeT-Amel elements are not completely overlapping, a probe containing all HeT-Amel coding regions would detect HeT-Ayak more effectively than a probe made from any one of those coding sequences. This suggests that adding HeT-Ayak sequences to our HeT-Amel probe will increase the possibility of finding HeT-A in other species. Preliminary results (O.N.D., K. Haynes, and M.-L.P., unpublished results) with this mixed probe show cross-hybridization to Southern blots of DNA from all Drosophila species examined. Proof that the hybrids are HeT-A elements will require cloning and characterization of this DNA.
Acknowledgments
We thank P. G. DeBaryshe, K. Lowenhaupt, T. Orr-Weaver, and K. L. Traverse for comments on the manuscript. This work has been supported by a grant from the National Institutes of Health (GM 50315).
ABBREVIATION
- LTR
long terminal repeat
Footnotes
References
- 1.Henderson E. In: Telomeres. Greider C, Blackburn E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 11–34. [Google Scholar]
- 2.Pardue M-L. In: Telomeres. Greider C, Blackburn E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 339–370. [Google Scholar]
- 3.Pardue M-L, Danilevskaya O N, Lowenhaupt K, Slot F, Traverse K L. Trends Genet. 1996;12:48–52. doi: 10.1016/0168-9525(96)81399-0. [DOI] [PubMed] [Google Scholar]
- 4.Danilevskaya O N, Petrov D A, Pavlova M A, Koga A, Kurenova E V, Hartl D L. Chromosoma. 1992;102:32–40. doi: 10.1007/BF00352288. [DOI] [PubMed] [Google Scholar]
- 5.Biessmann H, Kasravi B, Bui T, Fujiwara G, Champion L E, Mason J M. Chromosoma. 1994;103:90–98. doi: 10.1007/BF00352317. [DOI] [PubMed] [Google Scholar]
- 6.Pardue M-L, Danilevskaya O N, Lowenhaupt K, Wong J, Erby K. J Mol Evol. 1996;43:572–583. doi: 10.1007/BF02202105. [DOI] [PubMed] [Google Scholar]
- 7.Danilevskaya O N, Lowenhaupt K, Pardue M-L. Genetics. 1998;148:233–242. doi: 10.1093/genetics/148.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lachaise D L, Cariou M-L, David J R, Lemueunier F, Tsacas L, Ashburner M. Evol Biol. 1988;22:159–225. [Google Scholar]
- 9.Pardue M L, Dawid I B. Chromosoma. 1981;83:29–43. doi: 10.1007/BF00286014. [DOI] [PubMed] [Google Scholar]
- 10.Danilevskaya O, Lofsky A, Kurenova E V, Pardue M-L. Genetics. 1993;134:531–543. doi: 10.1093/genetics/134.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powell J R. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford: Oxford Univ. Press; 1997. pp. 267–298. [Google Scholar]
- 14.Beverley S M, Wilson A C. J Mol Evol. 1985;21:1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- 15.Danilevskaya O N, Arkhipova I R, Traverse K L, Pardue M-L. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 16.Jacks T. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- 17.Wills J W, Craven R C. AIDS. 1992;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Covey S N. Nucleic Acids Res. 1986;14:623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summers M F, South T I, Kim B, Hare D R. Biochemistry. 1990;29:329–340. doi: 10.1021/bi00454a005. [DOI] [PubMed] [Google Scholar]
- 20.Mammano F, Ohagen A, Hoglund S, Gottlinger H G. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craven R C, Leure-duPree A E, Weldon R A, Jr, Wills J W. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okazaki S, Tsuchida K, Maekawa H, Ishikawa H, Fujiwara H. Mol Cell Biol. 1993;13:1424–1432. doi: 10.1128/mcb.13.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y J, Kamnert I, Lopez C C, Cohn M, Edstrom J-E. Mol Cell Biol. 1994;14:8028–8036. doi: 10.1128/mcb.14.12.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez C C, Nielsen L, Edstrom J-E. Mol Cell Biol. 1996;16:3285–3290. doi: 10.1128/mcb.16.7.3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pardue M-L, Danilevskaya O N, Traverse K L, Lowenhaupt K. Genetica. 1997;100:73–84. [PubMed] [Google Scholar]
- 26.Biessmann H, Donath J, Walter M. Insect Mol Biol. 1996;5:11–20. doi: 10.1111/j.1365-2583.1996.tb00035.x. [DOI] [PubMed] [Google Scholar]
- 27.Counter C M, Meyerson M, Eaton E N, Weinberg R A. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ligner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Ligner J, Harley C B, Cech T R. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 30.Muller H J. Collect Net. 1938;13:181–195. [Google Scholar]
- 31.Danilevskaya O, Slot F, Pavlova M, Pardue M L. Chromosoma. 1994;103:215–224. doi: 10.1007/BF00368015. [DOI] [PubMed] [Google Scholar]
- 32.Morris D K, Lundblad V. Curr Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]