Abstract
We describe here the identification of eight polymorphic microsatellite loci with (CA)n repeats in the Trypanosoma cruzi genome based on the affinity capture of fragments using biotinylated (CA)12 attached to streptavidin-coated magnetic beads. The presence of two peaks in PCR amplification products from individual clones confirmed that T. cruzi is diploid. Hardy–Weinberg and linkage disequilibrium analyses suggested that sexual reproduction is rare or absent and that the population structure is clonal. Several strains, especially those isolated from nonhuman sources, showed more than two alleles in many loci demonstrating that they were multiclonal. The phylogenetic analysis of T. cruzi based on microsatellites revealed a great genetic distance among strains, although the strain dispersion profile in the Wagner network was in general agreement with the species dimorphism found by PCR amplification of the divergent region of the rRNA 24Sα gene.
The protozoan Trypanosoma cruzi is the causative agent of Chagas’ disease, which is estimated to affect 18 million people in Latin America (1). The clinical features of the chronic phase of the disease are quite variable, taking an asymptomatic indeterminate course or evolving as chronic cardiomyopathy and/or gastrointestinal pathology. It is believed that such clinical pleotropism is the product of a complex interaction of environmental and genetic factors from both host and parasite (reviewed in ref. 2). Thus, studies of the genetic structure of natural populations of T. cruzi will be important for a better understanding of the pathogenesis of Chagas’ disease.
Characterization of natural populations of T. cruzi stocks, initially performed using a small number of isoenzyme markers, suggested a classification into three distinct groups that were called zymodemes (3, 4). Afterward, more extensive studies with 15 enzyme markers in T. cruzi clones from the whole ecogeographical range of the parasite and from various hosts increased significantly the number of natural isoenzyme strains or “clonets” (5). Although the isoenzyme studies provided convincing evidence that T. cruzi is diploid, there were significant deviations from Hardy–Weinberg expectations and strong linkage disequilibrium, suggesting that sexual reproduction is rare or absent and that the population structure is that of independently evolving clonal lineages (reviewed in ref. 6 and 7).
The advent of DNA studies based on multilocal markers such as DNA fingerprinting (8), random-amplified polymorphic DNA (9, 10), simple sequence repeat-PCR (11), and molecular karyotyping (12) showed a much greater level of intraspecific genetic variability of T. cruzi than isoenzymes, suggesting an absolute genetic individuality of clonal lineages. More recently, single locus analysis of the ribosomal RNA genes (LSU-rDNA) and mini-exon genes (13, 14) has suggested a division of T. cruzi strains into two major clusters, which probably represent subspecies (14).
The study of microsatellite markers has had an revolutionary impact in human molecular genetics (see, for instance, ref. 15). We reasoned that examination of microsatellite markers from the T. cruzi genome should likewise provide genetic information about the parasite. We report here the isolation of eight (CA)n polymorphic microsatellites from T. cruzi by using a magnetic capture affinity technique. Analysis of strains and clones of T. cruzi with these microsatellites provided valuable new information about the population structure of the parasite.
MATERIALS AND METHODS
Parasites and Nucleic Acids Isolation.
The strains and clones of T. cruzi examined are listed in Table 1. Epimastigote forms were grown in liver infusion tryptose medium. The parasites were collected and stored −70°C until standard DNA extraction with phenol/chloroform was performed (8).
Table 1.
List of species and strains used
| Species | Strain | Source | Geographical origin | rRNA typing |
|---|---|---|---|---|
| T. cruzi | 1001 | D. marsupialis | MG, Brazil | 2 |
| T. cruzi | Gambá 3 | D. marsupialis | MG, Brazil | 1 |
| T. cruzi | 1009 | P. megistus | MG, Brazil | 2 |
| T. cruzi | 1014 | P. megistus | MG, Brazil | 1 |
| T. cruzi | 1005 | T. infestans | MG, Brazil | 1 |
| T. cruzi | 1023 | T. infestans | MG, Brazil | 1/2 |
| T. cruzi | 1030 | T. infestans | MG, Brazil | 1/2 |
| T. cruzi | CL Brener | T. infestans | RGS, Brazil | 1 |
| T. cruzi | 1017 | Human | Brazil | 2 |
| T. cruzi | JM | Human | MG, Brazil | 1 |
| T. cruzi | Bas | Human | MG, Brazil | 1 |
| T. cruzi | Y | Human | SP, Brazil | 1 |
| T. cruzi | Y P1 clone | Human | SP, Brazil | 1 |
| T. cruzi | Y P2 clone | Human | SP, Brazil | 1 |
| T. cruzi | Y P3 clone | Human | SP, Brazil | 1/2 |
| T. cruzi | 138 | Human | MG, Brazil | 1 |
| T. cruzi | 222 | Human | MG, Brazil | 2 |
| T. cruzi | 226 | Human | MG, Brazil | 1/2 |
| T. cruzi | LPS | Human | MG, Brazil | 1 |
| T. cruzi | 115 | Human | MG, Brazil | 1/2 |
| T. cruzi | 167 | Human | MG, Brazil | 1 |
| T. cruzi | 229 C1 clone | Human | MG, Brazil | 1 |
| T. cruzi | 239 | Human | MG, Brazil | 1 |
| T. cruzi | Colombiana | Human | Colombia | 2 |
| T. cruzi | DL | Human | MG, Brazil | 1 |
| T. cruzi | JG | Human | SP, Brazil | 1 |
| T. cruzi | NHR | Human | MG, Brazil | 1 |
| T. cruzi | 182 | Human | MG, Brazil | 1 |
| T. cruzi | 182 A3 clone | Human | MG, Brazil | nd |
| T. cruzi | 231 | Human | MG, Brazil | 2 |
nd, not determined; Didelphis marsupialis, D. marsupialis; Panstrongylus megistus, P. megistus; Triatoma infestans, T. infestans. MG, Minas Gerais; RGS, Rio Grande do Sul; SP, São Paulo.
Enrichment for (CA)n Repeats Containing Fragments.
The protocol followed in this paper has been described (16). The clone library was constructed with genomic DNA from the CL Brener clone (17). Genomic DNA was digested with MseI or Sau3AI, and the fragments (500 ng) were ligated with 50 pmols of the corresponding adapters at 14°C overnight with 40 units of T4 DNA ligase in a 20 μl of reaction volume. An MseI adapter was formed with MseI-20 (TCTCCAGCCTCTCACCGCAT) and MseI-11 (TAATGCGGTGA), and Sau3AI adapter was formed with Sau3AI-24 (CGGGAATTCTGGCTCTGCGACATG) and a Sau3AI-10 (GATCCATGTC). After the ligation, the 3′ ends of the fragments were repaired with 0.5 unit of Taq polymerase and 100 μM dNTPs for 10 min at 72°C. To capture the fragments containing (CA)n repeats, 100 ng of the ligation product was mixed with 20 pmols of the biotinylated oligonucleotide GATGATCCGACGCAT(CA)12 in 50 μl of TE buffer (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA, pH 8.0) containing 2 μM of the MseI-20 or Sau3AI-24 to avoid hairpin formation. Mineral oil was added, and the mixture was heated to 95°C for 10 min and then annealed at 60°C for 1 min. Forty-five microliters were collected, added to 100 μg of the prewashed streptavidin-coated magnetic beads M-280 (Dynal, Oslo), and resuspended in 500 μl of TE/1 M NaCl. After a 30 min incubation with gentle rotation at room temperature, the beads were collected by magnet, washed three times with 3X SSC/0.5% SDS at 60°C for 15 min, once each with TE/1 M NaCl at room temperature, and resuspened in 50 μl of TE. To obtain double-stranded fragments, the captured DNA was PCR-amplified in 50 μl containing 67 mM Tris⋅HCl (pH 8.8), 4 mM MgCl2, 16 mM (NH4)2SO4, 10 mM 2-mercaptoethanol, 300 μM of each dNTPs, 2 units of Taq polymerase, and 10 μM oligonucleotide MseI-20 or Sau3AI-24. The samples were incubated at 94°C for 3 min and subjected to 23 cycles consisting of 1 min at 94°C for denaturation and 3 min at 72°C for annealing and extension and then incubated for a final extension for 5 min at 72°C.
Characterization of the Fragments Enriched for (CA)n Repeats.
PCR products before and after the affinity capturing of (CA)n repeats were eletrophoresed in 1.5% agarose gel and transferred to nylon filter Biodyne B (GIBCO/BRL). Approximately equal amounts of DNA (100 ng) were applied per lane. The filter was hybridized with alkaline phosphatase-conjugated (CA)n probe following the directions of the manufacturer (Lifecodes, Stamford, CT). After developing the x-ray film, the degree of enrichment for (CA)n repeats was analyzed on a Macintosh computer using the public domain NIH image program, version 1.52 (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image).
Construction of Genomic Library Enriched for (CA)n Repeat.
The PCR fragments enriched for (CA)n were cloned into the pCRII vector using the TA cloning kit (Invitrogen). The recombinant colonies were screened with the alkaline phosphatase-conjugated (CA)n probe. DNA plasmids from 25 colonies were isolated and sequenced using the AutoRead kit and ALF sequencer (Pharmacia LKB). Eight clones containing microsatellite longer than eight repeat units were selected for PCR analysis.
PCR Amplification and Detection of Microsatellite Alleles.
Primers for each microsatellite locus were designed, and one oligonucleotide of each pair was 5′-labeled with fluorescein. Table 2 shows the primer sequences for each marker. After PCR, the amplified microsatellites were loaded on a 6% denaturing polyacrylamide gel and analyzed on ALF sequencer using the Fragment Manager software (Pharmacia). To determine the allele size, the samples were compared directly with band sizes from an allelic ladder prepared by amplification of an artificial mixture of DNA from 60 T. cruzi strains.
Table 2.
Microsatellites markers isolated from T. cruzi
| Marker | Size range alleles, bp | Repeat sequence | PCR primers 5′ → 3′ | Number of alleles found | Observed heterozygosity | Expected heterozygosity |
|---|---|---|---|---|---|---|
| MCLE01 | 110–150 | (CA)9 | 1 CTGCCATGTTTGATCCCT 2 CGTGTACATATCGGCAGTG | 11 | 0.60 | 0.84 |
| MCLE08 | 124–130 | (CA)2AA(CA)12 | 1 ATGGACAACAAATGGGAG 2 TGGGTATGCCAAATGTGAT | 4 | 0.20 | 0.71 |
| SCLE10 | 237–291 | (GT)2(TG)10 | 1 GATCCCGCAATAGGAAAC 2 GTGCATGTTCCATGGCTT | 12 | 0.80 | 0.87 |
| SCLE11 | 139–157 | (AC)9 | 1 ACGACCAAAGCCATCATT 2 GATGCTAACTGCTCAAGTGA | 9 | 0.60 | 0.83 |
| MCLF10 | 182–194 | (CA)2A(CA)14 | 1 GCGTAGCGATTCATTTCC 2 ATCCGCTACCACTATCCAC | 5 | 0.35 | 0.53 |
| MCLG10 | 151–187 | (CA)8 | 1 AGGAGTCAAATATAATGAGGCA 2 ACGTGTGAAAGGCATCTATC | 7 | 0.20 | 0.69 |
| MCL03 | 257–319 | (CT)4(GT)2CT AT(GT)15 | 1 GGAGCAAGAATGAAGGCA 2 TCAGAAAAAGCACGCCTC | 13 | 0.65 | 0.91 |
| MCL05 | 194–228 | (TC)9(GT)4 | 1 TTAAACGACCTCTATGTCTCTC 2 CCTGAGCAAGATACAAGGAC | 12 | 0.55 | 0.90 |
Construction of a Phylogenetic Tree.
To make phylogenetic inferences, we assumed a stepwise mutation model for the microsatellites and used as a measure of genetic distance between any two strains the minimum number of mutational steps necessary to transform one into the other. The microsatellite multilocus genotypes were transformed into 0 and 1 characters by using factor program from phylip package version 3.57c (18). These data were collected and used next in the mix program also from phylip with which the evolutionary relationships were estimated based in the Wagner parsimony method. The significance of the branching in the Wagner network was done by bootstrapping (19). We used the program seqboot of the phylip package to perform 1,000 bootstraps. The number of times that a given branching was observed was used to evaluate the robustness of the tree.
RESULTS
Preparation of an Enriched Library and Characterization of (CA)n Repeats from T. cruzi Genome.
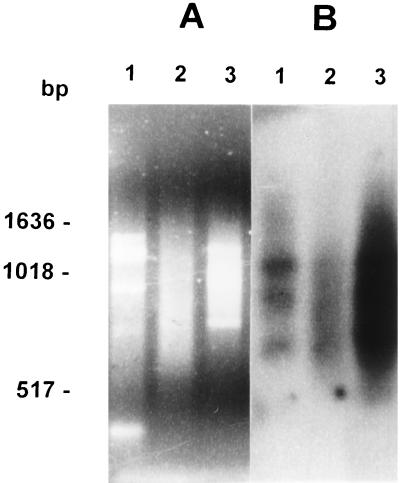
To obtain a library enriched in (CA)n repeats, we employed affinity capture of genomic restriction fragments by hybridizing them with a biotinylated (CA)12 oligonucleotide and binding the complexes to streptavidin-coated magnetic beads. This resulted in a 10- to 15-fold enrichment of (CA)n containing fragments, as revealed by Southern blotting (Fig. 1B). A clone library of these fragments was then constructed, from which we randomly selected 44 recombinant colonies. After hybridization with the (CA)n probe, 39 colonies were positive and 25 of them were randomly selected and sequenced. Seventeen colonies (68%) showed CA-repeat lengths greater than six, and from these, we chose 12 for which we designed specific primer pairs. A single set of PCR cycling parameters was chosen to facilitate the amplification of several markers simultaneously. We have been successful in amplifying 10 of the microsatellite markers, and eight of the markers that showed size polymorphism (Table 2) were studied further.
Figure 1.
Evaluation of enrichment for (CA)n-containing fragments after affinity capture. We show an ethidium bromide-stained agarose gel (A) and a Southen blot hybridized with an alkaline phosphatase-labeled (CA)8 probe (B). Lanes: 1, T. cruzi DNA digested with Sau3AI; 2, T. cruzi DNA digested and ligated with Sau3AI adapters; and 3, T. cruzi DNA digested with Sau3AI with adapters after capturing the (CA)n-containing fragments. Molecular size markers were from a 1-Kb ladder (Life Technologies, Gaithersburg, MD) are indicated.
Polymorphism of CA Repeats in T. cruzi.
To test the variability of these eight microsatellite markers, 30 different T. cruzi strains and clones were typed (Table 1). An important distinction to be made here is between strains and clones. Strains or isolates are obtained from triatomid insect vectors or mammalian hosts of T. cruzi, including humans. Patients in endemic areas may be infected by multiple contacts with different triatomids, and these triatomids, in turn, may feed on different infected individuals. This promiscuity propitiates the formation of multiclonal populations in hosts and vectors, leading to the isolation of correspondingly multiclonal strains when grown in culture. Molecular studies had confirmed that some T. cruzi strains were multiclonal (8, 20), but the general prevalence of multiclonality was not known.
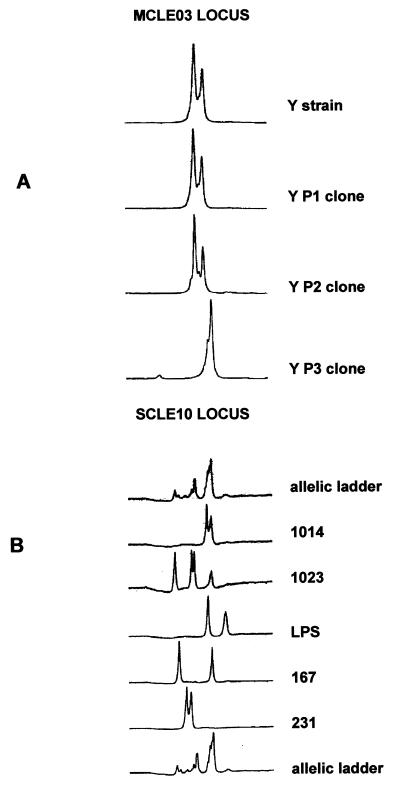
To establish the ploidy of T. cruzi, we initially typed six different clones by PCR with primers for all microsatellite loci. All of the clones showed amplification of one or two fragments of different size (Fig. 2A). We interpreted this result as an indication that T. cruzi is diploid, the two-peak patterns indicating heterozygosity at the respective loci and the one-peak patterns denoting homozygosity. We next typed 24 different strains at all the eight loci. Again, we observed amplification of one- or two-fragment peaks for most strains. However, six strains (Gambá 3, 1009, 1023, 1030, 1017, and JM) consistently showed amplification of three or four major fragments of different size, implicating that these strains are multiclonal (Fig. 2B). It was interesting to observe that in the initial experiments, none of the strains showed more than two alleles isolated from chronic patients (data not shown). To further verify this point, we examined another 25 different strains isolated from chronic patients with one single microsatellite (MCLE01). Two strains (520/5 and JJS) showed four and three peaks respectively, showing that multiclonality also can be observed in chronic patients. However, when data from the 24 strains analyzed with all eight loci summarized, there seemed to be an important difference in the prevalence of multiclonality from strains obtained from the insects, wild animal, or acute patients (6/11) and chronic patients (0/13).
Figure 2.
(A) Tracing of electrophoresis of alleles of the MCLE03 microsatellite locus of the Y strain and three of its clones. (B) Tracing of electrophoresis of alleles of the SCLE10 locus of different T. cruzi strains. Strain 1023 shows four different alleles, indicating multiclonality.
Tests for Hardy–Weinberg Proportions and Linkage Disequilibrium.
Phenotypes for the eight microsatellite repeats were established for 20 of the T. cruzi strains or clones (excluded from this analysis were the strains with more than two bands, the clones from the Y strain—YP1, YP2, YP3, and the clone from the 182 strain), and genotypes were inferred. The observed heterozygosity of the microsatellite markers for this set of strains ranged from 0.20 to 0.80 (Table 2). Statistical tests showed highly significant departures from Hardy–Weinberg proportions (χ2 values ranged from 25.3 to 118.3; all were significant at P < 0.001). Indeed, as can be seen in Table 2, expected heterozygosities (H = 1-Σxi2, where xi is the frequency of the ith allele) calculated assuming Hardy–Weinberg were very different from the observed ones. Statistical tests for all of the 28 pairwise comparisons possible for all of the eight microsatellite loci showed that all of these loci are associated in very strong linkage disequilibrium (P < 0.001).
Phylogenetic Relationships.
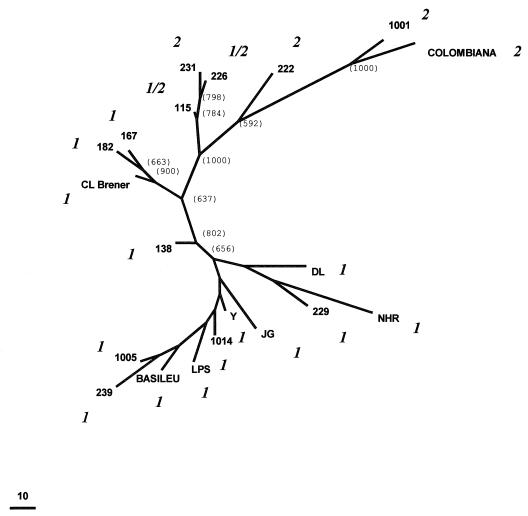
T. cruzi multilocus genotypes were constructed based on the eight microsatellites repeats studied. Each strain produced a highly individual genotype. We utilized these genotypes to investigate the phylogenetic relationships and T. cruzi population structure by using the maximal parsimony approach. It is believed that microsatellites evolve by DNA strand slippage, with most mutations involving the gain or loss of only a single repeat unit (21). Considering each microsatellite allele as one state from a multistate character and assuming that the most frequent allele was the ancestral state, we can establish a genetic distance measure between any two strains as being the number of mutational steps necessary to transform one into the other. We then used the Wagner parsimony algorithm from the mix computer program of the phylip package phylogenetic tree (18) to obtain one most parsimonious unrooted Wagner network that is shown in Fig. 3, where the figures in parenthesis indicate the number of times that the branch was observed in 1,000 bootstraps. The minimal distance between two T. cruzi strains was 17 mutational events, and the overall dispersion pattern based on microsatellite genotypes shows that the T. cruzi strains are very distant from each other. This is especially significant because most strains were from the same geographic area. The only significant clusters observed were one involving the 115, 226, and 231 strains, another with 167, 182, and CL Brener, and a third one with 1001 and Colombiana. These same strains also formed clusters when studied with other approaches used to characterize genetic variability of anonymous DNA sequences in T. cruzi strains (11). On the other hand, we could observe a clear separation of strains based on rRNA typing (14). Strains classified as belonging to group 1 were found at one extremity of the network whereas strains belonging to groups 1/2 and 2 were found at the other end (Fig. 3). This clear separation was observed in 100% of the bootstraps.
Figure 3.
Unrooted Wagner network of 20 T. cruzi strains based on the amplification of eight polymorphic loci with (CA)n repeats. Considering each microsatellite allele as one state from a multistate character, the genetic distance between any two strains was estimated as the number of mutational steps necessary to transform one into the other. The figures in parenthesis indicate the number of times that the branch was observed in 1,000 bootstraps. Bootstrap values inside the lower cluster (below strain 138) were all low (below 300) and are not given individually. The classification of each strain indicated in italics (1, 2, or 1/2) was based on the rRNA 24Sα amplification.
DISCUSSION
Our studies with CA-repeat microsatellites provide several insights in the genetic structure of T. cruzi populations. First, the fact that all clones showed patterns of one or two microsatellite bands with different loci gives strong support to the notion that T. cruzi is diploid in most, if not all, of its genome. Most of the previous information supporting the diploidy was derived from isoenzyme studies or on hybridization on restricted DNA or chromosomal bands separated by pulse-field gel electrophoresis (5, 12, 22, 23).
Second, our microsatellite studies showed drastic departures from Hardy–Weinberg expectations and strong linkage disequilibrium, suggesting that sexual reproduction is rare or absent in T. cruzi and that the population structure is clonal, as had been suggested on the basis of isoenzyme studies (5, 6, 7). Thus, each clone represents a lineage that reproduces by binary division and remains unaltered for a large number of generations until mutations occur.
Third, we observed several strains, especially those obtained from insects, wild animals, and acute patients with more than two bands in a single microsatellite. This observation is best compatible with a multiclonal composition of these strains. Previously, a few studies already had demonstrated that clones obtained from some strains could display different restriction fragment length polymorphism patterns of kinetoplast DNA minicircles (20) or were heterogeneous in DNA fingerprinting (8). However, our microsatellite studies provide the first direct demonstration of frequent multiclonality of strains, which indeed was to be expected on theoretical grounds. We also have to consider that we are probably only observing the tip of the multiclonality iceberg because growth in culture may entail the loss of some clones incapable of adapting to the new in vitro environment. Actually, we also should expect that the infection of the human host also might entail reduction of multiclonality. T. cruzi has existed as a species for ten or hundreds of millions of years but has only met the human host after the colonization of the American continent 10,000–30,000 years ago (24). Thus, it is quite conceivable that some clonal lineages found in wild animals and insect hosts are incapable of infecting humans. Wild strains of T. cruzi presumably represent swarms of clones that may present symbiotic relationships but also certainly compete fiercely for available resources. The patterns of interactions within the swarm may change drastically in different environments, especially in the passage of the multiclonal population from the invertebrate vector to man. One would expect that some clones would be eliminated by their inability to compete and propagate in the human environment or by the action of the host defense mechanisms, decreasing the complexity of the infecting strain. Thus, it is not surprising that we found that the prevalence of multiclonality was much higher in strains obtained from insects, wild animals, or patients with acute disease than from chronic patients. Studies with microsatellites also provide a very useful means to test whether a given strain contains one or several clones. If a strain consistently displays one or two bands on amplification with a battery of several microsatellites, as we have seen for several of the strains that we studied, monoclonality may be safely assumed. This is particularly relevant in some experimental studies because occasionally some strains with interesting biological properties are incapable of being cloned in culture. Furthermore, these microsatellites markers may have a major impact in the understanding of the pathogenesis of the Chagas’ disease through the genetic typing of T. cruzi present directly in the host tissue.
Fourth, with the polymorphic CA-repeat microsatellites, we found a remarkable genetic diversity in T. cruzi. Furthermore, no other previous method of genetic analysis illustrated so cogently this genetic individuality in T. cruzi because no single repeated multilocus genotype was detected among the strains analyzed. Indeed, with the study of 15 isoenzymes, identical phenotypes were observed in geographically unrelated clones (5). When we used parsimony to display the genetic relationship among the clonal lineages in an unrooted Wagner network (Fig. 3), we did not observe any geographical clustering of the clones and strains and the minimal genetic distance separating them was 17 mutational steps. However, when we typed the clones and strains according to a ribosomal DNA restriction fragment length polymorphism (14), there was an excellent correlation of types 1 and 2 with the tree topography (Fig. 3). The only two strains that belonged to 1/2 (115 e 226) clustered with group 2, suggesting that conceivably the latter may be the ancestral state.
In summary, we show in this article that microsatellite polymorphisms typed by PCR amplification represent a powerful tool for the understanding of the population structure and dynamics of T. cruzi. The simple and fast strategy used here is applicable to broad range of parasites permitting the discovery of CA repeats and other microsatellites. This strategy will be fundamental for physical mapping in the parasites included in the Parasite Genome Initiative supported by the World Health Organization (25). We have provided evidence that T. cruzi is diploid, that it has a genetically extremely diverse clonal population structure, and that clones associate to form multiclonal strains. Indeed, microsatellite typing is a direct and useful means of testing the monoclonality of a strain. Our next goal will be to achieve the genetic typing of single parasites isolated by cell sorters, dispensing cloning, and permitting considerable refinement of T. cruzi population studies.
Acknowledgments
This work was supported by Conselho Nacional de Pesquisas (Brazil), Comissão de Aperfeiçoamento de Pessoal de Nível Superior, and Programa de Núcleos de Excelencia. We thank Prof. Egler Chiari from the Department Parasitology of the Universidade Federal de Minas Gerais for providing the strains and clones of T.cruzi used in this study. We are grateful to Anna Izabel R. Melo for helping in the microsatellite typing.
Footnotes
A commentary on this article begins on page 3346.
References
- 1.World Health Organization. WHO Tech. Rep. Series 811. Geneva, Switzerland: WHO; 1991. [Google Scholar]
- 2.Macedo A M, Pena S D J. Parasitol Today. 1998;14:119–124. doi: 10.1016/s0169-4758(97)01179-4. [DOI] [PubMed] [Google Scholar]
- 3.Miles M A, Souza A, Povoa M, Shaw J J, Lainson R, Toye P J. Nature (London) 1978;272:819–821. doi: 10.1038/272819a0. [DOI] [PubMed] [Google Scholar]
- 4.Miles M A, Lanham S M, de Souza A A, Póvoa M. Trans R Soc Trop Med Hyg. 1980;74:221–237. doi: 10.1016/0035-9203(80)90251-5. [DOI] [PubMed] [Google Scholar]
- 5.Tibayrenc M, Ward Pl, Moya A, Ayala F J. Proc Natl Acad Sci USA. 1986;83:293–296. doi: 10.1073/pnas.83.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayala F J. Biol Res. 1993;26:47–63. [PubMed] [Google Scholar]
- 7.Tibayrenc M. Adv Parasitol. 1995;36:47–115. doi: 10.1016/s0065-308x(08)60490-x. [DOI] [PubMed] [Google Scholar]
- 8.Macedo A M, Martins M S, Chiari E, Pena S D J. Mol Biochem Parasitol. 1992;55:147–154. doi: 10.1016/0166-6851(92)90135-7. [DOI] [PubMed] [Google Scholar]
- 9.Steindel M, Dias Neto E, Menezes C L P, Romanha A J, Simpson A J G. Mol Biochem Parasitol. 1993;60:71–80. doi: 10.1016/0166-6851(93)90030-2. [DOI] [PubMed] [Google Scholar]
- 10.Tibayrenc M, Neubauer K, Barnabé C, Guerrini F, Skarecky D, Ayala F. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira R P, Macedo A M, Chiari E, Pena S D J. Parasitol Today. 1997;13:196–200. doi: 10.1016/s0169-4758(97)01044-2. [DOI] [PubMed] [Google Scholar]
- 12.Henriksson J, Petterson U, Solari A. Exp Parasitol. 1993;77:334–348. doi: 10.1006/expr.1993.1091. [DOI] [PubMed] [Google Scholar]
- 13.Souto R P, Zingales B. Mol Biochem Parasitol. 1993;62:45–52. doi: 10.1016/0166-6851(93)90176-x. [DOI] [PubMed] [Google Scholar]
- 14.Souto R P, Fernandes O, Macedo A M, Campbell D A, Zingales B. Mol Biochem Parasitol. 1996;83:141–152. doi: 10.1016/s0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- 15.Pena S D J, Prado V F, Epplen J T. J Mol Med. 1993;73:555–564. doi: 10.1007/BF00195140. [DOI] [PubMed] [Google Scholar]
- 16.Broude N E, Chandra A, Smith C L. Proc Natl Acad Sci USA. 1997;94:4548–4553. doi: 10.1073/pnas.94.9.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zingales B, Pereira M E S, Oliveira R P, Almeida K A, Umezawa E S, Souto R P, Vargas N, Cano M I, da Silveira J F, Nehme N S, et al. Acta Trop. 1997;68:159–173. doi: 10.1016/s0001-706x(97)00088-0. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein J. phylip, Phylogeny Inference Package. University of Washington, Seattle: Department of Genetics; 1993. , Version 3.5c. [Google Scholar]
- 19.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 20.Morel C, Chiari E, Camargo E P, Mattei D M, Romanha A J, Simpson L. Proc Natl Acad Sci USA. 1980;77:6810–6814. doi: 10.1073/pnas.77.11.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber J L, Wong C. Hum Mol Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- 22.Gibson W C, Miles M A. EMBO J. 1986;5:1299–1305. doi: 10.1002/j.1460-2075.1986.tb04359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henriksson J, Porcel B, Rydaker M, Ruiz A, Sabaj V, Galanti N, Cazzulo J J, Frasch A C C, Petterson U. Mol Biochem Parasitol. 1995;73:63–74. doi: 10.1016/0166-6851(95)00096-j. [DOI] [PubMed] [Google Scholar]
- 24.Maslov D A, Simpson L. Parasitol Today. 1995;11:30–32. [Google Scholar]
- 25.Zingales B, Rondinelli E, Degrave W, da Silveira J F, Levin M, Le Paslier D, Modabber F, Dobrokhotov B, Swindle J, Kelly J M, et al. Parasitol Today. 1997;13:16–22. [Google Scholar]