Structural informatics and modelling correctly predicted that substrate was required to obtain diffracting crystals of the first characterized nitrile oxidoreductase: the homododecameric QueF.
Keywords: bioinformatics, YkvM, YqcD, RNA, post-transcriptional modification
Abstract
QueF (MW = 19.4 kDa) is a recently characterized nitrile oxidoreductase which catalyzes the NADPH-dependent reduction of 7-cyano-7-deazaguanine (preQ0) to 7-aminomethyl-7-deazaguanine, a late step in the biosynthesis of the modified tRNA nucleoside queuosine. Initial crystals of homododecameric Bacillus subtilis QueF diffracted poorly to 8.0 Å. A three-dimensional model based on homology with the tunnel-fold enzyme GTP cyclohydrolase I suggested catalysis at intersubunit interfaces and a potential role for substrate binding in quaternary structure stabilization. Guided by this insight, a second crystal form was grown that was strictly dependent on the presence of preQ0. This crystal form diffracted to 2.25 Å resolution.
1. Introduction
Prior to its use in translation, transfer RNA (tRNA) undergoes extensive post-transcriptional processing and maturation in all cells (for reviews, see Wolin & Matera, 1999 ▶; Hopper & Phizicky, 2003 ▶). Nucleoside modification is perhaps the most elaborate step in tRNA processing (Björk, 1995 ▶). Over 90 modified nucleosides have been characterized (McCloskey & Crain, 1998 ▶), many of which are conserved across broad phylogenetic boundaries (Grosjean et al., 1995 ▶). Their modifications vary from simple methylation of the base or ribose ring to extensive hypermodification of the canonical bases, resulting in structural changes requiring multiple enzymatic steps. Here, we present the results of attempts to crystallize a complex protein assembly that is essential for synthesis of one of the most prevalent of these modifications.
Queuosine (Q) is a hypermodified guanosine found exclusively in the wobble position (position 34) of tRNAGUN coding for the amino acids asparagine, aspartate, histidine and tyrosine in eukarya and bacteria (Kersten & Kersten, 1990 ▶). The presence of Q in the anticodons of these tRNAs (including mitochondrial tRNAs) is compatible with a fundamental role in translation, specifically in modulating fidelity. For example, the −1 frameshifting events essential for correct translation of the retroviral Gag-Pol-Pro polypeptides of human T-cell lymphotropic virus and bovine leukemia virus appear to be dependent on (Q−)-tRNAAsn (Jacks et al., 1988 ▶; Hatfield et al., 1989 ▶; Carlson et al., 1999 ▶, 2000 ▶). Disruption of Q biosynthesis in the pathogenic bacterium Shigella flexneri results in loss of pathogenicity, making several enzymes in its biosynthesis pathway attractive targets for anti-shigellosis drugs. This loss is caused by mistranslation of virF, a transcription factor responsible for up-regulation of a suite of virulence-associated proteins (Durand et al., 2000 ▶). Similarly, Q is essential in the biosynthesis of tyrosine in animals by its preventing a (Q−)-tRNA-dependent mistranslation of mRNA coding for the tyrosine-biosynthesis enzyme phenylalanine hydroxylase (Marks & Farkas, 1997 ▶).
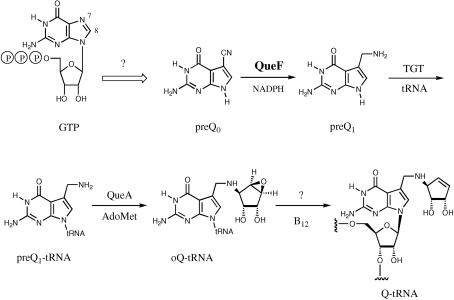
Q is characterized by a cyclopentenediol ring appended to 7-aminomethyl-7-deazaguanosine (Kasai et al., 1975 ▶; Ohgi et al., 1979 ▶). In some mammalian tRNAs, Q is glycosylated with galactose or mannose at the C5 hydroxyl (Okada & Nishimura, 1977 ▶). While Q is acquired as a nutrient factor in eukarya, it is synthesized de novo in bacteria. Bacterial biosynthesis of Q starts with the conversion (by an unknown pathway) of GTP to 7-cyano-7-deazaguanine (preQ0), followed by reduction to 7-aminomethyl-7-deazaguanine (preQ1). PreQ1 is then attached to tRNA by tRNA-guanine transglycosylase and further modified to Q in two in situ steps (Fig. 1 ▶). Using comparative genomics, QueF was identified as an enzyme family involved in Q biosynthesis (Reader et al., 2004 ▶). QueF is an NADPH-dependent oxidoreductase that carries out the unprecedented reduction of a nitrile group (preQ0) to a primary amine (preQ1; Van Lanen et al., 2005 ▶).
Figure 1.
Biosynthetic pathway to queuosine in bacteria.
Based on their amino-acid sequences, QueF family members fall into two structural subfamilies (Van Lanen et al., 2005 ▶). The YkvM subfamily is comprised of ∼160-amino-acid unimodular proteins with a characteristic QueF motif, i.e. E(S/L)K(S/A)hK(L/Y)(Y/F/W) (where h is a hydrophobic amino acid) bracketed on the N- and C-terminal sides by an invariant Cys and Glu, respectively. The YqcD subfamily of QueF enzymes is characterized by ∼280-amino-acid bimodular proteins in which the QueF motif and the invariant Cys and Glu are located separately in the weakly homologous N- and C-terminal halves (modules) of the polypeptide chain, respectively. Functional analysis of an enzyme from each subfamily, YkvM (Bacillus subtilis QueF) and YqcD (E. coli QueF), showed that YkvM enzymes function as homododecamers, while the YqcD enzymes are homodimers.
Homology between QueF and GTP cyclohydrolase I (GTP-CH-I) has been noted (Van Lanen et al., 2005 ▶) and resulted in initial functional misannotation of the QueF gene family as GTP-CH-I in genomic databases. However, we used this homology to build a three-dimensional working model of B. subtilis QueF (19.4 kDa, 165 amino acids) based on the crystal structure of GTP-CH-I. The model allowed functional predictions that aided in understanding the crystallization properties of this YkvM subfamily member and enhanced the common prediction that the presence of substrate may improve crystal quality. The results and the general case of QueF discovery provide a powerful example of the use of bioinformatics tools to aid in proper functional assignment and consequent structural characterization of a new enzyme family.
2. Materials and methods
2.1. Three-dimensional homology modeling of B. subtilis QueF
A pairwise alignment of B. subtilis QueF and E. coli CTP-CH-I was extracted from a multiple sequence alignment of 30 QueF sequences and 30 GTP-CH-I sequences and used to build a three-dimensional model of a monomer of B. subtilis QueF in MODELLER-6 (v.1; John & Sali, 2003 ▶). The X-ray crystal structure of Escherichia coli GTP cyclohydrolase I (Nar et al., 1995 ▶; GTP-CH-I; PDB code 1fbx) was used as a template. The B. subtilis QueF sequence was obtained from GenBank (accession No. NP_389258, GeneID 939296). The sequence similarity and identity between the two proteins is 26 and 14%, respectively. Using the symmetry of the GTP-CH-I decamer, a homodimer of the QueF monomeric model was generated. Using the coordinates of bound GTP in the GTP-CH-I structure, a 7-cyano-7-deazaguanine molecule was docked onto the putative active site that was located at the intersubunit interface. The final model was energy-minimized in CNS (Brünger et al., 1998 ▶) and validated in PROCHECK (Vaguine et al., 1999 ▶). The model had a standard Ramachandran plot with 90.2% of residues falling in favored regions and 8.3 and 1.5% in the allowed and generously allowed regions, respectively. The r.m.s. deviations of bonds and angles from standard values are 0.019 Å and 2.33°, respectively. A least-squares superposition with the GTP-CH-I structure yielded a good fit with an r.m.s. deviation of 0.64 Å over 318 Cα atoms.
2.2. Crystallization of B. subtilis QueF
N-terminal His6-tagged B. subtilis QueF (GenBank NP_389258) was overexpressed, purified and processed for His6-tag removal as described in Van Lanen et al. (2005 ▶). Prior to use in crystallization, the enzyme was dialyzed against 100 mM Tris pH 7.5, 100 mM KCl and 2 mM dithiothreitol. The substrate preQ0 was synthesized as described previously (Van Lanen et al., 2003 ▶).
B. subtilis QueF (15 mg ml−1, apoenzyme) was subjected to high-throughput sparse-matrix and grid crystallization screens using the vapor-diffusion method. 200 nl sitting drops were set up using the Mosquito crystallization robot (Molecular Dimensions Ltd) in 96-well low-profile Greiner microplates (Greiner BioOne, FL, USA) and imaged with the CrystalPro imaging system (Tritek Corp., VA, USA). Crystallization experiments (1536) were performed at 293.15 and 277.15 K. Initial thick hexagonal plate crystals appeared in seven drops of various conditions. Crystals were reproduced manually at 293.15 K in hanging drops (1 µl) containing 4–15 mg ml−1 enzyme, 15–22%(v/v) polyethylene glycol 550 monomethyl ether (PEG 550 MME), 100 mM HEPES, Bis-Tris-propane or imidazole pH 7.2–7.8, 50 mM CaCl2 and 0.05%(w/v) NaN3 as a preservative. For crystallization in the presence of substrate, preQ0 was dissolved in DMSO and directly added to the enzyme. The enzyme (4 mg ml−1, ∼0.2 mM) was pre-incubated for 30 min on ice with 1.2–5 mM preQ0 (enzyme:substrate molar ratio of 1:6–1:25). 1 µl sample was mixed with 1 µl reservoir solution containing 16–24%(v/v) PEG 550 MME, 100 mM HEPES or imidazole pH 7.2–7.8, 30 mM CaCl2 and 0.05%(w/v) NaN3. The mixed drop was equilibrated against 0.5 ml reservoir solution at 293.15 K until rod-shaped crystals grew in 2–3 weeks to a final size of 0.2 × 0.2 × 0.5 mm. Crystals were harvested and flash-cooled in liquid nitrogen without need for cryoprotection.
2.3. X-ray diffraction analysis
Crystals were screened for diffraction quality at beamline 11-1 of the Stanford Synchrotron Research Laboratory (SSRL), using a robot for mounting of crystals. A single-wavelength data set was collected from a crystal of the enzyme–preQ0 complex on an ADSC Quantum 315 CCD detector at SSRL beamline 1-5 (crystal-to-detector distance 240 mm). Data were processed using the HKL package (Otwinowski & Minor, 1997 ▶).
3. Results
All attempts to crystallize the apoenzyme led to showers of single or clustered hexagonal crystal plates (0.05 × 0.1 × 0.1 mm, space group P61, unit-cell parameters a = b = 81.6, c = 200.0 Å) that grew from 15 mg ml−1 enzyme using PEG 3350, PEG 2000, PEG 1000 or PEG 550 MME as the precipitant, a variety of buffers in the pH range 6.0–9.0 and 50 mM CaCl2. After refinement of conditions, the showering and clustering effects were controlled by lowering the CaCl2 and protein concentrations to 30 mM and 4 mg ml−1, respectively (Fig. 2 ▶ a, left). However, the crystals remained highly mosaic (2.0–3.0°) and their diffraction quality poor as assessed by a lack of detectable diffraction beyond 7–8 Å resolution (data not shown). Previously reported crystals of B. subtilis YkvM (Midwest Center for Structural Genomics, ID APC35752) also diffracted poorly and were not pursued for structure determination (Anderson, 2005 ▶).
Figure 2.
Crystals, three-dimensional model and X-ray diffraction of B. subtilis QueF. (a) Crystals of apoenzyme (hexagonal form, left) and of enzyme grown under the same conditions but pre-incubated with a sixfold excess of preQ0 (trigonal form, right). (b) Putative active site of B. subtilis QueF based on the homology model built from the crystal structure of E. coli GTP-CH-I. The active site is located at the interface between two monomers shown in light and dark grey. The putative binding mode of the substrate’s guanine is shown. Potential active-site interactions are indicated. The conserved QueF motif region is highlighted in green. The conserved Glu characteristic of T-fold enzymes and the invariant Cys shared between QueF and GTP-CH-I are shown in black. (c) A 0.5° oscillation image of the synchrotron X-ray diffraction of trigonal B. subtilis QueF. The image was taken at the SSRL beamline 1-5, with a 60 s exposure time. The crystal lasted 9 h in the beam before it denatured suddenly from radiation damage. The resolution at the edge of the detector was 2.15 Å.
The similarity between B. subtilis QueF and GTP-CH-I in sequence and multimeric quaternary structure prompted us to predict a similar location for the active site of B. subtilis QueF, i.e. at the intersubunit interface. Based on the crystal structure of E. coli GTP-CH-I, a three-dimensional homology model of two adjacent QueF monomers was built and a preQ0 molecule was docked in the putative active site (Fig. 2 ▶ b). In one monomer, the docked substrate interacts with the two invariant side chains of Glu97 and Cys55 (two interactions also found in the structure of the GTP-CH-I–GTP complex; residue numbers are those of B. subtilis YkvM), the conserved Phe95 and the backbone NH of His96. In the other monomer, the side chain of Ser97 and the backbone CO of Val77 interact with substrate. Significantly, the QueF motif lies in a nearby α-helix. This model suggests that preQ0 plays an important role in stabilizing and tying together the functional multimeric enzyme structure, bridging the two halves of the active site; that is, the QueF motif from one monomer and the invariant Glu and Cys from the other. This observation prompted us to test the effect of preQ0 on the crystallization properties of QueF.
We used the refined conditions for crystal growth obtained for the apoenzyme [4 mg ml−1 protein, 20%(v/v) PEG 550 MME, 100 mM HEPES, 30 mM CaCl2] and applied them to samples containing enzyme pre-incubated with preQ0 (concentrations of 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 2 and 5 mM, i.e. enzyme:preQ0 molar ratio of 1:1–1:25). To rule out the effect of DMSO, control experiments were set up simultaneously using enzyme pre-incubated with the same added volume of DMSO. A second trigonal crystal form (space group P3121) appeared as the sole form only in drops containing ≥1.2 mM preQ0 (enzyme:preQ0 molar ratio ≥ 1:6, Fig. 2 ▶ a, right). PreQ0 concentrations of ≤ 0.8 mM (molar ratio ≤ 1:4) yielded the hexagonal form and intermediate concentrations yielded a mixture of both forms in the same drop. The trigonal form diffracted synchrotron X-rays to 2.25 Å with an apparent mosaicity of 0.29°. Analysis of the solvent content with the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶) gave a unique solution consisting of half a dodecamer in the asymmetric unit (Fig. 2 ▶ c; Table 1 ▶). Growth of larger crystals was aided by inclusion of 1%(w/v) dextran sulfate or 100 mM imidazole in the crystallization buffer. Pre-incubation of enzyme with DMSO alone or with the cofactor for catalysis NADPH failed to produce the trigonal crystals (data not shown). A self-rotation search in CNS detected sixfold non-crystallographic symmetry around an axis parallel to the c axis of the unit cell. Because their unit-cell parameters differ significantly, it is unlikely that the two crystal forms are related. It is well known that use of substrates or substrate analogs in the crystallization process increases the chances of obtaining crystals or improving crystal quality. A dodecameric member of the tunnel-fold structural superfamily, the QueF quaternary arrangement is predicted to be a dimer of hexamers. Binding of substrate at the intersubunit interfaces may shift a hexamer–dodecamer equilibrium toward the dodecameric form, increasing sample homogeneity and allowing crystal growth around the twofold axis of the dodecamer as in the trigonal form. Although structure determination may be performed by molecular replacement, crystals of the selenomethionine-labeled protein have been produced and await collection of seleno-multiwavelength anomalous diffraction X-ray data.
Table 1. X-ray data-collection parameters.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 0.97944 |
| Space group | P3121 |
| Unit-cell parameters (Å) | a = b = 93.52, c = 193.76 |
| Crystal mosaicity (°) | 0.29 |
| Resolution range (Å) | 2.25–50.0 (2.25–2.33) |
| No. of observations | 278757 (17116) |
| No. of unique reflections | 46429 (3890) |
| Completeness (%) | 97.8 (83.3) |
| Redundancy | 6.0 (4.3) |
| Rmerge | 0.078 (0.39) |
| 〈I/σ(I)〉 | 9.9 (7.9) |
| Matthews coefficient (Å3 Da−1) | 2.1 |
| Solvent content (%) | 40.1 |
| Asymmetric unit content | 6 monomers |
Acknowledgments
We thank Drs Clyde Smith and Aina Cohen at SSRL beamlines 11-1 and 1-5, respectively, for help in X-ray data collection. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, National Institutes of Health (NIH) and the National Institute of General Medical Sciences. This work was supported by grant No. GM15539 from the NIH and by a fellowship from the National Foundation for Cancer Research.
References
- Anderson, W. (2005). Personal communication.
- Björk, G. R. (1995). tRNA: Structure, Biosynthesis and Function, edited by U. L. RajBhandary, pp. 165–206. Washington DC: ASM Press.
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carlson, B. A., Kwon, S. Y., Chamorro, M., Oroszlan, S., Hatfield, D. L. & Lee, B. J. (1999). Virology, 255, 2–8. [DOI] [PubMed] [Google Scholar]
- Carlson, B. A., Kwon, S. Y., Lee, B. J. & Hatfield, D. (2000). Mol. Cell, 10, 113–118. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Durand, J. M., Dagberg, B., Uhlin, B. E. & Björk, G. R. (2000). Mol. Microbiol.35, 924–935. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Sprinzl, M. & Steinberg, S. (1995). Biochimie, 77, 139–141. [DOI] [PubMed] [Google Scholar]
- Hatfield, D., Feng, Y.-X., Lee, B. J., Rein, A., Levin, J. G. & Oroszlan, S. (1989). Virology, 173, 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper, A. K. & Phizicky, E. M. (2003). Genes Dev.17, 162–180. [DOI] [PubMed] [Google Scholar]
- Jacks, T., Madhani, H. D., Masiarz, F. R. & Varmus, H. F. (1988). Cell, 55, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, B. & Sali, A. (2003). Nucleic Acids Res.31, 3982–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai, H., Ohashi, Z., Harada, F., Nishimura, S., Oppenheimer, N. J., Crain, P. F., Liehr, J. G., von Minden, D. L. & McCloskey, J. A. (1975). Biochemistry, 14, 4198–4208. [DOI] [PubMed] [Google Scholar]
- Kersten, H. & Kersten, W. (1990). Chromatography and Modification of Nucleosides Part B, edited by K. C. T. Kuo, pp. B69–B108. Amsterdam: Elsevier.
- McCloskey, J. A. & Crain, P. F. (1998). Nucleic Acids Res.26, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, T. & Farkas, W. R. (1997). Biochem. Biophys. Res. Commun.230, 233–237. [DOI] [PubMed] [Google Scholar]
- Nar, H., Huber, R., Meining, W., Schmid, C., Weinkauf, S. & Bacher, A. (1995). Structure, 3, 459–466. [DOI] [PubMed] [Google Scholar]
- Ohgi, T., Kondo, T. & Goto, T. (1979). J. Am. Chem. Soc.101, 3629–3633.
- Okada, N. & Nishimura, S. (1977). Nucleic Acids Res.4, 2931–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Reader, J., Metzgar, D., Schimmel, P. & de Crécy-Lagard, V. (2004). J. Biol. Chem.279, 6280–6285. [DOI] [PubMed] [Google Scholar]
- Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999). Acta Cryst. D55, 191–205. [DOI] [PubMed] [Google Scholar]
- Van Lanen, S. G., Kinzie, S. D., Matthieu, S., Link, T., Culp, J. & Iwata-Reuyl, D. (2003). J. Biol. Chem.278, 10491–10499. [DOI] [PubMed] [Google Scholar]
- Van Lanen, S. G., Reader, J. S., Swairjo, M. A., de Crécy-Lagard, V., Lee, B. & Iwata-Reuyl, D. (2005). Proc. Natl Acad. Sci. USA, 102, 4264–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin, S. L. & Matera, A. G. (1999). Genes Dev.13, 1–10. [DOI] [PubMed] [Google Scholar]