Crystallization of the human CHP2–NHE1 binding domain complex.
Keywords: calcineurin homologous proteins, Na+/H+ exchangers
Abstract
Calcineurin homologous protein (CHP) is a Ca2+-binding protein that directly interacts with and regulates the activity of all plasma-membrane Na+/H+-exchanger (NHE) family members. In contrast to the ubiquitous isoform CHP1, CHP2 is highly expressed in cancer cells. To understand the regulatory mechanism of NHE1 by CHP2, the complex CHP2–NHE1 (amino acids 503–545) has been crystallized by the sitting-drop vapour-diffusion method using PEG 3350 as precipitant. The crystals diffract to 2.7 Å and belong to a tetragonal space group, with unit-cell parameters a = b = 49.96, c = 103.20 Å.
1. Introduction
The Na+/H+ exchangers (NHEs) are electroneutral transporters that catalyze the countertransport of Na+ and H+ through the plasma membrane and other intracellular organellar membranes in various animal species (Wakabayashi et al., 1997 ▶; Orlowski & Grinstein, 2004 ▶). Nine different NHE isoforms (NHE1–NHE9) have been identified in mammalian tissues. Although they have been shown to exhibit similar membrane topology, these isoforms are thought to play different roles in various tissues (Counillon & Pouysségur, 2000 ▶; Orlowski & Grinstein, 2004 ▶). The isoform NHE1 is ubiquitously expressed in all tissues and cell types and plays a major role in maintaining intracellular pH and cell-volume homoeostasis (Putney et al., 2002 ▶). The activity of NHE1 is controlled by various extrinsic factors, including growth factors, hormones and mechanical stimuli (Wakabayashi et al., 1997 ▶; Counillon & Pouysségur, 2000 ▶; Orlowski & Grinstein, 2004 ▶). NHE1 is regulated by a variety of signalling molecules including calcineurin B homologous protein (CHP; Lin & Barber, 1996 ▶; Pang et al., 2001 ▶) and Ca2+/calmodulin (Bertrand et al., 1994 ▶; Wakabayashi et al., 1994 ▶). Despite intensive studies on NHE1 and its regulation, structural information is extremely limited, especially for the cytoplasmic C-terminal domain which contains most of the binding domains for the regulatory proteins.
CHP was initially identified as a protein (p22) involved in vesicular transport (Barroso et al., 1996 ▶) and also as a molecule that interacts with NHE (Lin & Barber, 1996 ▶). CHP has also been reported to be involved in various cell functions, such as inhibition of calcineurin phosphatase activity (Lin et al., 1999 ▶) and interaction with microtubules (Timm et al., 1999 ▶), DRAK2 (death-associated protein kinase-related apoptosis-inducing protein kinase 2; Matsumoto et al., 2001 ▶) and KIF1Bβ2 (kinesin family 1Bb2; Nakamura et al., 2002 ▶). We have previously reported that CHP is an essential cofactor for supporting the physiological activity of the Na+/H+ exchanger by interacting with its juxtamembrane cytoplasmic domain (Pang et al., 2001 ▶). Furthermore, we demonstrated that in contrast to the ubiquitous CHP1 isoform, CHP2 (61% amino-acid identity with CHP1) is highly expressed in malignantly transformed cells and may be involved in maintaining the high intracellular pH (pHi) in cancer cells (Pang et al., 2002 ▶). NHE1 mutants lacking the CHP-binding region (amino acids 515–530) exhibited low exchange activity (5–10% of the wild-type level; Pang et al., 2001 ▶), suggesting that this region is essential for normal exchange activity of NHE1. This region with bound CHP would therefore function as a key structure maintaining the physiologically active conformation of NHE1 (Pang et al., 2001 ▶). Consequently, more detailed structural information including the crystal structure of CHP complexed with its binding domain is of great importance to reveal the mechanism by which CHP is involved in this important regulation pathway of NHE1.
Here, we report the first crystallization and preliminary crystallographic studies of the human CHP2 complexed with the C-terminal cytoplasmic region (amino acids 503–545) of NHE1. Hereafter, the protein complex is referred to as CHP2–NHE1-peptide.
2. Materials and methods
2.1. Protein expression and purification
Human CHP2 cDNA (GenBank accession No. AF146019) corresponding to amino-acid residues 1–196 cloned into pET11 vector (Novagen) as a fusion protein with a C-terminal His6 tag was coexpressed in Escherichia coli (BL21-Star; Invitrogen) with the human cDNA encoding the cytoplasmic binding-domain region of NHE1 peptide cloned into pET24 vector (Novagen). Six histidine residues were inserted after Lys196 of CHP2, while a stop codon was incorporated just after the sequence coding for the NHE1 peptide to eliminate the His6 tag from the vector. Using this coexpression system, as also described previously for CHP1 (Pang et al., 2004 ▶), we were able to obtain CHP2 in a complex form with its binding domain of NHE1. Cells were cultured in 2×YT medium containing 100 µg ml−1 ampicillin and 100 µg ml−1 kanamycin at 310 K. At an optical density of 0.6 at 600 nm, protein expression was induced by the addition of IPTG to a final concentration of 1 mM and cells were grown overnight at 291 K. The cells were harvested and resuspended in PBS buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and disrupted by sonication at 277 K. After centrifugation at 277 K, the supernatant containing the complex CHP2–NHE1-peptide was applied onto an Ni–NTA agarose affinity column (Invitrogen) equilibrated with PBS buffer. The column was washed with buffer A (20 mM sodium phosphate, 500 mM NaCl and 2 M KCl pH 6.0), buffer B (20 mM sodium phosphate and 500 mM NaCl pH 4.7) and then buffer C (20 mM sodium phosphate and 500 mM NaCl pH 6.0). The adsorbed protein complex was eluted with buffer C containing 500 mM imidazole, dialyzed overnight against buffer D (20 mM Tris–HCl pH 8.5) and further purified using a DEAE-Sepharose column (HiTrap DEAE FF 5 ml; Amersham Biosciences) eluted with a gradient from 0 to 1 M NaCl in 20 mM Tris–HCl buffer pH 8.5. A final purification step was carried out using gel-filtration chromatography (Superdex 200; Amersham Bioscience). The gel-filtration column was eluted with a buffer solution containing 100 mM NaCl and 20 mM Tris–HCl pH 7.5. The fraction containing CHP2–NHE1-peptide was pooled, dialyzed against 20 mM Tris–HCl pH 7.5, concentrated (20–25 mg ml−1) using Amicon Ultra (Millipore) and subjected to crystallization without removing the His6 tag.
2.2. Crystallization
Preliminary screening of crystallization conditions was performed using various commercial kits (Hampton Research Crystal Screen kits, Emerald BioSystems Screen kits, Sigma–Aldrich Crystallization kits) and carried out using the sitting-drop vapour-diffusion method at 293, 287 and 277 K. 1 µl aliquots of protein-complex solution (20–25 mg ml−1) were mixed with 1 µl reservoir solution to form the droplet, which was equilibrated against 100 µl reservoir solution. The initial screening, involving about 1440 individual trials, was unsuccessful. Additives from Hampton Research were used together with the above screening kits in a second trial involving about 4320 individual trials and very small and thin needle-shape crystals were finally obtained with a crystallization solution containing 200 mM ammonium acetate, 100 mM Bis-Tris pH 5.5, 25%(w/v) polyethylene glycol 3350 (PEG 3350) and 5 mM yttrium chloride as an additive at 277 K. Refinement of the crystallization conditions to 200 mM ammonium acetate, 100 mM Bis-Tris pH 5.5, 25%(w/v) PEG 3350 and 10 mM yttrium chloride at 293 K improved the size of the crystals. The resultant crystals are mostly in clusters, with the occasional appearance of single crystals. Single crystals or dissected parts from the clusters were used for data collection.
2.3. Crystallographic data collection
Prior to data collection, single crystals were soaked in a solution containing 200 mM ammonium acetate, 100 mM Bis-Tris pH 5.5, 35%(w/v) PEG 3350 and 10 mM yttrium chloride and flash-frozen under a nitrogen flow at 100 K. The crystals were evaluated in-house with Cu Kα radiation (λ = 1.5418 Å) generated by an RA-Micro 7 rotating-anode X-ray generator with R-AXIS VII imaging-plate detector (Rigaku). High-resolution data sets were collected using an ADSC Quantum 315 CCD detector installed on the BL41XU beamline at SPring-8. The data collection was performed at a wavelength of 1.000 Å over a total range of 180°, with individual frames of 1° and an exposure time of 4 s. The crystal-to-detector distance was 350 mm. The collected images were processed using HKL2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
CHP is an important regulatory factor that maintains the physiologically active conformation of NHE1. In this study, in order to clarify the regulatory mechanism of NHE1 by CHP, we coexpressed CHP2 and its binding domain in NHE1 (amino acids 503–545) in E. coli and crystallized the complex. Firstly, we confirmed that the purified complex CHP2–NHE1-peptide was retained as a single peak on gel-filtration chromatography, indicating that the stable complex exists as a monomer (M r = 28 000) in solution. In addition, using a 4–12% polyacrylamide gradient gel we confirmed that the purity of the complex is suitable for crystallization assay and that the purified sample contained equimolar amounts (1:1 molar ratio) of CHP2 and NHE1-peptide (Fig. 1 ▶).
Figure 1.
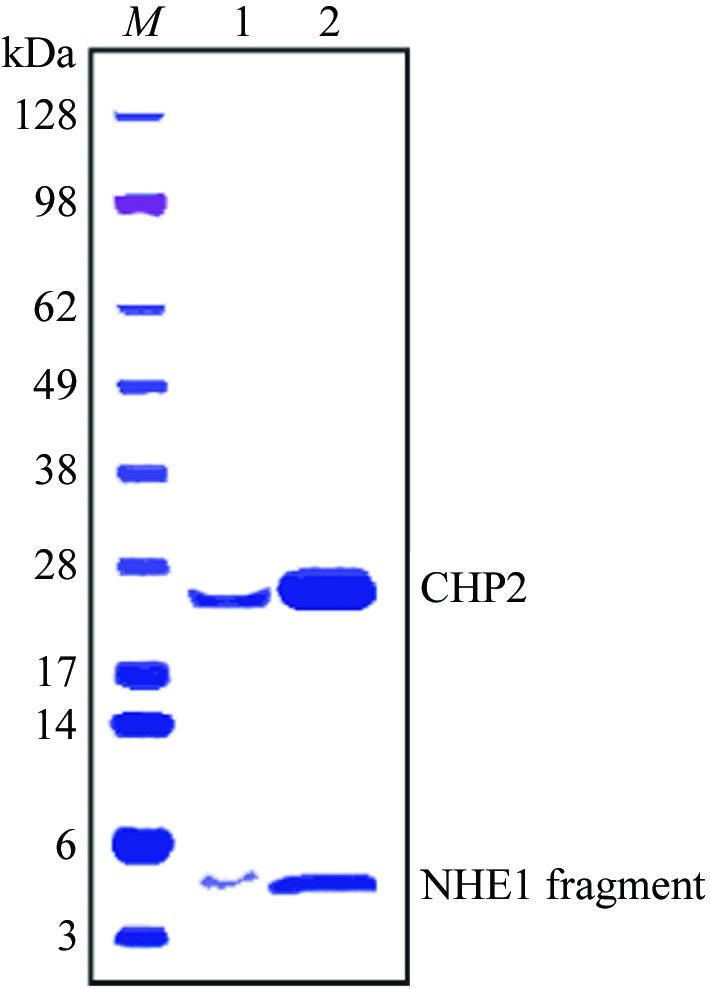
Polyacrylamide gel-electrophoresis pattern of the complex CHP2–NHE1-peptide. Crystals were collected and washed with the cryoprotectant solution. Collected crystals and 10 µg of the purified complex were applied to 4–12% gradient gel for lanes 1 and 2, respectively. Proteins were stained with Coomassie Brilliant Blue.
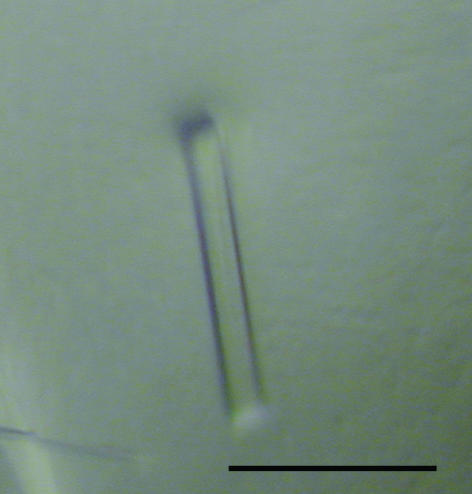
Crystals suitable for X-ray crystallographic analysis were obtained within 2–3 d at 293 K using the sitting-drop vapour-diffusion method (Fig. 2 ▶). A previous attempt to collect crystallographic data at beamline BL44B2 (SPring-8) gave a maximum resolution of 3.0 Å owing to the small size of crystals. To obtain higher resolution data, we used the undulator beamline BL41XU. Crystals diffracted to 2.5 Å resolution along the c axis of the crystal, but the data set was only qualitatively useful to 2.7 Å because of anisotropic diffraction.
Figure 2.
Crystal of human CHP2–NHE1-peptide as grown by the sitting-drop method. The scale bar indicates 0.1 mm.
The tetragonal crystal of CHP2–NHE1-peptide was determined to be P41 or P43, with unit-cell parameters a = b = 49.96, c = 103.20 Å. Assuming the presence of one CHP2–NHE1-peptide complex molecule in the asymmetric unit, the Matthews coefficient V M was calculated to be 2.5 Å3 Da−1, indicating a solvent content of approximately 49.5% in the unit cell. These values are within the typical range for protein crystals (Matthews, 1968 ▶).
The native data set has 6807 unique reflections, giving a data-set completeness of 97.3% in the resolution range 50.0–2.7 Å, with an R(I)merge of 4.8% (Table 1 ▶). Although CHP2 shows about 36% sequence identity with human CNB (PDB code 1aui; Kissinger et al., 1995 ▶), molecular replacement using CNB as a search model with MOLREP (Vagin & Teplyakov, 1997 ▶) was unsuccessful. Further crystallization refinement and structural analysis by multi-wavelength anomalous dispersion methods using selenomethionine and also taking advantage of the presence of yttrium as an additive are in progress.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell (2.8–2.7 Å).
| X-ray source | SPring-8 BL41XU |
| Space group | P41 or P43 |
| Unit-cell parameters (Å, °) | a = b = 49.96, c = 103.20, α = β = γ = 90 |
| Wavelength (Å) | 1.0000 |
| Resolution range (Å) | 50.00–2.70 (2.80–2.70) |
| Total reflections | 26984 |
| Unique reflections | 6807 |
| Rmerge† (%) | 4.8 (25.1) |
| Completeness (%) | 97.3 (83.1) |
| 〈I/σ(I)〉 | 17.3 (4.5) |
| Redundancy | 4.0 (3.1) |
| Crystal mosaicity (°) | 0.458 |
R
merge = 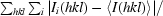
 , where I
i(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is its average.
, where I
i(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is its average.
Acknowledgments
We thank the staff at beamlines BL44B2 and BL41XU, SPring-8 for data-collection support and Dr Tianxiang Pang for initial participation in this study. This work was supported by grant Nano-001 for Research on Advanced Medical Technology from the Ministry of Health, Labour and Welfare of Japan and Grant-in-Aid for Priority Areas 13142210 for Scientific Research from the Ministry of Education, Science and Culture of Japan. YBA was supported by a Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship.
References
- Barroso, M. R., Bernd, K. K., DeWitt, N. D., Chang, A., Mills, K. & Sztul, E. S. (1996). J. Biol. Chem.271, 10183–10187. [DOI] [PubMed] [Google Scholar]
- Bertrand, B., Wakabayashi, S., Ikeda, T., Pouysségur, J. & Shigekawa, M. (1994). J. Biol. Chem.269, 13703–13709. [PubMed] [Google Scholar]
- Counillon, L. & Pouysségur, J. (2000). J. Biol. Chem.275, 1–4. [DOI] [PubMed] [Google Scholar]
- Kissinger, C. R., Parge, H. E., Knighton, D. R., Lewis, C. T., Pelletier, L. A., Tempczyk, A., Kalish, V. J., Tucker, K. D., Showalter, R. E., Moomaw, E. W., Gastinel, L. N., Habuka, N., Chen, X., Maldonado, F., Barker, J. E., Bacquet, R. & Villafranca, J. E. (1995). Nature (London), 378, 641–644. [DOI] [PubMed] [Google Scholar]
- Lin, X. & Barber, D. L. (1996). Proc. Natl Acad. Sci. USA, 93, 12631–12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X., Sikkink, R. A., Rusnak, F. & Barber, D. L. (1999). J. Biol. Chem.274, 36125–36131. [DOI] [PubMed] [Google Scholar]
- Matsumoto, M., Miyake, Y., Nagita, M., Inoue, H., Shitakubo, D., Takemoto, K., Ohtsuka, C., Murakami, H., Nakamura, N. & Kanazawa, H. (2001). J. Biochem.130, 217–225. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Nakamura, N., Miyake, Y., Matsushita, M., Tanaka, S., Inoue, H. & Kanazawa, H. (2002). J. Biochem.132, 483–491. [DOI] [PubMed] [Google Scholar]
- Orlowski, J. & Grinstein, S. (2004). Pflugers Arch.447, 549–565. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pang, T., Hisamitsu, T., Mori, H., Shigekawa, M. & Wakabayashi, S. (2004). Biochemistry, 43, 3628–3636. [DOI] [PubMed] [Google Scholar]
- Pang, T., Su, X., Wakabayashi, S. & Shigekawa, M. (2001). J. Biol. Chem.276, 17367–17372. [DOI] [PubMed] [Google Scholar]
- Pang, T., Wakabayashi, S. & Shigekawa, M. (2002). J. Biol. Chem.277, 43771–43777. [DOI] [PubMed] [Google Scholar]
- Putney, L. K., Denker, S. P. & Barber, D. L. (2002). Annu. Rev. Pharmacol. Toxicol.42, 527–552. [DOI] [PubMed] [Google Scholar]
- Timm, S., Titus, B., Bernd, K. & Barroso, M. (1999). Mol. Biol. Cell, 10, 3473–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, A. A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]
- Wakabayashi, S., Bertrand, B., Ikeda, T., Pouysségur, J. & Shigekawa, M. (1994). J. Biol. Chem.269, 13710–13715. [PubMed] [Google Scholar]
- Wakabayashi, S., Shigekawa, M. & Pouyssegur, J. (1997). Physiol. Rev.77, 51–74. [DOI] [PubMed] [Google Scholar]