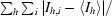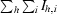Crystallization of HLA-B*2704 in complex with two peptides.
Keywords: HLA-B27 subtypes, HLA-B*2704, subtype-dependent peptide-binding modes, ankylosing spondylitis, polymorphism
Abstract
The product of the human leukocyte antigen (HLA) gene HLA-B*2704 differs from that of the prototypical subtype HLA-B*2705 by three amino acids at heavy-chain residues 77 (Ser instead of Asp), 152 (Glu instead of Val) and 211 (Gly instead of Ala). In contrast to the ubiquitous HLA-B*2705 subtype, HLA-B*2704 occurs only in orientals. Both subtypes are strongly associated with spondyloarthropathies and the peptides presented by these subtypes are suspected to play a role in disease pathogenesis. HLA-B*2704 was crystallized in complex with a viral peptide and with a self-peptide using the hanging-drop vapour-diffusion method with PEG as a precipitant. Both crystals belong to space group P212121. Data sets were collected to 1.60 Å (complex with the self-peptide pVIPR) or to 1.90 Å (complex with the viral peptide pLMP2) resolution using synchrotron radiation. With HLA-B*2705 complexed with pVIPR as a search model, unambiguous molecular-replacement solutions were found for the complexes of HLA-B*2704 with both peptides.
1. Introduction
One of the hallmarks of the human major histocompatibility (HLA) complex is its association with a multitude of diseases, among them most or even all autoimmune diseases (Horton et al., 2004 ▶). For example, the class I allele HLA-B27 is very strongly associated with spondyloarthropathies, including ankylosing spondylitis (AS; Khan & Ball, 2002 ▶; Ramos & López de Castro, 2002 ▶). HLA class I molecules consist of a highly polymorphic heavy chain (HC) which is non-covalently associated with β2-microglobulin (β2m). The HC forms a groove which carries a peptide that may be derived from self- or nonself-proteins within the cell (Madden, 1995 ▶). A large number of HLA class I molecules have already been investigated by X-ray crystallography, but the pairwise structural comparison of the products of very closely related alleles, which additionally may differ in their association with diseases, has only recently been accomplished (Hülsmeyer et al., 2002 ▶, 2004 ▶, 2005 ▶; Macdonald et al., 2003 ▶; Webb et al., 2004 ▶; Zernich et al., 2004 ▶; Fiorillo et al., 2005 ▶; Loll et al., 2005 ▶). Cytotoxic T lymphocytes directed against the self-antigen pVIPR [RRKWRRWHL, derived from vasoactive intestinal peptide type 1 receptor (residues 400–408)] have been found in individuals with the AS-associated HLA-B27 subtype HLA-B*2705 (all HLA-B27 subtypes will be refered to as ‘B*2705’ etc. for simplicity) and these cells increase in number during the development of AS (Fiorillo et al., 2000 ▶). About one-sixth of these T cells cross-react with the viral pLMP2 peptide [RRRWRRLTV, derived from latent membrane protein 2 (residues 236–244) of Epstein–Barr virus; Brooks et al., 1993 ▶; Fiorillo et al., 2000 ▶].
The B*2704 subtype is restricted to orientals, in particular individuals from China and Southeast Asia, and it has been demonstrated that, like B*2705, B*2704 is associated with AS (López-Larrea et al., 1995 ▶; Nasution et al., 1997 ▶; Ren et al., 1997 ▶; Chen et al., 2002 ▶; Ramos & López de Castro, 2002 ▶; Dhaliwal et al., 2003 ▶). Its product differs from that of B*2705 by three amino acids (Ser77, Glu152 and Gly211 in B*2704; Asp77, Val152 and Ala211 in B*2705; Vega et al., 1986 ▶; Rudwaleit et al., 1996 ▶). While the first two amino-acid replacements are expected to alter the properties of the peptide-binding groove, the Gly211Ala substitution might affect the interaction of B*2704 with accessory ligands on effector cells. The consequences of these exchanges for peptide binding (Garcia et al., 1997 ▶; Sesma et al., 2002 ▶; López de Castro et al., 2004 ▶) or T-cell responses (Sesma et al., 2002 ▶) have been investigated but are not fully understood.
Our structural studies of the B*2704 subtype address the following questions. How are peptides such as pVIPR and pLMP2, whose binding modes had already been determined in the B*2705 and B*2709 subtypes (Hülsmeyer et al., 2004 ▶; Fiorillo et al., 2005 ▶) as well as in B*2703 (Loll et al., 2005 ▶; Zawacka et al., in preparation), bound to B*2704? How do the HC amino-acid replacements in positions 77 and 152 affect the binding mode of the peptide and is there any influence of the exchange at HC residue 211 on the structure of the α3-domain of the HC or the interaction with β2m? Furthermore, can the peptide conformation which characterizes pVIPR and pLMP2 binding in the B*2705 subtype (p6α; i.e. main-chain ϕ/ψ torsion angles in α-helical conformation at peptide position p6 instead of the common p4; Hülsmeyer et al., 2004 ▶; Fiorillo et al., 2005 ▶) also be observed in B*2704? Our study is the first to determine the structural properties of the disease-associated B*2704 subtype.
2. Materials and methods
2.1. Protein preparation
The nonapeptides pVIPR (RRKWRRWHL) and pLMP2 (RRRWRRLTV) were synthesized by the solid-phase method and purified by Alta Bioscience (Birmingham, UK). The cDNA clone for the B*2704 heavy chain containing the three extracellular domains was generated by in vitro mutagenesis from a B*2705 clone. Human β2m and the B*2704 HC were separately expressed in Escherichia coli as inclusion bodies. They were solubilized by treatment with 50%(w/v) urea. According to a previously described reconstitution protocol (Garboczi et al., 1992 ▶; Menssen et al., 1999 ▶; Hülsmeyer et al., 2002 ▶), the HLA-B27–peptide complexes (B*2704–pVIPR or B*2704–pLMP2) were reconstituted for 14 d at 277 K, starting from 12 mg unfolded HC, 10 mg β2m and 4 mg of either pVIPR or pLMP2. All three components were rapidly injected into 400 ml refolding buffer [400 mM arginine–HCl, 2 mM EDTA, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 20%(v/v) glycerol, 100 mM Tris–HCl pH 7.5]. Following reconstitution, the entire mixture was dialyzed overnight to remove glycerol and Amicon Ultra-15 concentrators were used to concentrate the complexes prior to size-exclusion chromatography. Fractions containing the trimeric complexes were pooled and their composition assessed by SDS–PAGE. For crystallization experiments, the protein concentration was adjusted to 13–15 mg ml−1 in a buffer system containing 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.01% sodium azide.
2.2. Crystallization and data collection
B*2704–pVIPR and B*2704–pLMP2 were crystallized using the hanging-drop vapour-diffusion method at 291 K. 1.5 µl protein solution was mixed with 1.5 µl precipitant solution, employing previously described conditions (Hülsmeyer et al., 2002 ▶, 2004 ▶, 2005 ▶; Fiorillo et al., 2005 ▶). Crystal formation for both complexes was optimized by varying the polyethyleneglycol (PEG) concentration in the precipitant solution [18–28%(w/v) PEG 8000 or PEG 6000, 100 mM Tris–HCl pH 7.0). As the initially obtained microcrystals were too small for X-ray data collection, streak-seeding was applied by passing a cat whisker through each crystallization drop in the screens. After 4 d, B*2704–pVIPR crystallized from a precipitant solution composed of 28%(w/v) PEG 8000, 100 mM Tris–HCl pH 7.0 and crystals of B*2704–pLMP2 were obtained from a solution composed of 26%(w/v) PEG 6000, 100 mM Tris–HCl pH 7.0. Crystals of both subtypes grew to a maximum size of about 80 × 20 × 10 µm and had a similar size and morphology. They were soaked in a cryoprotectant solution by stepwise increase of glycerol to a final concentration of 10%. Using a nylon loop, they were picked up and subsequently frozen in liquid nitrogen.
All data sets were collected at beamline ID14-2 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France at a wavelength of λ = 0.933 Å at 100 K. The novel automatic sample changer at the beamline allowed rapid screening of different crystals. In addition, the MD2M mini-diffractometer installed at the beamline permitted the precise centring of the small crystals. Data were collected with 0.5° oscillation steps over a range of 100° using an ADSC-Q4 (Area Detector Systems Cooperation) CCD detector. Initially elongated diffraction spots could be improved using flash-annealing by shielding the cryostream for about 5 s.
Data were autoindexed using the program DENZO (Otwinowski & Minor, 1997 ▶), indicating that both subtypes crystallized in an orthorhombic lattice. The diffraction data were processed and scaled with DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶). The crystals of B*2704–pVIPR belong to space group P212121, with unit-cell parameters a = 50.8, b = 82.2, c = 109.9 Å, and show diffraction beyond 1.6 Å. Assuming the presence of one HC–peptide–β2m complex (MW = ∼44 000 Da) per asymmetric unit, the V M value is 2.5 Å3 Da−1, indicating a solvent content of 53%. The crystals of the other subtype (B*2704–pLMP2) also belong to space group P212121. They diffracted to 1.9 Å and exhibit unit-cell parameters a = 50.7, b = 82.3, c = 109.1 Å. The V M value of 2.6 Å3 Da−1 is also similar, corresponding to a water content of 52%. Data-collection statistics are summarized in Table 1. Molecular replacement was performed with the program PHASER (Storoni et al., 2004 ▶) using coordinates of the high-resolution crystal structure of B*2705–pVIPR (PDB code 1ogt; Hülsmeyer et al., 2004 ▶) as a search model (peptide and water molecules as well as the residues distinguishing B*2705 from B*2704 were omitted). Unambiguous solutions for both structures were found, limiting the diffraction data to the resolution range 20–3 Å. Initial F o − F c difference maps provide clear-cut evidence for the presence of the nonapeptides as well as polymorphic amino-acid residues.
3. Results
Purrifed B*2704–pVIPR and B*2704–pLMP2 complexes (Fig. 1 ▶) were crystallized and crystal formation was optimized using streak-seeding techniques, resulting in well ordered crystals (Fig. 2 ▶) that diffracted to 1.6 Å (B*2704–pVIPR) and 1.9 Å (B*2704–pLMP2) resolution at the ESRF. The inferior quality of the B*2704–pLMP2 data set is a consequence of handling problems with the new automatic sample changer at the ID 14-2 beamline. Contrary to our expectation from the experiments with the B*2705 and B*2709 subtypes, which crystallized in the P21 space group when complexed with pVIPR and pLMP2 (Hülsmeyer et al., 2004 ▶; Fiorillo et al., 2005 ▶), the B*2704–pVIPR and B*2704–pLMP2 complexes crystallized in space group P212121, which has previously been observed in crystals of B*2705 and B*2709 in complex with two peptides: m9 (GRFAAAIAK; Hülsmeyer et al., 2002 ▶) or TIS (RRLPIFSRL; Hülsmeyer et al., 2005 ▶). This might indicate that the nonapeptides adopt a conformation in B*2704 similar to those of the latter two peptides, i.e. the conventional p4α conformation and not the unorthodox p6α binding mode (Hülsmeyer et al., 2004 ▶; Fiorillo et al., 2005 ▶). Refinement of both B*2704 high-resolution structures has been initiated, confirming the space-group assignment (Table 1 ▶). The structure of the B*2704 complexes will permit comparison with other previously solved crystal structures of B2705, B*2709 and recently also of B*2703 (Loll et al., 2005 ▶). Determination of the influence of selected polymorphisms on peptide presentation by HLA-B27 subtypes will aid in understanding subtype-dependent differential disease associations (Ramos & López de Castro, 2002 ▶; López de Castro et al., 2004 ▶).
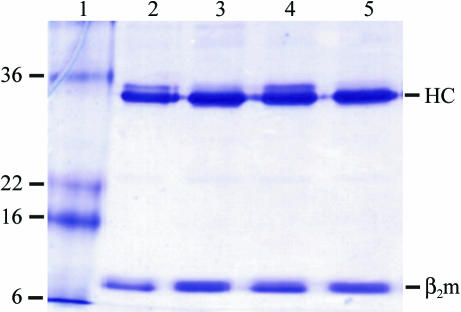
Figure 1.
Analysis of the purity of the refolded B*2704 complexes. Samples were subjected to SDS–PAGE under reducing (lanes 2 and 4) or non-reducing (lanes 3 and 5) conditions and stained with Coomassie Brilliant Blue. Lane 1, molecular-weight markers (kDa), lanes 2 and 3, B*2704–pLMP2; lanes 4 and 5, B*2704–pVIPR. The positions of HLA-B27 HC and β2m are indicated. The impurity with slightly higher molecular weight than the HC is occasionally found in preparations of HLA class I molecules. We have so far not observed it to interfere with the crystallization of the complexes.
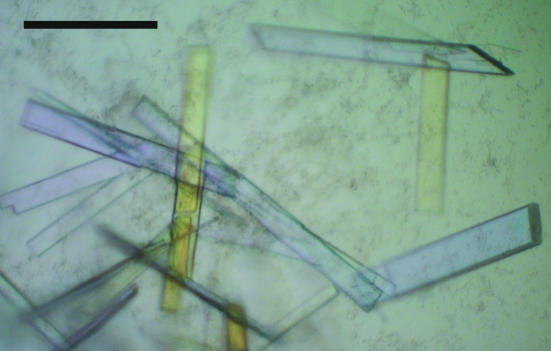
Figure 2.
Crystals of B*2704–pLMP2. The black bar indicates a length of 50 µm. Crystals of B*2704–pVIPR exhibited comparable size and morphology.
Table 1. Data-collection statistics of HLA-B*2704–pVIPR and HLA-B*2704–pLMP2.
Values in parentheses refer to the highest resolution shell.
| Data collection | HLA-B*2704–pVIPR | HLA-B*2704–pLMP2 |
|---|---|---|
| Space group | P212121 | P212121 |
| Unit-cell parameters (Å) | a = 50.8, b = 82.2, c = 109.9 | a = 50.7, b = 82.3, c = 109.1 |
| Solvent content (%) | 53 | 52 |
| Matthews coefficient† (Å3 Da−1) | 2.7 | 2.6 |
| Resolution (Å) | 30.0–1.6 (1.63–1.60) | 30.0–1.9 (1.93–1.90) |
| Unique reflections | 61196 (2949) | 36793 (1805) |
| Completeness (%) | 99.5 (97.4) | 100.0 (100.0) |
| Redundancy | 4.9 (4.8) | 7.9 (7.5) |
| 〈I〉/〈σ(I)〉 | 29.1 (4.5) | 16.5 (3.5) |
| Rsym‡ | 0.045 (0.391) | 0.131 (0.583) |
| Rmerge§ | 0.053 (0.391) | 0.131 (0.585) |
| Rr.i.m.§ | 0.059 (0.436) | 0.140 (0.628) |
| Rp.i.m.§ | 0.026 (0.188) | 0.050 (0.227) |
Acknowledgments
Financial support for this work was provided by the Deutsche Forschungsgemeinschaft (to BL and JB as well as to WS, BU-Z and Andreas Ziegler through SFB 449/B6), and the Fonds der Chemischen Industrie [to Anna Zawacka (Kekulé-Fellowship) and WS). We thank U. Gruber for excellent technical assistance and are grateful for allocation of beam time and support at ESRF (Grenoble).
References
- Brooks, J. M., Murray, R. J., Thomas, W. A., Kurilla, M. G. & Rickinson, A. B. (1993). J. Exp. Med.178, 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, I. H., Yang, K. L., Lee, A., Huang, H. H., Lin, P. Y. & Lee, T. D. (2002). Eur. J. Immunogenet.29, 435–438. [DOI] [PubMed] [Google Scholar]
- Dhaliwal, J. S., Too, C. L., Lisut, M., Lee, Y. Y. & Murad, S. (2003). Tissue Antigens, 62, 330–332. [DOI] [PubMed] [Google Scholar]
- Fiorillo, M. T., Maragno, M., Butler, R., Dupuis, M. L. & Sorrentino, R. (2000). J. Clin. Invest.106, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorillo, M. T., Rückert, C., Hülsmeyer, M., Sorrentino, R., Saenger, W., Ziegler, A. & Uchanska-Ziegler, B. (2005). J. Biol. Chem.280, 2962–2971. [DOI] [PubMed] [Google Scholar]
- Garboczi, D. N., Hung, D. T. & Wiley, D. C. (1992). Proc. Natl Acad. Sci. USA, 89, 3429–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, F., Marina, A. & López de Castro, J. A. (1997). Tissue Antigens, 49, 215–221. [DOI] [PubMed] [Google Scholar]
- Horton, R., Wilming, L., Rand, V., Lovering, R. C., Bruford, E. A., Khodiyar, V. K., Lush, M. J., Povey, S., Talbot, C. C. Jr, Wright, M. W., Wain, H. M., Trowsdale, J., Ziegler, A. & Beck, S. (2004). Nature Rev. Genet.5, 889–899. [DOI] [PubMed]
- Hülsmeyer, M., Fiorillo, M. T., Bettosini, F., Sorrentino, R., Saenger, W., Ziegler, A. & Uchanska-Ziegler, B. (2004). J. Exp. Med.199, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülsmeyer, M., Hillig, R. C., Volz, A., Rühl, M., Schröder, W., Saenger, W., Ziegler, A. & Uchanska-Ziegler, B. (2002). J. Biol. Chem.277, 47844–47853. [DOI] [PubMed] [Google Scholar]
- Hülsmeyer, M., Welfle, K., Pöhlmann, T., Misselwitz, R., Alexiev, U., Welfle, H., Saenger, W., Uchanska-Ziegler, B. & Ziegler, A. (2005). J. Mol. Biol.346, 1367–1379. [DOI] [PubMed] [Google Scholar]
- Khan, M. A. & Ball, E. J. (2002). Best Pract. Res. Clin. Rheumatol.16, 675–690. [PubMed] [Google Scholar]
- Loll, B., Zawacka, A., Biesiadka, J., Rückert, C., Volz, A., Saenger, W., Uchanska-Ziegler, B. & Ziegler, A. (2005). Acta Cryst. F61, 372–374. [DOI] [PMC free article] [PubMed]
- López de Castro, J. A., Alvarez, I., Marcilla, M., Paradela, A., Ramos, M., Sesma, L. & Vazquez, M. (2004). Tissue Antigens, 63, 424–445. [DOI] [PubMed] [Google Scholar]
- López-Larrea, C., Sujirachato, K., Mehra, N. K., Chiewsilp, P., Isarangkura, D., Kanga, U., Dominguez, O., Coto, E., Pena, M., Setien, F. & Gonzalez-Roces, S. (1995). Tissue Antigens, 45, 169–176. [DOI] [PubMed] [Google Scholar]
- Macdonald, W. A., Purcell, A. W., Mifsud, N. A., Ely, L. K., Williams, D. S., Chang, L., Gorman, J. J., Clements, C. S., Kjer-Nielsen, L., Koelle, D. M., Burrows, S. R., Tait, B. D., Holdsworth, R., Brooks, A. G., Lovrecz, G. O., Lu, L., Rossjohn, J. & McCluskey, J. (2003). J. Exp. Med.198, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden, D. R. (1995). Annu. Rev. Immunol.13, 587–622. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Menssen, R., Orth, P., Ziegler, A. & Saenger, W. (1999). J. Mol. Biol.285, 645–653. [DOI] [PubMed] [Google Scholar]
- Nasution, A. R., Mardjuadi, A., Kunmartini, S., Suryadhana, N. G., Setyohadi, B., Sudarsono, D., Lardy, N. M. & Feltkamp, T. E. (1997). J. Rheumatol.24, 1111–1114. [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Ramos, M. & López de Castro, J. A. (2002). Tissue Antigens, 60, 191–205. [DOI] [PubMed] [Google Scholar]
- Ren, E. C., Koh, W. H., Sim, D., Boey, M. L., Wee, G. B. & Chan, S. H. (1997). Tissue Antigens, 49, 67–69. [DOI] [PubMed] [Google Scholar]
- Rudwaleit, M., Bowness, P. & Wordsworth, P. (1996). Immunogenetics, 43, 160–162. [DOI] [PubMed] [Google Scholar]
- Sesma, L., Montserrat, V., Lamas, J. R., Marina, A., Vazquez, J. & López de Castro, J. A. (2002). J. Biol. Chem.277, 16744–16749. [DOI] [PubMed] [Google Scholar]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed] [Google Scholar]
- Vega, M. A., Bragado, R., Ivanyi, P., Pelaez, J. L. & López de Castro, J. A. (1986). J. Immunol.137, 3557–3565. [PubMed] [Google Scholar]
- Webb, A. I., Borg, N. A., Dunstone, M. A., Kjer-Nielsen, L., Beddoe, T., McCluskey, J., Carbone, F. R., Bottomley, S. P., Aguilar, M. I., Purcell, A. W. & Rossjohn, J. (2004). J. Immunol.173, 402–409. [DOI] [PubMed] [Google Scholar]
- Weiss, M. (2001). J. Appl. Cryst.34, 130–135. [Google Scholar]
- Zernich, D., Purcell, A. W., Macdonald, W. A., Kjer-Nielsen, L., Ely, L. K., Laham, N., Crockford, T., Mifsud, N. A., Bharadwaj, M., Chang, L., Tait, B. D., Holdsworth, R., Brooks, A. G., Bottomley, S. P., Beddoe, T., Peh, C. A., Rossjohn, J. & McCluskey, J. (2004). J. Exp. Med.200, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]