A DEDDh-type 3′-5′ oligoribonuclease from the plant pathogen X. campestris pv. campestris has been overexpressed in E. coli, purified and crystallized. The crystals diffracted to 2.1 Å with good quality.
Keywords: 3′-5′ oligoribonucleases, Orn, DEDDh, Xanthomonas campestris
Abstract
Oligoribonucleases are essential components of RNA and DNA metabolism and close homologues of genes encoding them are found not only in prokaryotes but also in a wide range of eukaryotes, including yeast and humans. Inactivation of the oligoribonuclease gene (orn) can result in cellular lethality. Despite their important biological function, they have been studied little from a structural point of view. In this report, the cloning, expression, crystallization and preliminary X-ray analysis of XC847, a DEDDh-type 3′-5′ oligoribonuclease from the plant pathogen Xanthomonas campestris pv. campestris, a Gram-negative bacterium causing major worldwide disease of cruciferous crops, is described. The XC847 crystals diffracted to a resolution of at least 2.1 Å. They are tetragonal and belong to space group P43212, with unit-cell parameters a = b = 67.5, c = 89.8 Å. One molecule is present per asymmetric unit.
1. Introduction
RNA metabolism plays an important role in cell viability (Ghosh & Deutscher, 1999 ▶) and requires a wide variety of distinct ribonucleases (RNases) to encompass all known reactions. In the case of mRNA, a steady-state concentration was found to be critical in gene regulation; dysregulation of mRNA half-life is implicated in several human diseases, including cancer, inflammation and Alzheimer’s disease (Hollams et al., 2002 ▶). Both endoribonucleases and exoribonucleases are required to complete the turnover of aged RNA molecules. Based on extensive data mining, six exoribonuclease superfamilies, as well as various subfamilies, have been identified (Zuo & Deutscher, 2001 ▶). Oligoribonuclease belongs to the DEDDh-type exoribonuclease superfamily and is responsible for degrading small oligoribonucleotides of two to five residues in length to mononucleotides (Zuo & Deutscher, 2001 ▶). It is distinct from the other known exoribonucleases of Escherichia coli (Yu & Deutscher, 1995 ▶) and knockout of its gene (orn) led to cellular lethality (Ghosh & Deutscher, 1999 ▶). Oligoribonucleases therefore constitute an important class of enzyme that deserve more thorough studies.
XC847 from the plant pathogen Xanthomonas campestris pv. campestris strain 17 (Xcc) is classified as belonging to the oligoribonuclease family in the Pfam database (Bateman et al., 2000 ▶). It contains 194 amino acids and shares a high degree of identity and similarity to most identified oligoribonucleases (Zuo & Deutscher, 2001 ▶). For example, it shares 76% identity with the oligoribonuclease from Xylella fastidiosa (gi|9106239), 58% with Ralstonia solanacearum (gi|17427954), 99.5% with X. campestris pv. campestris ATCC 33913 (gi|21113297), 52.6% with Escherichia coli (gi|67469688), 56% with Haemophilus influenzae (gi|1176352), 43.3% with Streptomyces griseus (gi|21221244), 37.1% with Mycobacterium tuberculosis (gi|54041582), 34.5% with human (gi|12643261) and 25.8% identity (37.5% similarity) with that from yeast (gi|1730818), respectively. This high degree of sequence conservation is present throughout the DEDDh domain including motif I, motif II and motif III and is unique to oligoribonucleases (Zuo & Deutscher, 2001 ▶)
To date, oligoribonucleases have been little studied from a structural point of view, although a crystal structure of H. influenzae oligoribonuclease was deposited several years ago (PDB code 1j9a) and a preliminary X-ray crystal study of E. coli oligoribonuclease was reported recently (Fiedler et al., 2004 ▶). Interestingly, a crystal structure of human ISG20 (PDB code 1wlj), an interferon-induced antiviral ribonuclease that shares only a low 14% sequence identity with XC847, has also been reported and found to contain a similar DEDDh domain (Horio et al., 2004 ▶). In addition, the crystal structure of the RNase domain of Saccharyomyces cerevisiae Pop2 (PDB code 1uoc) has also been determined recently (Thore et al., 2003 ▶). It also belongs to the DEDD superfamily, but contains two non-canonical active-site residues.
2. Materials and methods
2.1. Cloning, expression and purification
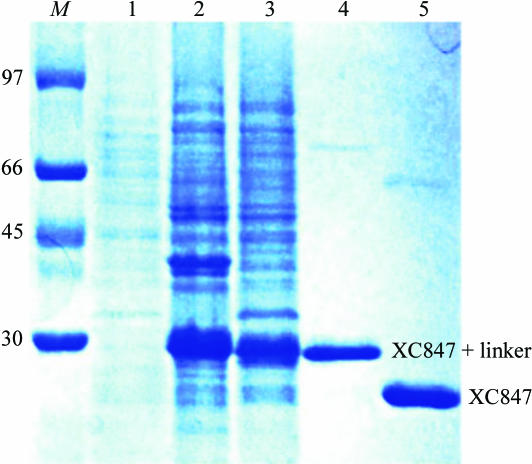
The XC847 gene fragment was PCR amplified directly from a local Xcc genome with a forward 5′-TACTTCCAATCCAATGCTATGGCAGACAACGTTGCGGGCA primer and a backward 5′-TTATCCACTTCCAATGTCAGTTCTGCACACCGCCGCCGTT primer. A ligation-independent cloning (LIC) approach (Aslanidis & de Jong, 1990 ▶) was carried out to obtain the desired construct. A pMCSG7 vector (Gao, unpublished results) was cut to completion with SspI (Novagen). For LIC, 100 ng of the linearized vector and 100 ng of the PCR product were treated with 2 units of T4 DNA polymerase (Novagen) in separate reactions in the presence of 2.5 mM dGTP or dCTP and 5 mM DTT. The reactions were carried out at 298 K for 30 min. The enzyme was subsequently heat-inactivated at 348 K for 20 min. The vector and the PCR product were then mixed and heated at 301 K for 3 min before cooling to room temperature. The mixture was directly transformed into the E. coli BL21 (DE3) host without ligation. The final construct codes for a N-terminal His6 tag, a 17-amino-acid linker and a XC847 target protein (194 amino acids) under the control of a T7 promoter. The transformed E. coli BL21 (DE3) host cell was grown in LB medium at 310 K until an OD600 of 0.8 was attained. Overexpression of the Hig6-tagged target protein was induced by the addition of 1 mM IPTG at 293 K for 20 h. The cells were harvested, resuspended in equilibration buffer (20 mM Na2HPO4, 70 mM NaCl pH 8.0) and lysed using a microfluidizer (Microfluidics). Most tagged target proteins were in the soluble fraction (Fig. 1 ▶). After centrifugation, the target protein was purified by immobilized metal-affinity chromatography (IMAC) on a nickel column (Sigma), which was eluted with 20 mM Tris pH 8.0, 70 mM NaCl and a gradient of 50–300 mM imidazole. The fractions containing XC847 were monitored by SDS–PAGE, recombined and dialyzed repeatedly against 50 mM Na2HPO4 pH 8.0, 10% glycerol and 500 mM NaCl. After buffer exchange, the His6 tag and linker were cleaved from XC847 by TEV (tobacco etch virus) protease at 283 K for 12 h to obtain the cleaved product. Without the His6 tag, the target XC847 protein cannot bind to the nickel column and was collected in the flowthrough fractions, with the His6 tag and the tagged TEV protease being retained on the nickel column. The purified protein was then dialyzed several times against 20 mM Tris pH 8.0 and 70 mM NaCl. For crystallization, XC847 was further purified on an anion-exchange column (AKTA, Pharmacia Inc.). The fractions eluted with 20 mM Tris pH 8.0, 400 mM NaCl were combined and dialyzed against 20 mM Tris pH 8.0 and 70 mM NaCl. The final target protein (194 amino acids) has a greater than 99% purity (Fig. 1 ▶) and contains only an extra tripeptide (SNA) at the N-terminal end. The overexpression and purification of XC847 was monitored by SDS–PAGE as shown in Fig. 1 ▶.
Figure 1.
SDS–PAGE monitoring of the overexpression and purification of XC847. Lane M, molecular-weight markers in kDa; lane 1, whole cell lysate before IPTG induction; lane 2, whole cell lysate after IPTG induction; lane 3, soluble fraction after IPTG induction; lane 4, purified XC847 before TEV cleavage; lane 5, purified XC847 after TEV cleavage. The positions of tagged and free XC1739 are also marked.
2.2. Crystallization
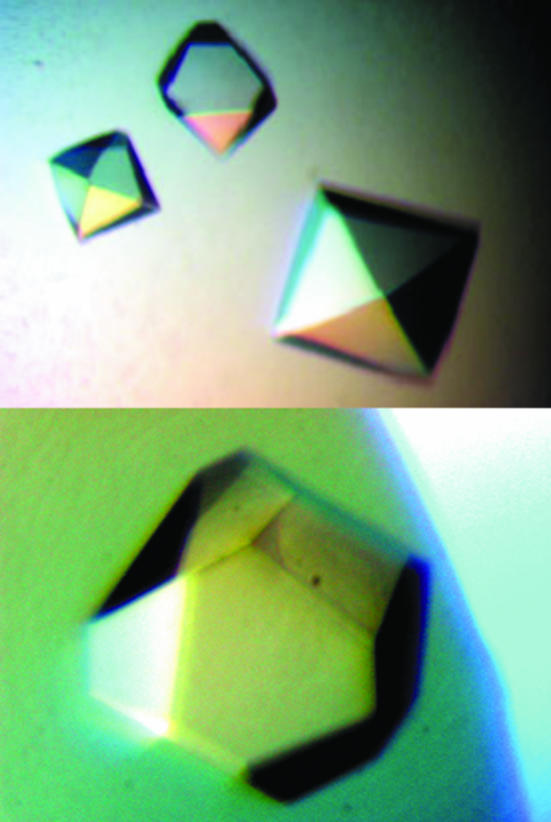
For crystallization, the protein was concentrated to 22 mg ml−1 in 20 mM Tris pH 8.0 and 70 mM NaCl using an Amicon Ultra-10 (Millipore). Crystallization screening was performed using the sitting-drop vapour-diffusion method in 96-well plates (Hampton Research) at 295 K by mixing 0.5 µl protein solution with 0.5 µl reagent solution. Initial screens included the Hampton sparse-matrix Crystal Screens 1 and 2, a systematic PEG–pH screen and a PEG/Ion screen and were performed using a Gilson C240 crystallization workstation. Pyramid-shaped crystals appeared in 2 d from a reservoir solution comprising 0.1 M Tris buffer pH 8.5, 0.2 M MgCl2 and 25%(v/v) PEG 4K MME (polyethylene glycol monomethyl ether). This initial condition was subsequently optimized by varying the concentrations of MgCl2 and PEG, the best concentrations of which were found to be 0.24 M and 25%(v/v), respectively. Pyramid-shaped crystals suitable for diffraction experiments were grown by mixing 1.5 µl protein solution with 1.5 µl reagent solution and reached maximum dimensions of 1 × 1 × 0.5 mm after 3 d (Fig. 2 ▶).
Figure 2.
Spindle-shaped crystals of XC847 from X. campestris obtained by the sitting-drop vapour-diffusion method. The final crystallization condition was 0.1 M Tris buffer pH 8.5, 0.24 M MgCl2 and 25% PEG 4K MME. These crystals reached approximate dimensions of 1.0 × 1.0 × 0.5 mm after 3 d.
2.3. Data collection
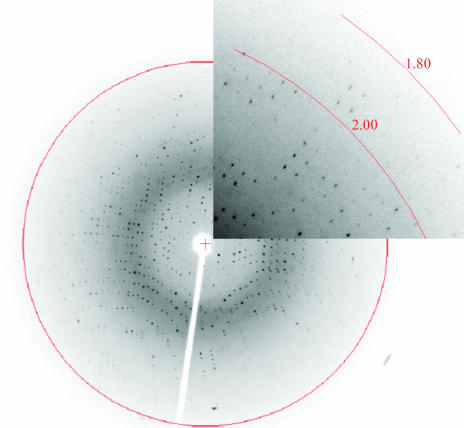
Although XC847 crystals diffracted to better than 2 Å resolution at 100 K, they seemed to suffer from cracking at this temperature using a wide variety of cryoprotectants. This resulted in a twinned diffraction pattern that could not be solved by flash-cooling, annealing or back-soaking. In order to eliminate this problem, the crystals were mounted in a capillary and data were collected at room temperature; a single set of high-quality diffraction data was obtained successfully in this way. The full data set was collected using Cu Kα radiation from an MSC/Rigaku RU300 rotating-anode generator equipped with Osmic mirror optics and an R-AXIS IV++ image-plate detector. A 2.1 Å resolution native data set was obtained. The data were indexed and integrated using the HKL processing software (Otwinowski & Minor, 1997 ▶), giving a data set that was 98.5% complete with an overall R merge of 6.7% on intensities. The crystals belong to the tetragonal space group P43212. The data-collection statistics are summarized in Table 1 ▶, with an X-ray diffraction image collected in-house shown in Fig. 3 ▶.
Table 1. Data-collection statistics for native XC847 crystals.
Values in parentheses are for the highest resolution shell.
| Space group | P43212 |
| Unit-cell parameters (Å) | a = b = 67.5, c = 89.8 |
| Data-collection temperature (K) | 300 |
| Wavelength (Å) | 1.5418 |
| Resolution range (Å) | 47.80–2.10 (2.18–2.10) |
| Mosaicity (°) | 0.5 |
| Unique reflections | 12608 (1248) |
| Redundancy | 4.5 (4.7) |
| Completeness (%) | 98.5 (100.0) |
| Rmerge (%) | 6.7 (35.6) |
| Mean I/σ(I) | 20.4 (6.6) |
| Solvent content (%) | 41.9 |
| Matthews coefficient† (Å3 Da−1) | 2.1 |
Matthews (1968 ▶).
Figure 3.
Picture of the diffraction pattern of XC847 collected in-house from a capillary-mounted crystal at room temperature. The exposure time was 12 min, with an oscillation range of 1.0° and a crystal-to-detector distance of 180 mm. The 2.0 Å resolution ring is also marked.
3. Results and discussion
The gene for XC847 consists of 585 bp coding for 194 amino-acid residues with an isoelectric point of 5.46. Purified XC847 showed a single band of approximately 22 kDa on SDS–PAGE (Fig. 1 ▶), consistent with the expected weight of 22 050 Da.
Although the DEDDh motif is very well conserved among all oligoribonucleases, many DNases were also found to contain this motif despite having very low sequence identity with XC847. For example, the E. coli DNA polymerase III ∊ subunit (∊ 186) has only 19.7% sequence identity (Hamdan et al., 2002 ▶), the E. coli exonuclease I (ExoI) 10% identity (Breyer & Matthews, 2000 ▶) and the human ISG20 antiviral ribonuclease 14% identity with XC847 (Horio et al., 2004 ▶), respectively. Yet they all share similar active-site residues, strongly suggesting that these enzymes share a common catalytic mechanism (Steitz & Steitz, 1993 ▶; Steitz, 1999 ▶). However, since oligoribonuclease is an exoribonuclease specific for small oligoribonucleotides of two to five residues in length (Yu & Deutscher, 1995 ▶), it remains to be seen how this specificity is achieved. Cocrystal studies of XC847 with oligoribonucleotides of 2–5 and 6–10 nucleotides in length in the absence of catalytic metal ions (Steitz & Steitz, 1993 ▶) may provide an answer. Such studies are now ongoing.
The high-resolution diffraction obtained from the native crystals establishes its suitability for X-ray structural analysis (Fig. 3 ▶). We have also examined the extent of crystal twinning using the twinning modules from the CNS software (Brünger et al., 1998 ▶). From the cumulative intensity statistics of 2.15 for the Stanley equation and 0.77 for the Wilson ratio, we conclude that our diffraction data are not subject to merohedral twinning (Yeates, 1997 ▶). Since a protein crystal structure from the H. influenzae genome sharing a high sequence identity of 56% with XC847 has been deposited (PDB code 1j9a), it may be possible to use the molecular-replacement method to solve the XC847 structure. Indeed, we have successfully obtained the preliminary XC847 structure using this method assuming a P43212 space group. The detailed refined structural data of XC847 will be reported later.
Acknowledgments
This work is supported by the Academic Excellence Pursuit grant from the Ministry of Education and by the National Science Council, Taiwan to SHC and PCL. We also thank the Core Facilities for Protein Production in the Academia Sinica, Taiwan for providing us with the original vectors used in this study (Shih et al., 2002 ▶) and the Core Facilities for Protein X-ray Crystallography in the Academia Sinica, Taiwan for assistance in preliminary X-ray analysis.
References
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res.18, 6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S. R., Howe, K. L. & Sonnhammer, E. L. L. (2000). Nucleic Acids Res.28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyer, W. A. & Matthews, B. W. (2000). Nature Struct. Biol.7, 1125–1128. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Fiedler, T. J., Vincent, H. A., Zuo, Y., Gavrialov, O. & Malhotra, A. (2004). Acta Cryst. D60, 736–739. [DOI] [PubMed] [Google Scholar]
- Ghosh, S. & Deutscher, M. P. (1999). Proc. Natl Acad. Sci. USA, 96, 4372–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan, S., Carr, P. D., Brown, S. E., Ollis, D. L. & Dixon, N. E. (2002). Structure, 10, 535–546. [DOI] [PubMed] [Google Scholar]
- Hollams, E. M., Giles, K. M., Thomson, A. M. & Leedman, P. J. (2002). Neurochemical Res.27, 957–980. [DOI] [PubMed]
- Horio, T., Murai, M., Inoue, T., Hamasaki, T., Tanaka, T. & Ohgi, T. (2004). FEBS Lett.577, 111–116. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Shih, Y.-P., Kung, W.-M., Chen, J.-C., Yeh, C.-H., Wang, A. H.-J. & Wang, T.-F. (2002). Protein Sci.11, 1714–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, T. A. (1999). J. Biol. Chem.274, 17395–17398. [DOI] [PubMed] [Google Scholar]
- Steitz, T. A. & Steitz, J. A. (1993). Proc. Natl Acad. Sci. USA, 90, 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore, S., Mauxion, F., Seraphin, B. & Suck, D. (2003). EMBO Rep.4, 1150–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates, T. O. (1997). Methods Enzymol.276, 344–358. [PubMed] [Google Scholar]
- Yu, D. & Deutscher, M. P. (1995). J. Biol. Chem.177, 4137–4139.
- Zuo, Y. & Deutscher, M. P. (2001). Nucleic Acids Res.29, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]