Recombinant cryptochrome 3 from A. thaliana with FAD and MTHF cofactors has been crystallized using the hanging-drop vapour-diffusion technique in the orthorhombic space group P212121 and X-ray diffraction data were collected to 1.9 Å resolution.
Keywords: cryptochrome 3, light receptors
Abstract
Cryptochromes are flavoproteins which serve as blue-light receptors in plants, animals, fungi and prokaryotes and belong to the same protein family as the catalytically active DNA photolyases. Cryptochrome 3 from the plant Arabidopsis thaliana (cry3; 525 amino acids, 60.7 kDa) is a representative of the novel cryDASH subfamily of UV-A/blue-light receptors and has been expressed as a mature FAD-containing protein in Escherichia coli without the signal sequence that directs the protein into plant organelles. The purified cryptochrome was found to be complexed to methenyltetrahydrofolate as an antenna pigment. Crystals of the cryptochrome–antenna pigment complex were obtained by vapour diffusion and display orthorhombic symmetry, with unit-cell parameters a = 76.298, b = 116.782, c = 135.024 Å. X-ray diffraction data were collected to 1.9 Å resolution using synchrotron radiation. The asymmetric unit comprises a cry3 dimer, the physiological role of which remains to be elucidated.
1. Introduction
Cryptochromes and DNA photolyases share a striking similarity in their amino-acid sequence and cofactor composition. Whereas DNA photolyases repair UV-damaged DNA by using light energy in the UV-A/blue region and are classified either as a cyclobutane pyrimidine dimer (CPD) or as a (6–4) photolyase, according to their substrate specificity, cryptochromes function as UV-A/blue-light receptors in plants, the fruit fly and bacteria or in mammals as central components of the circadian clock (Sancar, 2003 ▶).
Three crystal structures of microbial-type (class I) CPD photolyase have been solved. These structures from Escherichia coli (Park et al., 1995 ▶), Anacystis nidulans (Tamada et al., 1997 ▶) and Thermus thermophilus (Komori et al., 2001 ▶) share a common U-shaped FAD cofactor. This cofactor is essential for catalysis in its fully reduced state and after excitation transfers an electron to the DNA photoproduct, resulting in an unstable pyrimidine dimer or (6–4) photoproduct radical. This radical spontaneously splits the covalent bonds between the pyrimidines and thus directly reverts the DNA damage (Sancar, 2003 ▶). Excitation of the flavin either occurs via direct light absorption or is mediated through energy transfer from a second cofactor that acts as an antenna pigment. In E. coli and many other photolyases, the antenna pigment is methenyltetrahydrofolate (MTHF), which is located at a distance of 16.8 Å from the FAD (Park et al., 1995 ▶). The cyanobacterial Anacystis photolyase contains 8-hydroxy-5-deazaflavin (8-HDF) as an antenna pigment at a similar antenna–FAD distance of 17.5 Å (Tamada et al., 1997 ▶). For the T. thermophilus photolyase, no other cofactor than FAD has yet been found (Komori et al., 2001 ▶). Whereas the above-mentioned photolyase structures lack bound substrates, the crystal structure of the A. nidulans photolyase complexed with a synthetic CPD DNA lesion was recently solved (Mees et al., 2004 ▶), which mimics the bound substrate during photorepair. Based on these structures together with spectroscopic and biochemical data, the reaction mechanism of class I CPD photolyases is now well understood.
Recently, the structures of two cryptochromes were also solved, namely the Synechocystis cryDASH structure (Brudler et al., 2003 ▶) and the structure of the N-terminal photolyase-like domain of Arabidopsis cryptochrome 1 (cry1) (Brautigam et al., 2004 ▶). Both structures contain the flavin but no antenna pigment. The overall structures of both cryptochromes are very similar to those of the DNA photolyases. However, there are also several marked differences between the cryptochrome and photolyase structures, in the flavin and photolyase substrate-binding pocket for example, that could explain the lack of DNA-repair activity of cryptochromes (Brudler et al., 2003 ▶). In addition, there are differences between the two cryptochrome structures; for example, there are surface features that could be responsible for the different DNA-binding properties of Arabidopsis cry1 and Synechocystis cryDASH. The phylogenetic trees of the photolyase/cryptochrome family show that the animal cryptochromes, for which no structure has been solved, are closely related to (6–4) photolyases, while most of the plant crytochromes group together with class I CPD photolyase and the recently discovered cryDASH members form a separate subfamily (Daiyasu et al., 2004 ▶). Members of the cryDASH family have been identified in cyanobacteria (Brudler et al., 2003 ▶), plants (Kleine et al., 2003 ▶), eubacteria (Worthington et al., 2003 ▶; Daiyasu et al., 2004 ▶), fungi and aquatic vertebrates (Daiyasu et al., 2004 ▶). To elucidate the structural features of cryptochromes in general and of subfamily-specific aspects from which conclusions about structure–function relations and functional differences among DNA photolyases can be drawn, it is necessary to solve the structures of more than one member of each family.
2. Material and methods
2.1. Cloning and overexpression
The cloning of mature A. thaliana cryptochrome 3 (cry3; amino acids Met1–Pro526; GenBank protein accession No. NP 568461), which lacks the putative dual targeting signal for import into chloroplasts and mitochondria, into the vector pQE-60 (Qiagen, Hilden, Germany) has previously been described (Kleine et al., 2003 ▶). Cry3 was overexpressed in E. coli M15(pREP4) cells that were first grown in 50 ml LB medium containing 100 µg ml−1 ampicillin and 25 µg ml−1 kanamycin at 310 K overnight by shaking at 250 rev min−1. This overnight culture was used to inoculate 1.5 l of the same medium and cells were grown at 303 K to OD600 ≃ 0.7–0.8. Induction of cry3 expression was achieved by the addition of 1 mM IPTG and further incubation at 303 K for another 12 h. Cells were harvested by centrifugation at 4000g and 277 K for 20 min, washed by resuspending in PBS (∼1/50th of the original culture volume), recentrifuged as above and finally resuspended in the same volume of lysis buffer (50 mM sodium phosphate pH 7.5, 300 mM NaCl, 10 mM β-mercaptoethanol, 10 mM imidazole, 0.5% Triton X-100 and 10% glycerol) containing protease inhibitors (10 mM benzamidine, 5 µg ml−1 E-64, 6 µg ml−1 pepstatin A and 4.5 mM PMSF) and lysozyme (1 mg ml−1). Cells were disrupted using a French press and debris was removed by centrifugation at 40 000g and 277 K for 25 min. The supernatant was filtered through a 0.2 µm membrane filter (Sarstedt, Nümbrecht, Germany) and used for cry3 purification.
2.2. Purification
The recombinant cry3 was purified by three chromatographic steps on an ÄKTA purifier (Amersham Biosciences, Buckinghamshire, UK). As a first step, Ni2+-affinity chromatography on a HisTrap HP column was used. The column was pre-equilibrated with buffer containing 50 mM sodium phosphate pH 7.5, 200 mM NaCl, 10 mM β-mercaptoethanol, 10 mM imidazole and 10% glycerol and the bound protein was eluted with increasing concentration of imidazole in the above buffer in four steps: 40 mM [washing step, three column volumes (CV)], 100, 250 and 500 mM (all 5 CV). Fractions from the last step containing the main portion of cry3 in a relatively purified state were pooled and cry3 was further purified by heparin chromatography on a HiTrap Heparin HP column pre-equilibrated with buffer containing 50 mM sodium phosphate pH 7.5, 200 mM NaCl, 10 mM β-mercaptoethanol and 10% glycerol. The bound protein was eluted with a short step (2 CV) of 300 mM NaCl in the above buffer followed by a linear gradient of 0.3–2 M NaCl in 5 CV. Fractions containing cry3 were pooled, concentrated by Amicon ultracentrifugal filter concentrator with 30 kDa cutoff (Millipore, Bedford, MA, USA) and subjected to size-exclusion chromatography on a Superdex 200 GL column in crystallization buffer containing 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM β-mercaptoethanol and 10% glycerol. Purified cry3 was concentrated to about 10 mg ml−1 as estimated by the Bradford method and analyzed by SDS–PAGE under reducing conditions (Fig. 1 ▶ a). The identity of Cry3 was confirmed by MALDI–TOF MS analysis. Obtained fragments covered 42% of the whole Cry3 sequence (GenBank protein accession No. NP 568461) and were 100% identical to corresponding fragments from this sequence. The cry3 protein was either immediately used for crystallization (cry3 was stable only for 1–2 d in this buffer at 277 K) or divided into several aliquots, flash-frozen in liquid nitrogen and stored at 193 K. The yield of purified protein was 2–3 mg per litre of induced E. coli culture.
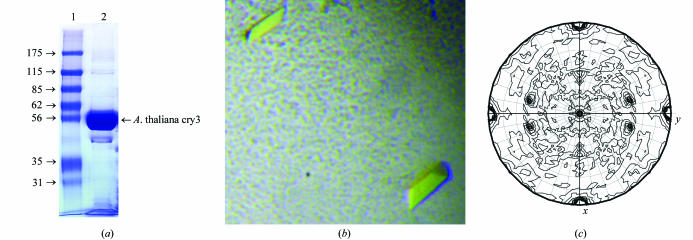
Figure 1.
(a) Polyacrylamide gel electrophoresis under denaturing conditions of purified A. thaliana cry3. Lane 1, Sigma molecular-weight markers 7B (kDa; Sigma-Aldrich, St Louis, MO, USA). Lane 2, purified cry3 (10 µg) used for crystallization. The gel was stained with Coomassie blue. (b) Typical morphology of A. thaliana cry3 crystals. Crystals were obtained by equilibration against a solution of 85 mM sodium citrate pH 4.6, 170 mM ammonium acetate, 21.5% PEG 4000 and 7.5% glycerol. The average dimensions of the cry3 crystals were about 0.2 × 0.1 × 0.1 mm. (c) Self-rotation function calculated with a native A. thaliana cry3 data set employing data between 20.0 and 2.5 Å resolution. A self-rotation search with the program MOLREP at the angle χ = 180° was used to identify the twofold rotation axis.
2.3. UV–Vis spectroscopy
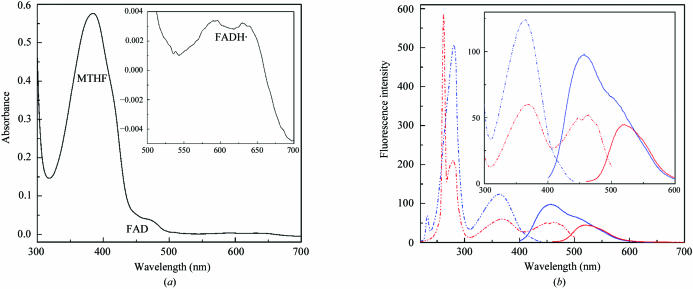
UV–Vis absorption spectra of purified cry3 (Fig. 2 ▶ a) were recorded using a UV-2401 PC spectrophotometer (Shimadzu, Kyoto, Japan) and were dominated by a 384 nm peak, which suggests the presence of MTHF as recently found for the Vibrio cholerae cryptochrome VcCry1 (Saxena et al., 2005 ▶). Shoulders at 450 and 480 nm are typical for an oxidized FAD species and some minor peaks at 590 and 640 nm indicate the presence of the neutral flavin radical FADH˙. Further evidence for the presence of both cofactors was provided by fluorescence spectroscopy. Emission and excitation spectra of cry3 (Fig. 2 ▶ b) were recorded using an RF-5301 PC spectrofluorophotometer (Shimadzu, Kyoto, Japan). Emission spectra obtained by excitation at 380 nm were dominated by the 460–480 nm peak, which corresponds to the excited MTHF* state and overlaps the flavin emission peak at 520 nm from emission spectra obtained by excitation at 440 nm, suggesting resonance energy transfer from MTHF to flavin. Three peaks were obtained in excitation spectra for emission at 460 nm: one at 380 nm that corresponds to MTHF absorption and two additional peaks in the UV-B region at 230 and 280 nm. In excitation spectra for emission at 520 nm, four peaks were found: two in the UV-B region (at 260 and 280 nm), one at 380 nm and one at 460 nm corresponding to flavin absorption alone or to the MTHF absorption and subsequent energy transfer to the flavin cofactor. The presence of both cofactors was further confirmed by TOF MS analysis. Observed peaks with weights of 455.2 and 784.2 Da corresponded to calculated MTHF (456.4 Da) and FAD (785.6 Da) weights, respectively. A peak with a weight of 472.9 Da could indicate some degradation of MTHF into N5-formyl-THF (calculated weight of 473.5 Da).
Figure 2.
(a) Absorption spectrum of purified A. thaliana cry3. The dominant absorption peak at 384 nm typical of MTHF absorption and the shoulders at 450 and 480 nm typical of absorption of two-electron oxidized form of FAD are clearly visible. Inset: absorption spectra in the 500–700 nm region shown at larger resolution with two peaks (at 590 and 640 nm) typical of the absorption of flavin neutral radical FADH˙. The concentration of cry3 was 1.3 mg ml−1. (b) Fluorescence emission and excitation (b) spectra of purified A. thaliana cry3. Emission spectra are shown as solid lines and excitation spectra as broken lines. Emission of excited flavin showing a peak at 520 nm and excitation spectra for this emission are shown in red. Emission of excited MTHF* with a peak at 460 nm overlapping the flavin emission peak and excitation spectra for emission at 460 nm are shown in blue. Inset: emission and excitation spectra in the 300–600 nm region shown at larger resolution. The concentration of cry3 was 50 µg ml−1.
2.4. Crystallization
Preliminary crystallization conditions were screened by the sitting-drop vapour-diffusion method using 96-well plates (Greiner, Frickenhausen, Germany) in combination with a Cartesian Microsys 4004 crystallization robot and several different crystallization screens at 291 K. Each drop was situated over 100 µl reservoir solution and contained 0.3 µl of cry3 sample (either 10 or 5 mg ml−1 in the above-mentioned crystallization buffer) and the same volume of reservoir solution. Crystals appeared in about a week in condition No. 9 of the Sigma Crystallization Cryo Kit (Sigma-Aldrich, St Louis, MO, USA), which comprised sodium citrate, PEG 4000 and glycerol. The crystallization condition was optimized by the hanging-drop vapour-diffusion method using 24-well plates (Nextal Biotechnologies, Montreal, Quebec, Canada) and systematic variations of sodium citrate (pH 3.8–5.6), PEG 4000 [21–28%(w/v)] and glycerol [5–15%(v/v)]. Each drop contained 1 µl of cry3 sample (10 mg ml−1) and the same volume of reservoir solution over 750 µl of the same reservoir solution. The best crystals were obtained after 3–7 d in 85 mM sodium citrate pH 4.6, 170 mM ammonium acetate, 21.5%(w/v) PEG 4000 and 7.5%(v/v) glycerol. Crystals grew usually as single column-shaped crystals with green–yellowish colour (Fig. 1 ▶ b) and were directly frozen from their mother liquor.
3. Data collection and processing
A complete 1.9 Å data set was collected from a single frozen crystal at beamline X11 at the DORIS III storage ring, DESY; Hamburg. Data processing was carried out in the orthorhombic system using the programs MOSFLM and SCALA (Collaborative Computational Project, Number 4, 1994 ▶). The mosaicity of the data set was estimated to be 0.27°. The presence of systematic absences clearly indicated space group P212121 (unit-cell parameters a = 76.3, b = 116.8, c = 135.0 Å). Data-collection and processing statistics are given in Table 1 ▶.
Table 1. Data-collection and processing statistics for native A. thaliana cry3.
Values in parentheses are for the highest resolution shell (2.00–1.90 Å).
| X-ray source and wavelength | X11, 0.8125 Å |
| Detector | MAR CCD 165 mm |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 76.30, b = 116.78, c = 135.02 |
| Resolution (Å) | 20–1.9 |
| Observations | 417113 |
| Unique reflections (F > 0) | 93259 |
| Mosaicity (°) | 0.27 |
| Wilson B factor (Å2) | 29 |
| I/σ(I) | 16.4 (2.2) |
| Rmerge | 0.051 (0.448) |
| Completeness (%) | 97.8 (85.9) |
4. Conclusions
Although the recombinant cry3 cryptochrome was not stable for long time periods, under low-pH conditions we could obtain an orthorhombic crystal form that diffracted to 1.9 Å resolution. Using the Matthews equation (Matthews, 1968 ▶), the solvent content of the crystal form was estimated to be 44% with two molecules per asymmetric unit (V M = 2.48 Å3 Da−1). The two cry3 molecules are related to each other by a twofold non-crystallographic symmetry axis (θ = 120.4°, ϕ = −73.6°) as shown by the self-rotation function calculated by MOLREP (Collaborative Computational Project, Number 4, 1994 ▶) using 20–2.5 Å data (Fig. 1 ▶ c).
A preliminary solution of the structure of the cry3 cryptochrome was found by molecular replacement using the crystal structure of Synechocystis cryDASH (Brudler et al., 2003 ▶; PDB code 1np7), with which the A. thaliana cry3 shares a sequence identity of 51% for residues Lys39–Leu485. As expected, the best result was obtained by MOLREP with two cryDASH molecules, giving a correlation coefficient of 35.5% and an R factor of 52.0% for data between 20 and 2.8 Å resolution. The resulting electron density is clear enough to build the regions of the cry3 dimer that were missing in the search model.
In contrast to the other crystallized cryptochromes, cry3 crystallized as a dimer and contains two cofactors: flavin and MTHF. The presence of two cofactors will allow us to draw conclusions on the distance, orientation and efficiency of energy transfer between MTHF and FAD in cryptochromes as soon as the structure is finally refined. Furthermore, dimerization of cryptochromes might be functionally important for the mediation of light-triggered conformational changes as recently exemplified for cry1 from A. thaliana (Sang et al., 2005 ▶).
Acknowledgments
This work was supported by DFG grants BA985/9-1 and BA985/7-3 to AB and DFG grant ES152/2-1 to L-OE. The authors are very grateful for support from Paul Tucker at the synchrotron beamline X11, EMBL, Hamburg and Julius Nyalwidhe and Uwe Linne for MALDI–TOF MS and TOF MS analyses.
References
- Brautigam, C. A., Smith, B. S., Ma, Z., Palnitkar, M., Tomchick, D. R., Machius, M. & Deisenhofer, J. (2004). Proc. Natl Acad. Sci. USA, 101, 12142–12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudler, R., Hitomi, K., Daiyasu, H., Toh, H., Kucho, K., Ishiura, M., Kanehisa, M., Roberts, V. A., Todo, T., Trainer, A. & Getzoff, E. D. (2003). Mol. Cell, 11, 59–67. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Daiyasu, H., Ishikawa, T., Kuma, K., Iwai, S., Todo, T. & Toh, H. (2004). Genes Cells, 9, 479–495. [DOI] [PubMed] [Google Scholar]
- Kleine, T., Lockhart, P. & Batschauer, A. (2003). Plant J.35, 93–103. [DOI] [PubMed] [Google Scholar]
- Komori, H., Masui, R., Kuramitsu, S., Yokoyama, S., Shibata, T., Inoue, Y. & Miki, K. (2001). Proc. Natl Acad. Sci. USA, 98, 13560–13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Mees, A., Klar, T., Gnau, P., Hennecke, U., Eker, A. P., Carell, T. & Essen, L. O. (2004). Science, 306, 1789–1793. [DOI] [PubMed] [Google Scholar]
- Park, H. W., Kim, S.-T., Sancar, A. & Deisenhofer, J. (1995). Science, 268, 1866–1872. [DOI] [PubMed] [Google Scholar]
- Sancar, A. (2003). Chem. Rev.103, 2203–2237. [DOI] [PubMed] [Google Scholar]
- Sang, Y., Li, Q. H., Rubio, V., Zhang, Y. C., Mao, J., Deng, X. W. & Yang, H. Q. (2005). Plant Cell, 17, 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, C., Wang, H., Kavakli, I. H., Sancar, A. & Zhong, D. (2005). J. Am. Chem. Soc.127, 7984–7985. [DOI] [PubMed] [Google Scholar]
- Tamada, T., Kitadokora, K., Higuchi, Y., Inaka, K., Yasui, A., de Ruiter, P. E., Eker, A. P. M. & Miki, K. (1997). Nature Struct. Biol.4, 887–891. [DOI] [PubMed] [Google Scholar]
- Worthington, E. N., Kavakli, I. H., Berrocal-Tito, G., Bondo, B. E. & Sancar, A. (2003). Biol. Chem.278, 39143–39154. [DOI] [PubMed]