Abstract
Cowpox virus Brighton red strain (CPV) contains a gene, crmD, which encodes a 320-aa tumor necrosis factor receptor (TNFR) of 44% and 22% identity, respectively, to the CPV TNFR-like proteins, cytokine response modifiers (crm) CrmB and CrmC. The crmD gene was interrupted in three other cowpox strains examined and absent in various other orthopoxviruses; however, four strains of ectromelia virus (ECT) examined contained an intact crmD (97% identity to CPV crmD) and lacked cognates of crmB and crmC. The protein, CrmD, contains a transport signal; a 151-aa cysteine-rich region with 21 cysteines that align with human TNFRII ligand-binding region cysteines; and C-terminal region sequences that are highly diverged from cellular TNFR C-terminal region sequences involved in signal transduction. Bacterial maltose-binding proteins containing the CPV or ECT CrmD cysteine-rich region bound TNF and lymphotoxin-α (LTα) and blocked their in vitro cytolytic activity. Secreted viral CrmD bound TNF and LTα and was detectable after the early stage of replication, using nonreducing conditions, as 60- to 70-kDa predominant and 90- to 250-kDa minor disulfide-linked complexes that were able to be reduced to a 46-kDa form and deglycosylated to a 38-kDa protein. Cells infected with CPV produced extremely low amounts of CrmD compared with ECT. Possessing up to three TNFRs, including CrmD, which is secreted as disulfide-linked complexes in varied amounts by CPV and ECT, likely enhances the dynamics of the immune modulating mechanisms of orthopoxviruses.
Poxviruses are able to acquire cellular components that interact with the cytokine network (1), including components that bind tumor necrosis factor (TNF) superfamily proteins. Insight into the concept that poxviruses can modulate host responses by using such proteins began with the discovery of a leporipoxvirus gene for a glycoprotein termed T2 that contains a region with four pseudo-repeat, cysteine-rich domains (CRDs) similar to those in cellular TNF receptor type II (TNFRII), a pleiotropic protein that modulates immune processes, including activating cell proliferation or apoptosis by signal transduction (2–4). T2 is secreted early after infection of cells as soluble 40.5-kDa monomers and 80.9-kDa dimers that can block rabbit TNF α cytolysis of mouse L cells and apoptosis of rabbit CD4+ lymphoma cells (5–7). Deleting the gene for T2, which is diploid because it is a genome inverted terminal repeat component, reduced myxoma lethality for European rabbits (8).
Of 14 vaccinia strains examined, only strains Lister and USSR produced TNF binding proteins (A. Alcami, personal communication). The protein of both strains is encoded by a protracted version of the reading frame A53R in the genome of vaccinia strain Copenhagen (9, 10). The genome inverted terminal repeat of the Copenhagen strain also contains another truncated and likely nonfunctional TNFR correlate encoded by the left-end reading frame C22L, and its duplicate, the right-end reading frame B28R (10). However, another orthopoxvirus, cowpox virus strain Brighton red (CPV) produces elongated, ligand-binding versions of the putative proteins specified by C22L/B28R and A53R—CPV cytokine response modifier (crm) CrmB and CrmC, respectively (11, 12). CrmB, which is 48% identical to T2, is an early-class, 355-aa protein that can block cytolysis by lymphotoxin α (LTα) and TNF. CrmC is a late-class 186-aa secreted protein that can bind TNF, but not LTα, and contains three CRDs.
In addition, variola (smallpox), monkeypox, and camelpox orthopoxviruses show at the genome right-end a 348-codon reading frame (termed G2R in variola) for a protein of 85% amino acid sequence homology to CrmB (13–15); however, these viruses appear to produce the CrmB-like analog late-in-infection (V.N.L., unpublished data). The cysteine-rich region of CrmB and the analogs is about 50% identical to the corresponding region of human TNFRII, and the cognate region of CrmC is 39% identical.
The present report describes a third TNFR-like glycoprotein of orthopoxviruses, termed CrmD, which is produced substantially less abundantly by CPV than the virulent ectromelia virus strain Moscow (ECT). CrmD is secreted from infected cells as disulfide-linked complexes that can bind and neutralize TNF and LTα. Possession of genes for up to three, perhaps more, TNFRs, that produce different amounts and distinctive secreted forms of ligand-binding proteins support the view that poxviruses have adapted such proteins into a dynamic array of survival components to counteract the wide assortment of host defense measures.
MATERIALS AND METHODS
Identification and Sequencing DNA Coding for CrmD.
A DNA segment near the right-end inverted terminal repeat of CPV DNA was cloned as 3.4-kb KpnI and 1.3-kb KpnI–HindIII fragments that we sequenced by primer walking, fluorescence-based methods (Applied Biosystems). Amplification of DNA encoding CrmD in other orthopoxviruses was done by using primers cpxaF15 (5′-ACCTAGTTAATGATATCTATG) and cpxbR1 (5′-TCACATCGATGATCTGAATATG); amplicons were resolved by gel electrophoresis, excised from gels, purified with the Wizard Prep kit (Promega), and then sequenced. Hitachi Software Engineering DNASIS, Genetics Computer Group (16), and blast (17) software were used to assemble and analyze sequences.
Cells, Viruses, and DNA Preparations.
Viruses were propagated in monkey kidney (BSC-1 or BSC-40), mouse L929, or Rat-2 cells grown in DMEM plus 5% fetal calf serum (FCS), or in human 143B cells grown in Eagle’s minimal essential medium plus 5% FCS. Genome DNA was prepared (18) from cells infected with CPV or cowpox strains Munich-89/5, -90/1, or -EP/2; ECT or ectromelia Munich-MP3, -MP4, or -SF; vaccinia Western Reserve (VAC) or Copenhagen; variola Bangladesh (VAR); monkeypox Copenhagen; or camelpox Somalia.
CrmD expression was examined by using ECT, CPV, and the white pock variant CPV-W2, which lacks crmD (19); recombinant CPV-A513, which lacks crmC expression (12); VAC, which lacks expression of crmB, crmC, and crmD (A. Alcami, personal communication; also see Fig. 3); and a coinfection system that uses VAC-VTF7–3 (20) and VAC-A607 to express CrmD via the phage T7 RNA polymerase promoter. VAC-A607 was constructed by using pTM1 (21) as follows: primers JFP4 (5′-GCTATCATGACACCATCATACATCTTGTTGGTATAT) and JFP5 (5′-TACGTGAATTCAATCTCTTTCACAATCATTTGGTGG) were used to amplify the crmD ORF from a chimeric plasmid, p1292 that contains a 4.5-kb EcoRI–PstI segment of the CPV genome PstI fragment L, which encompasses crmD. The 1-kb amplicon cleaved with BspHI and EcoRI and ligated into NcoI- and EcoRI-cleaved pTM1 created plasmid p1893, which was sequenced. The expression cassette in p1893 was recombined into VAC to create VAC-A607. CrmD was expressed by coinfecting cells with VAC-A607 and VAC-VTF7–3, which expresses the T7 RNA polymerase (20).
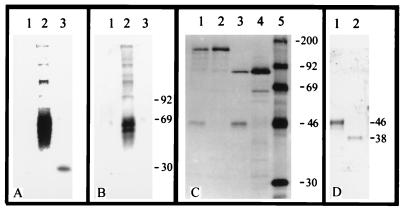
Figure 3.
Reactivity of TNF and anti-MBP-CPV serum with CrmD. (A and B) Ligand and Western blots, respectively, of proteins in ×20 concentrated medium from Rat-2 cell cultures 12 hpi with VAC (lane 1), ECT (lane 2), or CPV (lane 3). Proteins were resolved by 12% SDS/PAGE and nonreducing conditions, transferred to membranes, and then reacted with (A) 125I-TNF or (B) anti-MBP-CPV serum. (C) Radioimmunoprecipitation of anti-MBP-CPV serum reactivity with proteins in lysates of human 143B cells infected with CPV (lane 1), CPV-W2 (lane 2), VAC-VTF7–3 and VAC-A607 (lane 3), VAC-VTF7–3 (lane 4), or marker proteins (lane 5). (D) Western blot of anti-MBP-CPV serum reaction with proteins in medium of cell culture at 12 hpi with ECT; concentrated medium was reduced with β-mercaptoethanol (lane 1) or enzymatically deglycosylated (Bio-Rad Enzymatic Deglycosylation Kit) and then reduced (lane 2).
Escherichia coli Expression of CrmD Cysteine-Rich Region.
Sequences encoding 218 amino acids, including the 151-residue CRD region of CrmD, were amplified from CPV or ECT genome DNAs by using forward primers VLcrmD-B1 (5′-CGAATTCGGAGATGTTCCTTATG) for CPV and VLcrmD-B2 (5′-CGAATTCGGAGATGTTCCGTATAC) for ECT, and reverse primer VLcrmD-E3 (5′-GGTCGACTTAGCACTGCGTTGTGAGG). Methods from New England Biolabs were used to ligate amplicons into pMAL-p2; to express, using E. coli XL1-blue, a 610-residue maltose-binding protein (MBP) containing a factor-Xa cleavage site between the MBP and the ECT or CPV CRD; and to purify by amylose affinity chromatography the CPV CRD fusion protein (MBP-CPV) and the ECT CRD fusion protein (MBP-ECT).
Antiserum Against CPV CrmD CRD.
Methods from Boehringer-Mannheim were used to digest the MBP-CPV with biotinylated factor-Xa and remove factor-Xa by avidin-affinity chromatography. The digested MBP-CPV (100 μg of protein/100 μl PBS, pH 7.4) was emulsified 1:1 in TiterMax R-1 adjuvant (CytRx) and then injected intramuscularly into rabbits (New Zealand White) monthly for 4 months. Western blot assays (22) using 12% gels in the SDS/PAGE (23) and bovine lacto transfer technique optimizer (Blotto) buffer (50 mM Tris⋅HCl, pH 7.4/100 mM NaCl/5% nonfat dry milk) (24) containing 3 μg MBP/ml in the blocking and antibody reactions were used to determine the rabbit antibody reactivities against native, denatured, or factor-Xa-digested MBP-CPV.
Biosensor Analysis.
Binding of human or rat TNF or human LTα [10 μg/ml (BioSource International, Camarillo, CA)] by MBP-CPV, MBP-ECT, and components in culture fluid from virus-infected Rat-2 cells were examined by using Pharmacia methods for the BIAcore continuous flow apparatus (25) in which ligands were immobilized (Pharmacia Amine Coupling Kit) on CM5-dextran-coated sensors in separate flow cells. Fusion protein samples (100 μl provided 0.01 μg of protein/ml) were injected into the flow system running buffer (10 mM Hepes, pH 7.4/150 mM NaCl/3.4 mM EDTA/0.05% BIAcore surfactant P20) as were concentrates of serum-free samples of medium from virus-infected cell cultures that had been clarified (10,000 rpm/30 min at 4°C) and then concentrated 20× by Centrisep-10 filtration methods (Amicon). Sensors were regenerated by using 10 mM HCl; SDS and hypochlorite were used to decontaminate the system.
Immunologic Assays.
Centrisep-10 filtered 20× concentrates of clarified (10,000 rpm/30 min at 4°C) culture medium and PBS-washed virus-infected cells were assayed by Western blots (22) of proteins separated by SDS/PAGE (12% gels). Gel loading buffer (23) used was with or without 100 mM β-mercaptoethanol, and samples were heated at 100°C for 5 min. Blots of resolved proteins were blocked in Blotto, incubated for 16 hr at room temperature in Blotto plus 1:1,000 of anti-MBP-CPV serum, and then incubated for 2 hr in phosphatase-conjugated goat anti-rabbit-IgG (Kirkegaard & Perry Laboratories) before developing in 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (Bio-Rad).
Radioimmunoprecipitation assays were done as described before (26) by using subconfluent 143B cell monolayers infected with 5 pfu/cell of CPV, CPV-W2, or VAC-VTF7–3, or coinfected with VAC-A607 and VAC-VTF7–3. At 6 hr postinfection (hpi), infected cells were pulse-labeled for 3 hr in cysteine-free Eagle’s minimal essential medium containing 50 μCi (1,300 Ci/mmol) [35S]cysteine (Amersham), and then the cultures were harvested and the cells and medium separated at 1,000 rpm/10 min at 4°C. The medium was adjusted to 1 mM phenylmethylsulfonyl fluoride, 0.5 μg leupeptin/ml, and 1 μg aprotinin/ml, and the cells were dissolved in lysis buffer. Proteins in the lysates and the medium were precipitated as described (26) by using 5 μl of anti-MBP-CPV serum per sample. Precipitates were dissolved, reduced, and heated (100°C/10 min) in gel loading buffer, and the proteins were resolved in 12% SDS/PAGE gels (23). Amersham methods using Amplify radiography solution were used to visualize resolved proteins.
Ligand Binding.
Ligand binding was examined by reacting 125I-Bolton-Hunter-labeled human TNF (New England Nuclear) with preparations of fusion protein, infected cell lysates, or medium 20× concentrates dotted onto nitrocellulose filters (11, 12). Concentrates from virus- or mock-infected cells were untreated or treated by precipitating CrmD with anti-MBP-CPV serum and then binding the precipitates to Protein-G Sepharose (Pharmacia). Sepharose-bound precipitates were washed three times in PBS and then suspended in 100 μl of 100 mM glycine-HCl buffer (pH 2.8) to release antigen. After removing the Sepharose by centrifugation, released antigen (and background controls—mock-infected cell culture or MBP) was dotted onto filters and air-dried. Filters then were washed in Blotto for 1 hr and reacted with 125I-TNF.
For binding assays, filters were placed in Blotto with 1 nM 125I-TNF and incubated at 4°C for 4 hr. For binding competition, filters were incubated at 4°C for 4 hr in Blotto plus a solution of 1 nM 125I-TNF and 200 nM of either human, rat, or mouse TNF, human LTα, or a mixture containing 200 nM each of human β-nerve growth factor; human interleukin 1α, 1β, 2, and 7; human interferon α, β, and γ (BioSource International). Residual 125I-TNF was extensively washed from the filters with PBS before γ radioactivity assay.
Inhibition of TNF or LTα Cytotoxicity for L929 Cells.
TNF or LTα cytotoxicity for L929 cells was examined by using a crystal violet staining method (27) in which 105 cells per well in 24-well dishes were incubated at 37°C for 24 hr in 200 μl of DMEM plus 8 μg of actinomycin D/ml and various concentrations of human TNF or LTα, or incubated with these ligands that had been premixed on ice for 1 hr with a fusion protein preparation (10 μg of protein/ml). Dye taken up by surviving cells was released into 0.5 ml of methanol and then the dye-methanol OD565 was determined.
RESULTS
Nucleotide and Deduced Amino Acid Sequences.
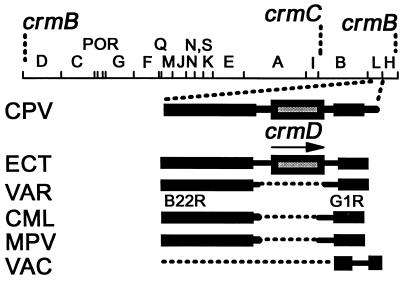
Fig. 1 shows the CPV HindIII map (18) position of the currently identified genes for TNFR-like proteins and the presence or absence of crmD in other orthopoxvirus DNAs examined. Initially, 1,456 bp of CPV DNA (GenBank accession no. U87234) were sequenced that contained an ORF for the CrmD 320-aa TNFRII isolog. Cognates in other orthopoxvirus DNAs then were identified by PCR and by Southern blot hybridizations using the crmD amplicon. The gene was identified in ECT DNA, sequenced (accession no. U87235), and further identified in three other strains of ectromelia from Munich (accession nos. U87581–83). However, three strains of cowpox virus from Munich (accession nos. U87578–80) showed frameshifts that would interrupt CrmD in the signal peptide, CRD-I, or CRD-II segments, likely rendering putative gene products functionless. The crmD gene was entirely absent from VAC and vaccinia strain Copenhagen DNAs and examined strains of monkeypox, camelpox, and variola. A CPV crmB cognate in ECT was determined to be absent (accession no. U86380), and a cognate of the CPV crmC in ECT was highly truncated (accession no. U93910).
Figure 1.
HindIII map of CPV (18) with locations of crmB, crmC, and crmD and cognate crmD regions in ECT, VAR, camelpox Somalia, monkeypox Copenhagen, and VAC DNAs [arrow = transcription direction; B22R and G1R reading frames flank the crmD region, which is absent in VAR DNA (13, 14) and DNA of CML, MPV, and VAC as indicated].
Scanning 300 nucleotides flanking the ECT and CPV crmD ORF did not enable prediction of the temporal class of crmD. Upstream sequences did not match promoters for early-, intermediate-, or late-class genes (28–30), though a motif, TAAAT at position −14, was noted that often specifies a late-class mRNA 5′-polyadenylation signal (31). On the other hand, an early-class mRNA termination signal, TTTTTAT (32), appeared 27 bp after the stop codon.
The deduced amino acid sequences of ECT and CPV CrmD denoted a 35-kDa protein of pI 5.2 and charge −8.7 at pH 7. Residues 32, 57, 76, and 81 signified N-linked glycosylation sites. The first 20 N-terminal amino acids indicated a cytoplasmic transport signal peptide, and residues 269–290 signified a hydrophobic region potentially for a putative transmembrane domain.
fasta and blast queries indicated 30–50% amino acid identity of CrmD with TNFR superfamily members, including previously reported poxvirus TNFR analogs. However, as shown in Fig. 2, alignment of ECT and CPV CrmD CRDs with corresponding domains in human and murine TNFRII, CPV CrmB and CrmC, VAR G2R, and myxoma T2 indicated that the CrmD ligand-binding region is similar to that in other TNFRs, particularly in the coincidence of the cysteines that likely provide disulfide bonds to form the ligand-binding structure (4, 33).
Figure 2.
Pairwise alignment of CrmD amino acid sequences of ECT and CPV CRDs and sequences of corresponding domains in VAR G2R (L22579), CPV CrmB (L08906), CPV CrmC (U55052), myxoma T2 (P29825), and human (P20333) and murine (P25119) TNFRII (GenBank accession numbers in parentheses). Domains are distributed as described for TNFRI pseudo-repeat elements (33). Underlined residues are conserved.
Cellular TNFRI and TNFRII contain four CRDs, and TNFRI CRD-II and CRD-III have been shown to form virtually all the contacts with TNF (4, 33). As Fig. 2 shows, CrmD residues 30–181 distribute into three CRDs that coincide with the TNFRI and TNFRII CRDs. Each CRD includes six cysteines that could provide three disulfide bonds; the fourth CRD contains three cysteines. In CRD-IV, sequences between the first and second cysteine diverge highly from counterparts in CrmB, G2R, T2, and TNFRII. The CrmC cysteine-rich region pairs with CRD-I, CRD-II, and CRD-III and has no counterpart to CRD-IV. There is moderate homology between the carboxyl half of CrmD CRD-IV and cognates in CrmB and T2. There is low homology between the CRD-IV segments of CrmD and TNFRII.
The C-terminal sequences following the ECT and CPV CrmD CRDs shown in Fig. 2 align very poorly with cellular TNFRII C-terminal sequences, which are the primary sequences involved in signal transduction. However, the CrmD C-terminal sequences are about 50% identical to C-terminal sequences of T2, CrmB, and G2R.
Characterization of CrmD.
CrmD from virus-infected cultures and recombinant maltose-binding CRD fusion proteins were examined by ligand and Western blot assays, radioimmunoprecipitation and biosensor methods, and by ligand dot blot and cytolytic inhibition tests. Initial examination of the MBP-ECT and MBP-CPV by SDS/PAGE using Coomassie blue staining showed expected 65-kDa migrating fusion proteins, and after factor-Xa digestion, 42-kDa MBP and 24-kDa CRD cleavage products were resolved (not shown).
CrmD Is a 46-kDa Glycoprotein.
The TNF-binding properties of proteins in the culture fluid of cells infected with either VAC, ECT, or CPV, were examined by ligand blot assays using nonreducing SDS/PAGE conditions (reduction inactivated ligand-binding activity) to separate viral components as shown in Fig. 3A. TNF-binding proteins were not detected in culture fluid from cells infected with VAC (lane 1), consistent with lack of functional TNFR genes in this vaccinia strain. The medium from ECT-infected cells (lane 2) showed two prominent TNF-binding components migrating with apparent molecular masses of 60–70 kDa, and several minor components in the 90- to 250-kDa range. Medium from CPV-infected cells showed a TNF-binding component of apparent molecular mass of 30 kDa (lane 3).
Western blot assay (Fig. 3B) of the same preparations separated under nonreducing conditions, and reaction of the blot with anti-MBP-CPV serum confirmed that ECT CrmD (lane 2) had the same electrophoretic characteristics as the TNF-binding components shown in Fig. 3A, lane 2. However, Western blots did not detect reactive components in culture fluid of VAC- or CPV-infected cells (Fig. 3B, lanes 1 and 3). The negative result shown in Fig. 3B, lane 3 suggested that the 30-kDa TNF-binding material shown in Fig. 3A, lane 3 was the CrmC protein. Consistent with this interpretation, the 30-kDa protein did not bind LTα (though the ECT multiple-migrating forms did), nor was it detected in medium or cell lysates of cultures infected with CPV-A513 (12), a crmC-minus mutant (data not shown).
To further examine CPV for expression of CrmD, metabolically radiolabeled proteins from CPV-infected cell cultures were immunoprecipitated with anti-MBP-CPV serum, reduced, heat-denatured, and then resolved by SDS/PAGE. As Fig. 3C shows, a 46-kDa protein was detected in lysates of CPV-infected cells (lane 1) that was absent from cells infected with CPV-W2 (lane 2), a virus that lacks crmD, but expresses CrmB and CrmC (19). A similar protein was detected in cells infected with recombinant VAC expressing the CPV crmD gene (lane 3), but not cells infected with the VAC vector alone (lane 4). These results confirmed that CPV expresses CrmD; however, failure to detect CrmD in culture fluid of CPV-infected human 143 cells suggested that CPV produces markedly lower amounts of CrmD than ECT in Rat-2 (Vero and BSC-40) cells. The reason for the difference is not clear.
After reduction, the ECT CrmD protein migrated as a single species with a molecular mass of 46 kDa (Fig. 3D, lane 1), identical to the mass of CPV CrmD (Fig. 3C). The molecular mass of ECT and CPV CrmD proteins is more than the 35-kDa mass predicted from the amino acid sequences, but consistent with predicted glycosylation of CrmD. In this regard, enzymatic deglycosylation and reduction of the ECT CrmD protein decreased the apparent molecular mass to 38 kDa (Fig. 3D, lane 2).
Collectively, the results shown in Fig. 3 indicated that CrmD is synthesized in infected cells as a 46-kDa glycoprotein capable of forming larger disulfide-linked complexes that can bind ligand. The precise composition of the different complex forms has not been determined.
Biosensor Analysis.
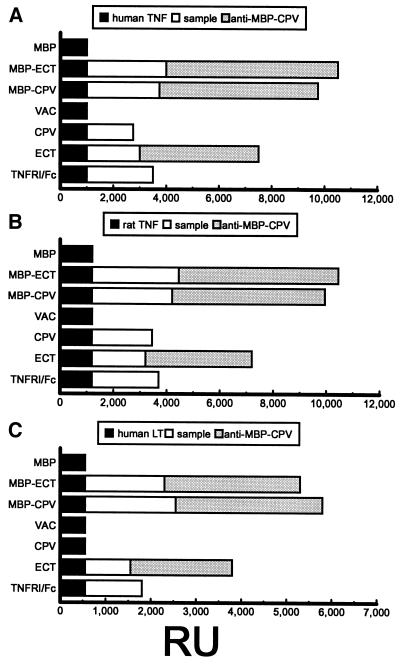
Fig. 4 shows that immobilization of human TNF, rat TNF, and human LTα onto flow cell sensors raised peak levels of plasmon resonance by 1,000, 1,200, and 550 resonance units (RUs) (1 RU = ≈1 pg protein/mm2 sensor surface), respectively. Further RU increases were detected by reacting ligands with MBP-CPV, MBP-ECT, human TNFRI/Fc fusion protein (34), or concentrated medium from cell cultures at 24 hpi with ECT or CPV. RU increases for the MBP-ECT and MBP-CPV paralleled increases for TNFRI/Fc. Slightly lower increases were noted when using concentrated medium from cell cultures harvested at a late time after infection with ECT or CPV. The ECT culture fluid reacted with the three ligands; however, consistent with SDS/PAGE results (Fig. 3A, lane 3) CPV culture fluid reacted with TNF, but not LTα, a feature of CrmC (12). Concentrated medium from cells infected with VAC or mock-infected (not shown) and MBP alone did not react with the ligands.
Figure 4.
Biosensor plasmon RUs determined for (A) human or (B) rat TNF, or (C) human LTα binding by MBP-ECT or MBP-CPV or concentrates of culture fluids from cells infected with VAC, CPV, or ECT. Peak RU values after immobilizing ligand (black), after sample ligand-binding (white), and after reaction with IgG from anti-MBP-CPV serum (gray) are shown (TNFRI/Fc and MBP, positive and negative controls).
The identity of the ligand-binding proteins then was determined by examining the reactivity of IgG prepared from anti-MBP-CPV serum. As Fig. 4 shows, the fusion proteins and components from culture fluid from ECT-infected cells had increased RU peak values, but no increases were detected after antibody reaction with ligand-bound material from culture medium of CPV- or VAC-infected cells. The CPV results supported the view that the binding component was CrmC, and that secreted CrmB and CrmD were below the detection level of the assay.
Ligand Blots.
Dot blot and dot blot competition assays were performed to examine the relative efficiency of ligand binding to various preparations of CrmD and provide further insight about which temporal class includes CrmD. Assays included fusion proteins, cell lysates, or concentrates of medium from cells at 4 or 12 hpi with ECT and after 12 hr of ECT growth in medium plus cytosine arabinoside, an inhibitor of DNA replication, which precedes transcription of poxvirus intermediate- and late-class genes (30).
Table 1 shows that 125I-TNF binds MBP-CPV and MBP-ECT and that 57- to 104-fold and 45- to 125-fold competitive reductions, respectively, in MBP-CPV and MBP-ECT binding resulted when 125I-TNF was mixed with excess human or murine TNF or human LTα. No competition was apparent when 125I-TNF was mixed with a solution of nerve growth factor and cytokines unrelated to TNF.
Table 1.
Competition with 125I-TNF binding by MBP-CPV, MBP-ECT, or freeze-thawed lysates and culture medium concentrates from cells infected with ECT
| 125I-TNF and competing ligand* | MBP-CPV | MBP-ECT | Lysates, 4 hpi | Medium, 4 hpi | Ara-C‡, 12 hpi | Lysates, 12 hpi | Medium, 12 hpi |
|---|---|---|---|---|---|---|---|
| None | 1,562 ± 32† | 1,254 ± 45 | 21 ± 19 | 32 ± 30 | 23 ± 10 | 675 ± 15 | 2,567 ± 27 |
| Human TNF | 20 ± 15 | 28 ± 27 | — | — | — | 13 ± 19 | 25 ± 27 |
| Human LTα | 15 ± 27 | 22 ± 23 | — | — | — | 32 ± 27 | 34 ± 21 |
| Mouse TNF | 17 ± 23 | 17 ± 21 | — | — | — | 21 ± 19 | 12 ± 16 |
| Rat TNF | 27 ± 20 | 10 ± 16 | — | — | — | 15 ± 17 | 23 ± 21 |
| Ligand mix | 1,598 ± 23 | 1,223 ± 12 | — | — | — | 618 ± 23 | 2,453 ± 32 |
Individual nitrocellulose filters dotted with 1 μg of recombinant fusion protein or lysates or culture fluid concentrates from ECT-infected cells were reacted with 1 nM 125I-TNF (human) or 125I-TNF mixed with 200 nM TNF or LTα from various animals, or with a mixture of eight different ligands described in the Materials and Methods section.
dpm γ radioactivity mean ± SD from three separate experiments. —, not tested.
Medium plus lysates of virus grown in the presence of cytosine arabinoside (Ara-C, 40 μg/ml).
Table 1 also shows the lack of 125I-TNF binding to immunoaffinity-purified material from cell lysates or culture fluid concentrates from ECT-infected cells harvested at 4 hpi, or a mixture of medium and lysates prepared after 12 hr of virus growth in the presence of cytosine arabinoside (similar data not shown here were obtained for material before immunoaffinity purification). However, the 125I-TNF did bind to cell lysates and medium collected at 12 hpi, either before (not shown) or, as shown in Table 1, after immunoaffinity purification. The TNF and LTα ligands competitively inhibited 125I-TNF binding to cell lysates 32- to 53-fold, and to culture medium 75- to 214-fold.
The data obtained by using cytosine arabinoside inhibition indicated that ECT CrmD is expressed after early gene expression. Moreover, results of time course experiments not included here indicated that the highest amount of ECT CrmD is detectable at 14 hpi. Though the data is highly suggestive that crmD is a late-class gene, confirming mRNA studies are being done. Finally, as shown in Table 1, no inhibition was detected with 125I-TNF in a mixture of nerve growth factor and cytokines unrelated to TNF.
Inhibition of Human TNF and LTα Cytotoxicity by Fusion Proteins.
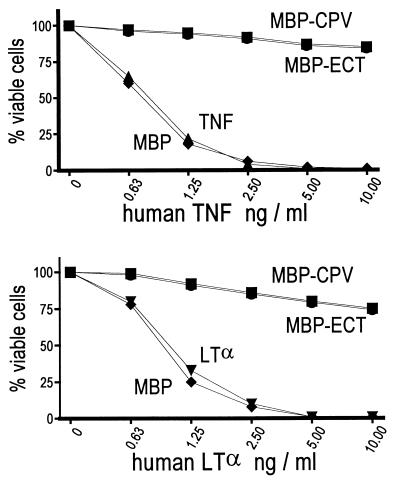
Fig. 5 shows that human TNF and LTα kills mouse L929 cells in a dose-dependent manner; however, cytolysis was blocked significantly by mixing either ligand with MBP-CPV or MBP-ECT.
Figure 5.
Purified bacterial recombinant fusion proteins block TNF (Upper) and LTα (Lower) cytolysis of mouse L cells. Crystal violet staining was used to determine the percent of cells viable after 24-hr treatment with human TNF (▴) or LTα (▾) and when human TNF or LTα are mixed with MBP-CPV (•), MBP-ECT (▪), or MBP (⧫).
DISCUSSION
The complexity and sophistication of viral countermeasures to TNF through the use of different versions of TNFRs are greater than first appreciated. Some poxviruses, such as most vaccinia strains, completely lack functional genes encoding TNFRs whereas other poxviruses, such as Shope fibroma virus and highly virulent myxoma, variola, and as shown here, ectromelia viruses, each have been shown to encode a single TNFR isolog. Remarkably, CPV, a strain of intermediate virulence among the orthopoxviruses has not one, but three genes (crmB, crmC, and crmD) for distinct TNFRs.
The newly discovered crmD gene is present in the genomes of both CPV and ECT in similar positions distinct from the crmB and crmC loci (Fig. 1). The overall structure of CrmD and CrmB is similar in that each possesses characteristic TNF-binding regions (Fig. 2), and an extensive C-terminal region that is rather diverged from corresponding portions of known TNFR superfamily members. The CPV and ECT CrmD proteins are 97% identical, but they have only 51% homology with CPV CrmB. Nonetheless, CrmB (11) and, as shown here, CrmD bind TNF and LTα. The crmD gene, like the crmC gene, but unlike crmB, is detected predominantly at late times during virus replication.
It is unclear why ECT possesses only one intact TNFR gene. Perhaps ECT partly compensates for the lack of crmB and crmC by somehow enhancing CrmD production, because at least in the cell cultures used in this study, ECT CrmD was much more abundant than CPV CrmD. Understanding the significance of differences between CPV and ECT efficiency of crmD gene expression awaits further study.
It is also unclear why secreted CrmD forms such novel disulfide-linked complexes (Fig. 3); perhaps the complexes increase ligand binding efficiency. In this regard, chimeric dimers of TNFRI have been shown to bind soluble TNF and LTα at 2,000-fold higher affinity compared with monomers, and the dimers had greater capacity to block cytolytic activity of TNF and LTα in vitro and in vivo (35–37). Greater blocking of TNF cytolysis in vitro by dimers of myxoma T2 protein also has been shown (5). Surely, CrmD endows a selective advantage in having the diversity to be produced in different amounts and in complexes.
The expression of three different TNFRs by CPV suggests that each of these modulates immune processes in different, perhaps complementary ways. The CPV CrmC protein is the most distinct of the three receptors because it binds TNF and not LTα. And, unlike CrmB and CrmD, CrmC is comprised of a ligand-binding domain that is not followed by an extensive C-terminal region (12); it is not clear whether CrmC is a vestige of a longer version of TNFR. The CrmB and CrmD proteins are more closely related in that each binds TNF and LTα. Each contains an approximate 160-aa C-terminal region that is not part of the TNF-binding region. The 44% amino acid sequence identity between CrmB and CrmD C-terminal regions and their similarity to the corresponding region of the more distantly related myxoma T2 protein suggests that this segment plays an important role during poxvirus infection (11), perhaps effecting functions related to, but distinct from, ligand binding. There may be less redundancy in CrmB and CrmD functions than simple analysis by ligand binding suggests and these functions may critically influence pathogenesis dynamics.
At this point it is unclear why CPV and ECT encode this third TNFR. However, the potential of orthopoxviruses to express several TNFRs suggests that they are providing the means to specifically modulate particular immune processes during virus replication. In this regard, viral TNFR-like proteins should provide tools to identify aspects of interactions not currently associated with TNF superfamily members. Understanding the mechanisms of action of such viral proteins may provide insight into the development of novel immune-modulating therapies.
Acknowledgments
We thank Robert Wohlheuter (Centers for Disease Control and Prevention Biotechnology Core Facility Branch) for guidance in biosensor assays, Linda Gooding (Emory University) for advice on cytolysis assays, and Hermann Meyer (Federal Armed Forces Medical Academy, Germany) for DNA from Munich strains of ectromelia and cowpox virus.
ABBREVIATIONS
- CPV
cowpox virus strain Brighton red
- ECT
ectromelia virus strain Moscow
- VAC
vaccinia virus strain Western Reserve
- VAR
variola virus strain Bangladesh
- crm
cytokine response modifier
- TNF
tumor necrosis factor
- TNFR
TNF receptor
- LTα
lymphotoxin-α
- CRD
cysteine-rich domain
- MBP
maltose-binding protein
- MBP-CPV
CPV CRD MBP fusion protein
- MBP-ECT
ECT CRD MBP fusion protein
- hpi
hours postinfection
- Blotto
bovine lacto transfer technique optimizer
- RU
resonance unit
Footnotes
References
- 1.Pickup D J. Infect Agents Dis. 1994;3:116–127. [PubMed] [Google Scholar]
- 2.Smith C A, Davis T, Anderson D, Solam L, Beckmann M P, Jerzy R, Dower S K, Cosman D, Goodwin R G. Science. 1990;248:1019–1023. doi: 10.1126/science.2160731. [DOI] [PubMed] [Google Scholar]
- 3.Smith C A, Davis T, Wignall J M, Din W S, Farrah T, Upton C, McFadden G, Goodwin R G. Biochem Biophys Res Commun. 1991;176:335–342. doi: 10.1016/0006-291x(91)90929-2. [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal B, Natarajan K. Eur Cytokine Netw. 1996;7:93–124. [PubMed] [Google Scholar]
- 5.Schreiber M, McFadden G. Virology. 1994;204:692–705. doi: 10.1006/viro.1994.1585. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber M, Rajarathnam K, McFadden G. J Biol Chem. 1996;271:13333–13341. doi: 10.1074/jbc.271.23.13333. [DOI] [PubMed] [Google Scholar]
- 7.Macen J L, Graham K A, Lee S F, Schreiber M, Boshkov L K, McFadden G. Virology. 1996;218:232–237. doi: 10.1006/viro.1996.0183. [DOI] [PubMed] [Google Scholar]
- 8.Upton C, Macen J L, Schreiber M, McFadden G. Virology. 1991;184:370–382. doi: 10.1016/0042-6822(91)90853-4. [DOI] [PubMed] [Google Scholar]
- 9.Howard S T, Chan Y S, Smith G L. Virology. 1991;180:633–647. doi: 10.1016/0042-6822(91)90077-o. [DOI] [PubMed] [Google Scholar]
- 10.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 11.Hu F-Q, Smith C A, Pickup D J. Virology. 1994;204:343–356. doi: 10.1006/viro.1994.1539. [DOI] [PubMed] [Google Scholar]
- 12.Smith C A, Hu F-Q, Smith T D, Richards C L, Smolak P, Goodwin R G, Pickup D J. Virology. 1996;223:132–147. doi: 10.1006/viro.1996.0462. [DOI] [PubMed] [Google Scholar]
- 13.Massung R F, Esposito J J, Liu L I, Qi J, Utterback T R, Knight J C, Aubin L, Yuran T E, Parsons J M, Loparev V N, et al. Nature (London) 1993;366:748–751. doi: 10.1038/366748a0. [DOI] [PubMed] [Google Scholar]
- 14.Massung R F, Liu L I, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J J. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 15.Shchelkunov S N, Blinov V M, Sandakhchiev L S. FEBS Lett. 1993;319:80–83. doi: 10.1016/0014-5793(93)80041-r. [DOI] [PubMed] [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Esposito J J, Knight J C. Virology. 1985;143:230–251. doi: 10.1016/0042-6822(85)90111-4. [DOI] [PubMed] [Google Scholar]
- 19.Pickup D J, Ink B S, Parsons B L, Hu W, Joklik W K. Proc Natl Acad Sci USA. 1984;81:6817–6821. doi: 10.1073/pnas.81.21.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuerst T R, Niles E G, Studier F W. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. Nature (London) 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 22.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Johnson D A, Gautsh J W, Sportsman J R, Elder J H. Gene Anal Tech. 1984;1:3–8. [Google Scholar]
- 25.Malmquist M. Nature (London) 1993;361:186–187. doi: 10.1038/361186a0. [DOI] [PubMed] [Google Scholar]
- 26.Pickup D J, Ink B S, Hu W, Ray C A, Joklik W K. Proc Natl Acad Sci USA. 1986;83:7698–7702. doi: 10.1073/pnas.83.20.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi G W, Montgomery R B, Stahl W L, Crittenden C A, Valentine M A, Thorning D R, Andrews D F, Lilly M B. Int J Immunopharmacol. 1994;16:723–736. doi: 10.1016/0192-0561(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 28.Davison A J, Moss B. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 29.Davison A J, Moss B. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 30.Moss B. In: Fields Virology. 3rd Ed. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 31.Bertholet C, Van Meir E, ten Heggeler-Bordier B, Wittek R. Cell. 1987;50:153–162. doi: 10.1016/0092-8674(87)90211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohrmann G, Yuen L, Moss B. Cell. 1986;46:1029–1035. doi: 10.1016/0092-8674(86)90702-6. [DOI] [PubMed] [Google Scholar]
- 33.Banner D W, D’Arcy A, Janes W, Gentz R, Schoenfeld H J, Broger C, Loetscher H, Lesslauer W. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 34.Crowe P D, VanArsdale T L, Walter B N, Dahms K M, Ware C F. J Immunol Meth. 1994;168:79–89. doi: 10.1016/0022-1759(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 35.Ashkenazi A, Marsters S A, Capon D J, Chamow S M, Figari I S, Pennica D, Goeddel D V, Palladino M A, Smith D H. Proc Natl Acad Sci USA. 1991;88:10535–10539. doi: 10.1073/pnas.88.23.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lesslauer W, Tabuchi I, Gentz R, Brockhaus M, Schlaeger E J, Grau G, Piguet P F, Pointaire P, Vassalli P, Loetscher H. Eur J Immunol. 1991;21:2883–2886. doi: 10.1002/eji.1830211134. [DOI] [PubMed] [Google Scholar]
- 37.Peppel K, Crawford D, Beutler B. J Exp Med. 1991;174:1483–1489. doi: 10.1084/jem.174.6.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]