Abstract
The placenta may play a critical role in inhibiting vertical transmission of HIV-1. Here we demonstrate that leukemia inhibitory factor (LIF) is a potent endogenous HIV-1–suppressive factor produced locally in placentae. In vitro, LIF exerted a potent, gp130-LIFRβ–dependent, HIV coreceptor–independent inhibition of HIV-1 replication with IC50 values between 0.1 pg/ml and 0.7 pg/ml, depending on the HIV-1 isolate. LIF also inhibited HIV-1 in placenta and thymus tissues grown in ex vivo organ culture. The level of LIF mRNA and the incidence of LIF protein–expressing cells were significantly greater in placentae from HIV-1–infected women who did not transmit HIV-1 to their fetuses compared with women who transmitted the infection, but they were not significantly different from placentae of uninfected mothers. These findings demonstrate a novel pathway for endogenous HIV suppression that may prove to be an effective immune therapy for HIV infection.
Introduction
Without antiretroviral therapy, the transmission of virus from HIV–infected mothers to their neonates occurs in 14.4%–39% of pregnancies (1, 2). With antiretroviral treatment of HIV-1–infected women, vertical transmission is reduced to less than 5% of pregnancies (3). Caesarian section in the absence of maternal antiretroviral treatment, however, reduces the rate of perinatal transmission to similar levels (4). This indirect evidence suggests that the majority of vertical transmission occurs during delivery (5) and that the placenta plays a role in inhibiting transmission during gestation.
Many mechanisms that inhibit vertical transmission have been proposed. First, studies suggest that maternal HIV-1–infected cells could be destroyed by MHC-incompatible cells in the placenta (6) or by interactions with TNF-related apoptosis-inducing ligand/Apo-2L (TRAIL) (7). Similarly, Fas ligand (FasL) expression on mouse trophoblasts has been shown to inhibit the trafficking of activated lymphocytes into the placenta (8), an activity that could protect against HIV-infected cells migrating into the placenta in humans. Second, selection for nontransmitted HIV-1 variants may occur in placentae from nontransmitting women (9). Third, maternal production of HIV-1–specific neutralizing Ab or cytotoxic T lymphocytes may inhibit HIV replication during pregnancy (10, 11). Last, expression of soluble factors that inhibit HIV replication has been described previously (12–14).
The search for HIV-suppressive factors has provided some insights into the role of the adaptive immune response in inhibiting a broad spectrum of HIV-1 strains, but expression of these factors in the placenta remain largely unidentified. Investigating conditions associated with transmission and nontransmission of HIV-1 from mothers to neonates suggests the presence of HIV-suppressive activity in the placenta. Studies in HIV-1 transgenic mice have demonstrated that the pregnancy-related hormone human chorionic gonadotropin (β-hCG) exerts an HIV-1–inhibitory effect (13). Subsequent studies propose that an unidentified factor associated with β-hCG inhibits HIV-1 replication whereas highly purified preparations of β-hCG demonstrated no HIV-1–inhibitory effects (14).
Leukemia inhibitory factor (LIF), well characterized for its role in blastocyst implantation (15) and maintenance of pregnancy (16), has until now not been documented for its potential role in inhibiting viral disease. Here, we show that LIF is upregulated in placentae from nontransmitting HIV-infected women compared with placentae from transmitting women. Moreover, LIF inhibits HIV-1 in a tropism-independent manner that is dependent upon the expression of gp130-LIFRβ on the surface of HIV-1–susceptible cells. Finally, LIF inhibits HIV-1 infection in in vivo explant models using placenta and thymus tissue.
Methods
Placenta-tissue processing.
Placenta samples were obtained from women at the time of delivery after informed consent was given. Transmitting women were matched with nontransmitting women for maternal CD4 count, maternal viral load (if available), placenta gestational age, and age. All women were off antiretroviral therapy during pregnancy. Tissue samples were washed extensively with RNase, DNase-free phosphate buffered, trisected, and either homogenized for RNA extraction or snap-frozen in OCT-embedding compound. RNA and DNA were extracted and purified as described previously (17).
Immunohistochemistry and immunofluorescence staining.
Immunohistochemistry was performed using the Vectastain ABC-HP kit (Vector Laboratories, Burlingame, California, USA) as described previously (18). Diaminobenzidine (DAB) was used as substrate with hematoxylin counterstain. Commercially available Ab’s to IL-2 (MQ1-17H12; BDPharMingen, San Diego, California, USA), IL-4 (8D4-8 and MP4-25D2; BDPharMingen), IL-10 (19.F.1; BDPharMingen), and LIF (R&D Systems, Minneapolis, Minnesota, USA) were used at concentrations optimized on control tissues. Flow-cytometric analysis was performed using phycoerythrin-conjugated (PE-conjugated) anti-CD130 (gp130-LIFRβ; BDPharMingen) on a FACScan instrument (Becton Dickinson Immunocytometry Systems, San Jose, California, USA).
Quantification of positive cells stained for cytokines including LIF.
Acquired computerized image analysis (ACIA) of digital images transferred from a DMR-X microscope (Leica Microsystems Holdings, Wetzlar, Germany) into a computerized image-analysis system, Quantimet (Q 550 IW; (Leica Cambridge Ltd., Cambridge, United Kingdom), was used for detection and separation of 16.7 million different colors. The total area of the sectioned placentae varied between 5 to 15 mm2/tissue, representing 0.6–6.5 × 104 cells/section. Calculation of the total number of nucleated cells stained by hematoxylin, per total section area, as well as the incidence of positively stained cells, was performed by using densitometric and morphometric criteria with a specific software program developed by Ola Noré, Leica Imaging System, Kista, Stockholm, Sweden.
Real-time quantitative PCR for HIV-1 gag and U5/R DNA.
Quantitative, kinetic DNA PCR was performed by adding 45 μl reaction mix (1× Taqman PCR buffer from Perkin-Elmer Applied Biosystems, Foster City, California, USA, 4.0 mM MgCl2, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 200 μM dTTP, 200 nM sense primer, 200 nM antisense primer, 100 nM internally conserved, sequence-specific, fluorogenic probe labeled at the 5′ end with FAM and at the 3′ end with TAMRA, 10 U AmpliTaq Gold polymerase) to approximately 500 ng DNA in 5 μl water. Thermal amplification was performed as described previously (17). The following previously published (19) primers and probes were used: LTR U5/R—sense-5′-GGCTAACTAGGGAACCCACTG-3′ and antisense-5′-CTGCTAGAGATTTTCCACACTGAC-3′, probe 5′-FAM-TGTGTGCCCGTCTGTTGTGTG-TAMRA-3′; LTR U3/gag—sense-5′-CAGATATCCACTGACCTTTGG-3′ and antisense-5′-GCTTAATACTGACGCTCTCGCA-3′, probe 5′-FAM-GAGGCTTAAGCAGTGGGTTC-TAMRA-3′.
Real-time quantitative RT-PCR panels.
Quantitative kinetic RT-PCR was performed by adding 45 μl of reaction mix (1× RT Taqman EZ buffer from Perkin-Elmer Applied Biosystems, 4.0 mM Mn (O)Ac2, 300 μM dATP, 300 μM dCTP, 300 μM dGTP, 300 μM dTTP, 200 nM upstream primer, 200 nM downstream primer, 200 nM internally conserved, sequence-specific, fluorogenic probes, and 10 U rTth polymerase) directly to 100 ng of total RNA in 5 μl RNase, DNase-free water (Ambion Inc., Austin, Texas, USA) (17). Input RNA was normalized using GAPDH mRNA quantification (Perkin-Elmer Applied Biosystems). Reverse transcription and thermal amplification was performed as described previously (17).
In vitro assays for anti-HIV effect.
Replication end-point concentration assays were performed using a tissue culture ID50 (TCID50) of 1,000 for HIV-1Bal, 10,000 for HIV-1Lai, 1,000 for HIV-1ME46, 2,000 for HIV-133074, 2,000 for HIV-133015, and 2,000 for HIV-193US151 to infect 107 phytohemagglutinin-stimulated (PHA-stimulated) PBMCs as described (20), except for the pretreatment of cells with LIF for 1 hour before the addition of virus. The concentrations of recombinant, ultra-pure (>99%) LIF (BDPharMingen) used were 0.01 pg/ml, 0.05 pg/ml, 1.0 pg/ml, and 10 pg/ml. All strains were tested in quadruplicate wells in three separate experiments. The mean p24 antigen produced in positive control PBMCs (without addition of LIF) averaged 843 ± 412 pg/ml, including all isolates tested.
Cell-proliferation assays.
PBMCs were isolated from heparinized blood and placed in quadruplicate wells in the presence or absence of LIF at 0, 0.1, 1.0, and 10.0 pg/ml. PHA was used at 0.25 μg/ml and 1 μCi [3H]thymidine was added per well after 72 hours of culture. The cultures were harvested and counted 18 hours later. Cytotoxicity was assessed using the soluble formazan assay as described previously (21).
Effects of LIF on the regulation of CD4, CCR5, and CXCR4 expression.
PBMCs were pretreated for 3 days with HIV-1–inhibitory concentration of LIF (0.1, 1.0, 10.0 pg/ml). Cells were washed one time with 1× PBS, pH 7.4. Cells were stained with anti–CD4-PerCP, anti–CD14-FITC, and anti–CCR5-PE or anti–CXCR4-PE (BDPharMingen) for 30 minutes at room temperature, washed in 1× PBS, pH 7.4, and fixed in 1% formaldehyde. Analysis was performed on a FACScalibur flow cytometer using Cell Quest software. Fluorochrome molecules per cell were determined using quantitative bead standards.
Placenta organ culture.
Fresh placenta tissue was obtained after informed consent was given. The placenta was cut into 2-cm × 2-cm pieces, and the fragments were washed extensively in PBS, pH 7.4, to remove any contaminating blood. The fragments were placed in MEM media with Earle’s salts and antibiotic/antimycotic solution as described previously (22). Placenta fragments were pretreated with LIF (BDPharMingen), IL-4 (R&D Systems), or polyclonal anti-LIF (R&D Systems), and infected with 3,000 TCID50 HIV-1Bal or the primary isolate HIV-193US151. After infection, total DNA and RNA were extracted using TriReagent per manufacturer’s instructions. HIV-1 DNA quantification was performed using real-time PCR as described previously. Duplicate placenta fragments were frozen in OCT freezing compound for in situ hybridization.
Thymus organ culture.
Fresh pediatric thymus tissue was dissected into pieces containing 3–6 million cells and cultured as described previously (23). The appropriate tissue fragments were treated with 1 pg/ml LIF (BDPharMingen) for 1 hour before infection. The tissue fragments were transferred into sterile tubes containing 500 μl of undiluted virus (3,000 TCID50). The tubes were gently rocked for 4 hours at room temperature. The tissue fragments were then transferred to 0.45-μm nucleopore filters atop gel-foam boats saturated in media (YSSL media: 1% human serum, 50 μg/ml streptomycin, 50 U/ml penicillin G, 1× MEM vitamin solution from Life Technologies Inc., Rockville, Maryland, USA) in six-well plates with a maximum of 16 pieces per raft. The tissue fragments were cultured fragments at 37°C, 5% CO2 for varying periods of time. At appropriate time intervals, tissue fragments were removed and passed through a 37-μm mesh with PBS, pH 7.4. Cells in suspension were used for immunophenotyping and dual immunophenotyping/ultrasensitive fluorescence in situ hybridization (UFISH).
UFISH.
Tissues were snap-frozen in OCT freezing compound (Fisher Scientific Co., Pittsburgh, Pennsylvania, USA). Cryostat-cut tissue sections (5-μm thick) were adhered to silanized slides, air dried, fixed briefly in cold acetone, and rehydrated in PBS, pH 7.4. UFISH was performed using ViroTect (INVIRION, Frankfort, Michigan, USA) as described previously (24). Image analysis was performed on a dual-laser confocal microscope (Olympus America Inc., Melville, New York, USA).
Statistical analysis.
Comparisons between transmitting and nontransmitting placentae were performed using either the Mann-Whitney rank sum test or paired t test. Comparisons yielding a two-tailed P value of less than 0.05 were considered to be significant.
Results
Quantification of HIV-1 DNA and mRNA in placentae.
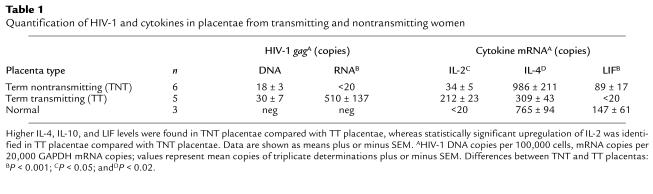
To investigate differences in HIV-1 load in placentae from transmitting and nontransmitting women, we quantified unspliced HIV-1 gag mRNA and DNA from 14 placentae including six HIV-seropositive term nontransmitting (TNT) placentae, five HIV-seropositive term transmitting (TT) placentae, and three HIV-seronegative “normal” placentae. Expression of HIV-1 gag mRNA was found in four of five (80%) TT placentae and only one of six TNT placentae (17%) (Table 1). The average number of HIV-1 gag mRNA copies in the TT placentae was 510/20,000 GAPDH copies whereas the average number of copies of HIV-1 mRNA in TNT placentae was less than the sensitivity of the assay (20 copies, P < 0.001). No significant difference was found in the number of placentae containing HIV-1 DNA+ cells; it was detected in five of five TT placentae (100%) and in eight of ten (80%) TNT placentae. The mean number of HIV-1 DNA copies in TT placentae (30 copies per 100,000 cells) was increased 1.5-fold, approaching statistically significant levels (P = 0.07) compared with TNT placentae (18 copies per 100,000 cells). These data provide further evidence for the low levels of HIV-1 RNA production in TNT placentae (25) and support the possibility of HIV-suppressive activity within these placentae.
Table 1.
Quantification of HIV-1 and cytokines in placentae from transmitting and nontransmitting women
Differential gene expression in HIV-infected placentae with high HIV-1 mRNA load versus low HIV-1 mRNA load.
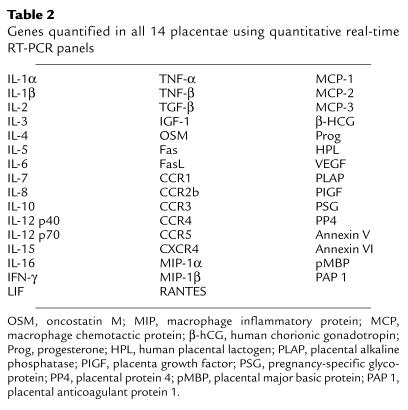
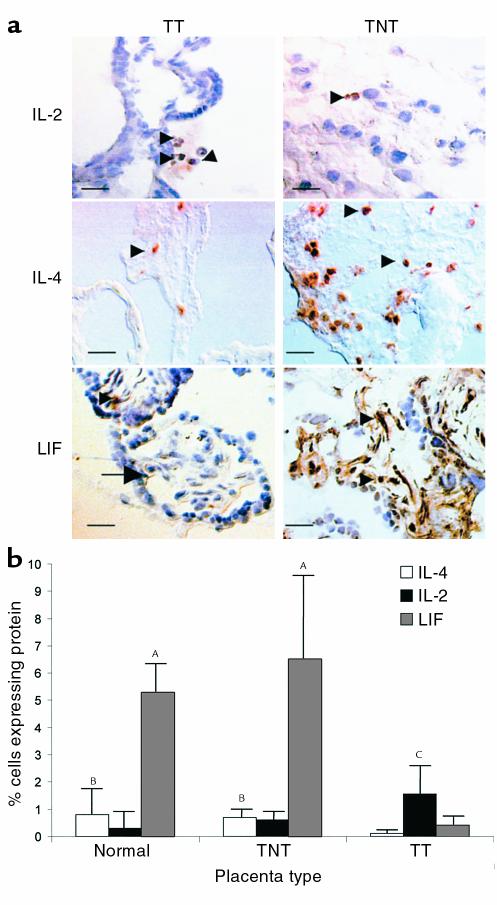
To identify potential inhibitory factors in low virus load (TNT) placentae, we performed real-time quantitative RT-PCR panels against 47 genes expressed in placentae, including cytokines, chemokines, and hormones (Table 2). Genes differentially expressed in gestational age–matched TNT and TT placentae were predominantly cytokines. Consistent with data from another cohort (B.K. Patterson, manuscript submitted for publication), type 2 cytokine (IL-4) mRNA and protein expression were significantly greater in TNT placentae compared with the expression in TT placentae (P < 0.02), but were not significantly different from expression in normal placentae (Table 1, Figure 1). In contrast, the type 1 cytokine IL-2 was upregulated at both the mRNA and protein level in transmitting placentae compared with both nontransmitting placentae and normal placentae (P < 0.05). IL-2 upregulation in TT placentae tended to be associated with superimposed chorioamnionitis, villitis, or funisitis in our cohort. As predicted in the presence of upregulated IL-4 (16), LIF mRNA and protein expression were significantly elevated in TNT placentae compared with TT placentae (P < 0.001) (Table 1, Figure 1). Though slightly decreased, LIF mRNA and protein expression (data not shown) were not statistically different in TNT placentae compared with normal placentae (Table 1). LIF protein expression, however, was below the limits of detection in PBMCs from TNT and TT women, indicating that LIF was not systemically upregulated in the nontransmitting mothers (data not shown).
Table 2.
Genes quantified in all 14 placentae using quantitative real-time RT-PCR panels
Figure 1.
Representative quantification and localization of LIF, IL-4, and IL-2 protein in placentae from transmitting and nontransmitting women using immunohistochemistry (a) and assisted computerized image analysis are seen (b). Cells staining with a brown precipitate express the protein indicated. Cells were counterstained blue with hematoxylin. Tissues from TNT placentae and normal placentae express significantly higher levels of IL-4 and LIF than TT placentae (AP < 0.001; BP < 0.02). IL-2 was significantly elevated in TT placentae compared with TNT placentae (CP < 0.05). IL-4, IL-2, and LIF are all expressed in placental lymphocytes (arrowheads), and LIF was also expressed in decidua and endothelial cells (arrow). Scale bar, 20 μm.
Identification of LIF as a potent HIV-1 suppressor.
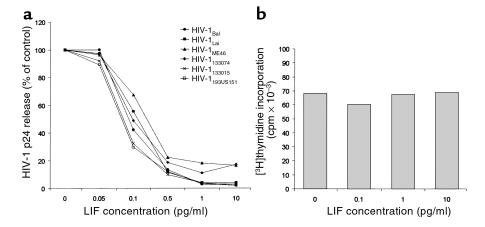
To test the effects of LIF on HIV-1, we performed triplicate dose-dependent inhibition experiments in quadruplicate wells with a range of LIF concentrations from 0.05 pg/ml to 10 pg/ml (Figure 2a). LIF inhibited the replication of CCR5-using (R5) HIV-1Bal, the CXCR4-using (X4) HIV-1Lai, the dual tropic (R5X4) HIV-1ME46, and the primary isolates HIV-133074/33015 with an IC50 between 0.3 and 0.7 pg/ml. The primary isolate HIV-193US151, taken from an infected, newborn child, was inhibited by LIF with the lowest IC50 of 0.1 pg/ml. LIF was not able, at the doses tested, to inhibit infection by Epstein-Barr virus strain B95-8 in human cord blood mononuclear cells nor cytomegalovirus strain AD-69 in human fibroblasts (data not shown).
Figure 2.
(a) Dose-dependent inhibition of HIV-1 infection by LIF is illustrated. Viral isolates were used at 1,000 TCID50 for the R5-using HIV-1Bal, 10,000 TCID50 for the X4-using HIV-1Lai, 3,000 TCID50 for the X4-using HIV-1Me40, and 2,000 TCID50 for the primary isolates 33074, 33015, and 93US151. The IC50 of LIF for all isolates tested was between 0.1 and 0.7 pg/ml. The mean p24 antigen produced in positive-control PBMCs (without addition of LIF) averaged 843 ± 412 pg/ml, including all isolates tested. (b) Effects of LIF on cell proliferation. LIF inhibited HIV-1 production without affecting cell proliferation in response to 0.25 μg/ml PHA.
Because significant cell-growth inhibition would affect HIV-1 production in culture, we assessed the proliferative response of PBMCs to PHA stimulation in the presence or absence of LIF. In multiple experiments of quadruplicate determinations using HIV-1–inhibitory concentrations, LIF had no inhibitory effect on [3H]-thymidine incorporation (Figure 2b).
To exclude the possibility that cytotoxicity due to LIF contributed to the results shown in Figure 2a, we tested the viability of LIF-treated cells using production of soluble formazan from a tetrazolium substrate. In triplicate colorimetric determinations of the soluble formazan product, we found less than 10% difference in values between LIF-treated and untreated control cells (data not shown). These experiments indicate that the inhibitory effects of LIF on HIV-1 production were not due to inhibition of proliferation or direct cytotoxicity.
LIF inhibits HIV replication but not HIV infection of target cells in vitro.
To elucidate the site of LIF action on HIV-1 replication, we first determined the effects of LIF on CD4 and the predominant HIV-1 coreceptors CCR5 and CXCR4. PBMCs grown in culture were treated with LIF for up to 3 days. Cells were removed from culture and stained with anti-CD4, anti-CCR5, anti-CXCR4 Ab’s, or control anti-IgG. The cell-surface staining was quantified using flow cytometry. LIF had no effect on CD4-, CCR5-, or CXCR4-staining intensity as compared with IgG or untreated controls. Additional control cells pretreated with IL-2 demonstrated that CCR5 and CXCR4 had regulatory responses similar to those reported previously by our laboratory (26). These data suggest that LIF neither blocks anti-CD4, anti-CXCR4, or anti-CCR5 Ab binding sites nor regulates the cell-surface expression of CD4, CCR5, or CXCR4, as do other cytokines (26).
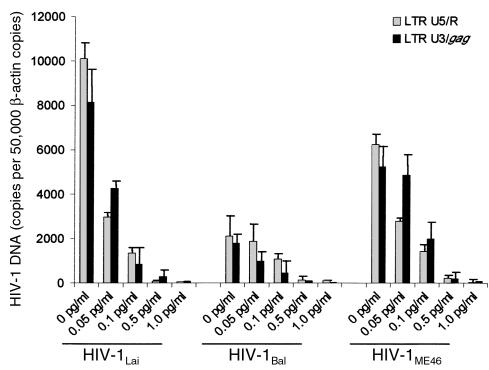
To determine if LIF inhibits the HIV-1 life cycle at the reverse transcription stage, we quantified early LTR U5/R and late LTR/gag reverse transcripts (27) in infected PBMCs treated with LIF, as well as in infected, untreated PBMCs as controls. In PBMCs treated with inhibitory concentrations of LIF and infected with HIV-1Bal, HIV-1Lai, or HIV-1ME46, early LTR U5/R reverse transcripts and late LTR-gag reverse transcripts were approaching the limits of detection (20 copies) (Figure 3). Untreated controls infected with all isolates expressed high levels of both early and late reverse transcripts. These data provide insight into the inhibitory pathway and site of action by suggesting that the antiviral activity of LIF takes place before reverse transcription: an activity that is distinct from CD8-derived antiviral factor (28).
Figure 3.
Dose-dependent effects of LIF on reverse transcription in PBMCs infected with HIV-1 are illustrated. Quantitative real-time PCR results from cells treated with increasing concentrations of LIF and infected with HIV-1Lai, HIV-1Bal, or HIV-1ME40. Both early (LTR U5/R) and late (LTR U3/gag) reverse transcripts were inhibited by LIF. Each determination was performed in triplicate.
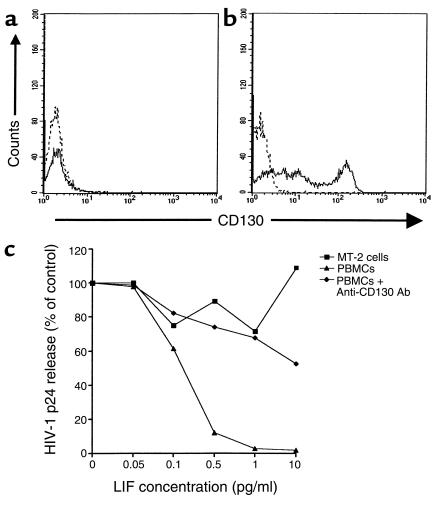
After binding to the gp130-LIFRβ receptor heterodimer (29), LIF upregulates Janus kinase (JAK), which ultimately phosphorylates tyrosine residues on the STAT family of transcription factors (30). To gain insight into the role of gp130-LIFRβ–mediated signal transduction in HIV-1 inhibition, we infected MT-2 cells and PBMCs in the presence and absence of LIF. MT-2 cells did not express detectable levels of gp130-LIFRβ when flow cytometric analysis with mAb’s specific for gp130-LIFRβ was performed (Figure 4a). In contrast, CD4+ T lymphocytes expressed 2 logs greater gp130-LIFRβ protein than internal negative control lymphocytes or isotype controls (Figure 4b). Viral production in MT-2 cells treated with inhibitory concentrations of LIF was equivalent to untreated control MT-2 cells, suggesting a lack of LIF-induced HIV-1 suppression in these gp130-LIFRβ–negative cells (Figure 4c). Accordingly, syncytia were formed in MT-2 cells cultured with up to 10 pg/ml of LIF (data not shown).
Figure 4.
Requirement of gp130-LIFRβ for HIV-1 inhibition by LIF is seen. (a) MT-2 cells do not express gp130-LIFRβ (CD130); however, (b) PBMCs express high levels of gp130-LIFRβ. Dotted lines represent PE-conjugated isotype controls for CD130-LIFRβ staining. (c) MT-2 cells are resistant to and PBMCs susceptible to the HIV-1–inhibitory effects of LIF. Anti-CD130 Ab’s abrogate the anti-HIV-1 effects of LIF. PBMCs and MT-2 cells were infected with 10,000 TCID50 of HIV-1Lai.
Anti–HIV-1 effects of LIF in ex vivo cultured placentae and thymus.
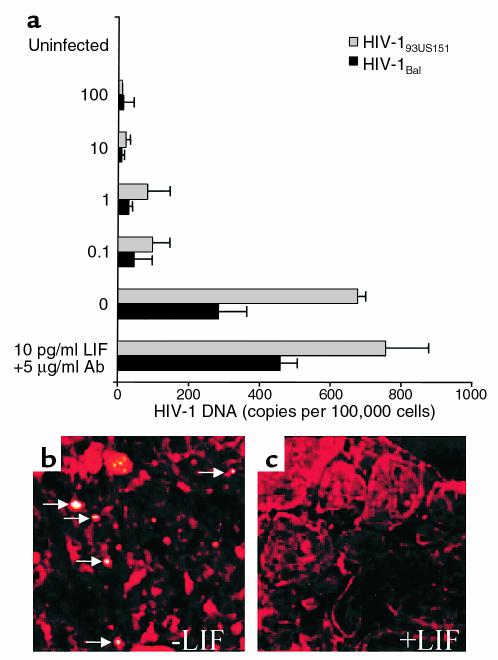
To test the effects of LIF in an in vivo model of HIV infection, we infected tissue fragments of placentae with either a common M-tropic isolate of HIV-1 (HIV-1Bal) or a primary isolate from an infected, newborn child (HIV-193US151) in the presence of HIV-inhibitory concentrations of LIF. As a specificity control, we infected placenta fragments in the presence of LIF and neutralizing, polyclonal anti-LIF Ab’s. In these experiments, we used concentrations of LIF known to inhibit HIV-1 in PBMCs and concentrations of anti-LIF Ab’s known to neutralize greater than 90% of LIF bioactivity. In preliminary experiments similar to those used to validate our other explant models (24), we found that placental explant cultures could be maintained for up to 7 days without loss of viability. Placenta explant cultures were pretreated for 1 hour with either LIF or a combination of LIF and anti-LIF. LIF inhibited infection of placental tissue by HIV-1Bal in a dose-dependent manner (Figure 5a). The IC50 of LIF in tissue was 3.2 pg/ml or roughly threefold higher than in PBMCs cultured in vitro. These data may reflect the accessibility of LIF to HIV-1 target cells in our ex vivo model though in situ hybridization experiments confirmed the inhibition of HIV-1 infection at the cellular level (Figure 5, b and c). Addition of polyclonal anti-LIF to our explant cultures unexpectedly increased infection of placental tissue by twofold compared with uninfected controls. These findings may reflect additional neutralization of endogenous LIF produced by the placental tissue.
Figure 5.
LIF inhibits HIV-1 by in placenta organ culture. Placenta tissue was infected with 3,000 TCID50 HIV-1Bal or 2,000 TCID50 of the primary newborn isolate HIV-193US151 after treatment with 0, 0.1, 1.0, 10.0, or 100 pg/ml LIF (a). Inclusion of 5 μg/ml neutralizing, polyclonal anti-LIF Ab’s increased infection of the placenta organ culture relative to the untreated control. These data may reflect additional neutralization of endogenously produced LIF. (b) Representative laser confocal UFISH images demonstrate numerous productively infected cells (arrows) confirming infection of the organ culture in the absence of LIF. (c)Treatment with 1 pg/ml LIF greatly inhibits the number of productively infected cells. Autofluorescence was quenched, and cells were labeled with a trypan blue counterstain (red).
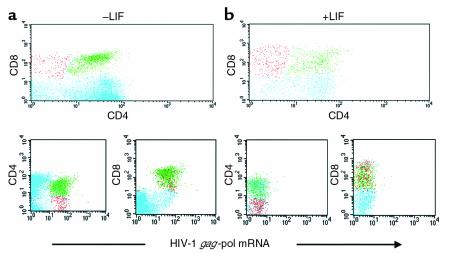
To further corroborate our findings in a second ex vivo model, thymus tissue grown in explant culture was infected by HIV-1Lai in the presence or absence of LIF. Using sensitive simultaneous immunophenotyping/UFISH (as demonstrated in Figure 6a), HIV-1 productively infected CD4, CD8 double-positive (DP), and CD4 or CD8 single-positive (SP) thymocytes on postinfection (PI) day 1. LIF at a concentration of 1 pg/ml inhibited infection of all but less than 0.1% of thymocytes at PI day 1 (Figure 6b). Furthermore, LIF promoted maturation of thymocytes as demonstrated by an increase in CD4, CD8 dim-positive cells (data not shown). Thus, LIF was able to inhibit HIV-1 in a tissue other than placenta, further supporting that LIF does not act in the placenta through an intermediate such as β-hCG–associated factor (HAF) or β-hCG (12, 14).
Figure 6.
LIF inhibits HIV-1 infection in human thymus organ culture. Human thymus tissue was infected with 3,000 TCID50 HIV-1Lai or 3,000 TCID50 HIV-1Bal with and without pretreatment with 1 pg/ml LIF. Representative flow-cytometric dot plots of simultaneous immunophenotyping (CD4 + CD8)/UFISH performed on thymic cell suspensions on PI day 1 are presented (a and b). HIV-1Lai infected predominantly the CD4, CD8 DP population of thymocytes and, to a lessor extent, the CD4 and CD8 SP populations (a). Treatment with LIF inhibited productive infection of the DP and SP thymocyte subsets with only a few (<0.1%) productively infected DP thymocytes identified (b).
Discussion
Our results demonstrate that LIF is a potent inhibitor of HIV-1 with an inhibitory concentration in PBMCs at least 1,000-fold less than the reportedly observed inhibitory concentration of the much-studied β-chemokines (RANTES, MIP-1α, MIP-1β) (31). We also demonstrated that LIF maintains HIV-1 inhibitory activity in three distinct tissue compartments: placenta, PBMCs, and thymus.
Since LIF is constitutively expressed throughout normal pregnancy in the placenta, but not the peripheral blood of pregnant women, the placenta may have an innate ability to inhibit HIV-1 infection. This is supported by the decrease in HIV-1 mRNA and DNA (though only approaching statistical significance) in placenta tissue from nontransmitting women compared with placenta tissue from transmitting women in the present study and by the relatively low placenta levels of HIV-1 DNA in the absence of HIV-1 p24 or gp160 antigen in nontransmitting women from a previous study (25). This pattern of decreased HIV-1 DNA and mRNA is consistent with the pre-reverse transcription mechanism of LIF anti–HIV-1 activity and consistent with the results seen in our placenta-explant culture model.
Here, we propose an HIV-inhibition model in the placenta based on the natural role of the placenta in preventing fetal allograft rejection. Establishment and maintenance of pregnancy is thought to be regulated at least in part by the balance of type 1 (Th1) and type 2 (Th2) cytokine responses (32). Strong Th2 responses inhibit fetal allograft rejection whereas Th1 responses are associated with recurrent abortions (16). Progesterone, one of the hormones essential for pregnancy, promotes the development of T-helper cells that produce Th2-type cytokines such as IL-4 and IL-10 (33, 34). IL-4, in turn, upregulates the production of LIF by T cells. Defective production of LIF by decidual T cells has also been linked to unexplained recurrent abortion (16). The high incidence of recurrent abortion in HIV-infected women (35) is consistent with a defect in IL-4, IL-10, and LIF production (16). Moreover, the identification of increased HIV-1–infected cells in abortuses from HIV-1–infected women (35) suggests an inverse relationship between LIF, Th2 cytokine expression, and HIV-1 infection.
Our findings demonstrate that placentae from nontransmitting women maintained a strong Th2 cytokine milieu whereas placentae from transmitting women showed a Th1 predominance. Consistent with previous reports, our data demonstrate a linkage between Th2 cytokine (IL-4) expression and LIF production. As reported here, LIF produces a potent, HIV coreceptor–independent inhibition of HIV-1 replication. Furthermore, the IC50 of LIF in PBMCs and in tissue-explant cultures is well below the level of LIF produced by CD4+ T-cell clones derived from deciduae of normal women (16).
Based on analysis of specific HIV-1 gene products, LIF-induced HIV-1 suppression is exerted before reverse transcription. LIF binds to gp130, a common receptor for a family of cytokines that also includes ciliary neurotropic factor (CNTF), IL-11, oncostatin M, and, paradoxically, the HIV-1–inducing cytokine IL-6 (36–38). The activities of these cytokines on HIV-1 most likely differ due to the dimerization of secondary components that transduce different signal cascades. Consequently, LIF binding to gp130-LIFRβ activates the JAK1/STAT signal transduction pathway. IL-6, however, induces homodimerization of gp130 and activates a signal transduction pathway different from LIF (39). Further study is necessary to elucidate the exact mechanism in this novel HIV-inhibitory pathway.
We cannot conclude, as yet, whether LIF is the CD8-derived (40), allo-induced (41), or the hCG-associated (14) unidentified antiviral factors described previously; however, cell types expressing LIF, including bone marrow stromal cells (42), CD4 and CD8 T cells (43), and monocytes/macrophages (44) are ubiquitous. The pre-reverse transcription site of LIF anti-HIV activity most closely resembles the site of allo-induced anti-HIV activity (41). This similarity is intriguing given that HLA class I concordance increases perinatal HIV-1 transmission (6). Though protection in HLA class I discordant pregnancies was suggested to be mediated by a protective allogeneic immune response, the findings from this previous study are consistent with allo-induced LIF expression.
Most importantly, our findings have significant therapeutic implications since progesterone, which can upregulate the positive LIF regulator IL-4, is commonly and safely administered to women with reduced fertility to increase the chances of implantation and term pregnancy. Progesterone has also been shown to be decreased in HIV-1–infected placenta (23), so progesterone therapy may also be important in order to restore critical hormonal levels in HIV-1–infected women.
Acknowledgments
The authors wish to acknowledge the gift of HIV-1 isolates from Phalguni Gupta and anti-LIF from Monica Sang (R&D Systems). The authors also wish to acknowledge Edward Barker, Stanford Shulman, Leon Epstein, Gene Shearer, and Judy Lieberman for critical review of the manuscript. This work was supported by American Foundation for AIDS Research grant 02633-26-RGI, the Women’s and Infants’ Transmission Study (WITS), the Women’s Interagency HIV Study (WIHS), NIH grant AI-47065, and the Swedish Research Foundation.
References
- 1.Ryder RW, et al. Perinatal transmission of the human immunodeficiency virus type 1 to infants of seropositive women in Zaire. N Engl J Med. 1989; 320:1637–1642. doi: 10.1056/NEJM198906223202501. [DOI] [PubMed] [Google Scholar]
- 2.The European Mode of Delivery Collaboration. Elective Caesarian-section versus vaginal delivery in prevention of vertical HIV-1 transmission: a randomized clinical trial. Lancet. 1999; 353:1035–1039. doi: 10.1016/s0140-6736(98)08084-2. [DOI] [PubMed] [Google Scholar]
- 3.Stiehm ER, et al. Efficacy of zidovudine and human immunodeficiency virus (HIV) hyperimmune immunoglobulin for reducing perinatal HIV transmission from HIV-infected women with advanced disease: results of Pediatric AIDS Clinical Trials Group protocol 185. J Infect Dis. 1999; 179:567–575. doi: 10.1086/314637. [DOI] [PubMed] [Google Scholar]
- 4.The International Perinatal HIV Group. The mode of delivery and the risk of vertical transmission of human immunodeficiency virus type 1: a meta-analysis of 15 prospective cohort studies. N Engl J Med. 1999; 340:977–987. doi: 10.1056/NEJM199904013401301. [DOI] [PubMed] [Google Scholar]
- 5.Rogers MF, Caldwell MB, Gwinn ML, Simonds RL. Epidemiology of pediatric human immunodeficiency virus infection in the United States. Acta Paediatr Suppl. 1994; 400:5–7. doi: 10.1111/j.1651-2227.1994.tb13324.x. [DOI] [PubMed] [Google Scholar]
- 6.MacDonald KS, et al. Mother-child class I HLA concordance increases perinatal human immunodeficiency virus type 1 transmission. J Infect Dis. 1998; 177:551–556. doi: 10.1086/514243. [DOI] [PubMed] [Google Scholar]
- 7.Phillips TA, et al. TRAIL (Apo-2L) and TRAIL receptors in human placentae: implications for immune privilege. J Immunol. 1999; 162:6053–6059. [PubMed] [Google Scholar]
- 8.Hunt JS, Vassmer D, Ferguson TA, Miller L. Fas ligand is positioned in mouse uterus and placenta to prevent trafficking of activated leukocytes between the mother and conceptus. J Immunol. 1997; 158:4122–4128. [PubMed] [Google Scholar]
- 9.Patterson, B.K., et al. 2000. Inhibition of and selection against vertical HIV-1 transmission by the placenta. The 7th Conference on Retroviruses and Opportunistic Infections. Chicago, Illinois, USA. January 31, 2000.
- 10.Rossi P, et al. Presence of maternal antibodies to human immunodeficiency virus 1 envelope glycoprotein gp120 epitopes correlates with the uninfected status of children born to seropositive mothers. Proc Natl Acad Sci USA. 1989; 86:8055–8058. doi: 10.1073/pnas.86.20.8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin X, et al. Longitudinal and cross-sectional analysis of cytotoxic T lymphocyte responses and their relationship to vertical human immunodeficiency virus transmission. J Infect Dis. 1998; 178:1317–1326. doi: 10.1086/314455. [DOI] [PubMed] [Google Scholar]
- 12.Bourinbaiar AS, Lee-Huang S. Anti-HIV effect of beta subunit of human chorionic gonadotropin (β-HCG) in vitro. Immunol Lett. 1995; 44:13–18. doi: 10.1016/0165-2478(94)00191-s. [DOI] [PubMed] [Google Scholar]
- 13.De SK, et al. Human chorionic gonadotropin hormone prevents wasting syndrome and death in HIV-1 transgenic mice. J Clin Invest. 1997; 99:1484–1491. doi: 10.1172/JCI119310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lunardi-Iskandar Y, et al. Effects of a urinary factor from women in early pregnancy on HIV-1, SIV and associated disease. Nat Med. 1998; 4:428–434. doi: 10.1038/nm0498-428. [DOI] [PubMed] [Google Scholar]
- 15.Stewart CL, et al. Blastocyst implantation depends on maternal expression of leukemia inhibitory factor. Nature. 1992; 359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 16.Piccinni M-P, et al. Defective production of both leukemia inhibitory factor and type 2 T-helper cytokines by decidual T cells in unexplained recurrent abortions. Nat Med. 1998; 4:1020–1024. doi: 10.1038/2006. [DOI] [PubMed] [Google Scholar]
- 17.Patterson BK, et al. Repertoire of chemokine receptor expression in the female genital tract: implications for human immunodeficiency virus transmission. Am J Pathol. 1998; 153:481–490. doi: 10.1016/S0002-9440(10)65591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson J, et al. Perforin is not co-expressed with granzyme A within cytotoxic granules in CD8 T lymphocytes present in lymphoid tissue during chronic HIV infection. AIDS. 1999; 13:1295–1303. doi: 10.1097/00002030-199907300-00005. [DOI] [PubMed] [Google Scholar]
- 19.Schmidtmayerova H, Alfano M, Nuovo G, Bukrinsky M. Human immunodeficiency virus type 1 T-lymphotropic strains enter macrophages via a CD4- and CXCR4-mediated pathway: replication is restricted at a postentry level. J Virol. 1998; 72:4633–4642. doi: 10.1128/jvi.72.6.4633-4642.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLeod GX, McGrath JM, Ladd EA, Hammer SM. Didanosine and zidovudine resistance patterns in clinical isolates of human immunodeficiency virus type 1 as determined by a replication endpoint concentration assay. Antimicrob Agents Chemother. 1992; 36:920–925. doi: 10.1128/aac.36.5.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winslow OS, et al. New soluble formazan assay for HIV-1 cytopathic effects: application to high flux screening of synthetic and natural products for AIDS-antimicrobial activity. J Natl Cancer Inst. 1989; 81:577–586. doi: 10.1093/jnci/81.8.577. [DOI] [PubMed] [Google Scholar]
- 22.Amirhessami-Aghili N, Spector SA. Human immunodeficiency virus type 1 infection of human placenta: potential route for fetal infection. J Virol. 1991; 65:2231–2236. doi: 10.1128/jvi.65.5.2231-2236.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonyhadi ML, Su L, Auten J, McCune JM, Kaneshima H. Development of a human thymic organ culture model for the study of HIV pathogenesis. AIDS Res Hum Retroviruses. 1995; 11:1073–1080. doi: 10.1089/aid.1995.11.1073. [DOI] [PubMed] [Google Scholar]
- 24.Collins KB, Patterson BK, Naus GJ, Landers DV, Gupta P. Development of an in vitro organ culture model to study transmission of HIV-1 in the female genital tract. Nat Med. 2000; 6:475–479. doi: 10.1038/74743. [DOI] [PubMed] [Google Scholar]
- 25.Tscherning-Casper C, et al. The trophoblastic epithelial barrier is not infected in full-term placentae of human immunodeficiency virus-seropositive mothers undergoing anti-retroviral therapy. J Virol. 1999; 73:9673–9678. doi: 10.1128/jvi.73.11.9673-9678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson BK, et al. Regulation of CCR5 and CXCR4 expression by type 1 and type 2 cytokines: CCR5 expression is downregulated by IL-10 in CD4-positive lymphocytes. Clin Immunol. 1999; 91:254–262. doi: 10.1006/clim.1999.4713. [DOI] [PubMed] [Google Scholar]
- 27.Bukrinsky MI, et al. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993; 90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackeiwicz CE, Blackbourn DJ, Levy JA. CD8+ T cells suppress human immunodeficiency virus replication by inhibiting viral transcription. Proc Natl Acad Sci USA. 1995; 92:2308–2312. doi: 10.1073/pnas.92.6.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis S, et al. LIFR beta and gp130 as heterodimerizing signal transducers of the tripartite CNTF receptor. Science. 1993; 260:1805–1808. doi: 10.1126/science.8390097. [DOI] [PubMed] [Google Scholar]
- 30.Fujio Y, Kunisada K, Hirota H, Yamauchi-Takihara K, Kishimoto T. Signals through gp130 upregulate bcl-x gene expression via STAT1-binding cis-element in cardiac myocytes. J Clin Invest. 1997; 99:2898–2905. doi: 10.1172/JCI119484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cocchi F, et al. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995; 270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 32.Wegmann TG, Lin H, Guilbert L, Mossman TR. Bidirectional cytokine interactions in the maternal-fetal relationship: is successful pregnancy a Th2 phenomenon? Immunol Today. 1990; 14:353–356. doi: 10.1016/0167-5699(93)90235-D. [DOI] [PubMed] [Google Scholar]
- 33.Szekenes-Bartho J, Wegmann TG. A progesterone-dependent immunomodulatory protein alters the Th1/Th2 balance. J Reprod Immunol. 1996; 31:81–95. doi: 10.1016/0165-0378(96)00964-3. [DOI] [PubMed] [Google Scholar]
- 34.Piccinni M-P, et al. Progesterone favors the development of human T helper cells producing Th2-type cytokines and promotes both IL-4 production and membrane CD30 expression in established Th1 cell clones. J Immunol. 1995; 155:128–133. [PubMed] [Google Scholar]
- 35.Shearer WT, et al. Early spontaneous abortions and fetal thymic abnormalities in maternal-to-fetal HIV infection. Acta Paediatr Suppl. 1997; 421:60–64. doi: 10.1111/j.1651-2227.1997.tb18322.x. [DOI] [PubMed] [Google Scholar]
- 36.Stahl N, et al. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995; 267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 37.Zhang XG, et al. Ciliary neurotropic factor, interleukin-11, leukemia inhibitory factor, and oncostatin M are growth factors for human myeloma cell lines using the interleukin 6 signal transducer gp130. J Exp Med. 1994; 179:1337–1342. doi: 10.1084/jem.179.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stahl N, et al. Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL6β receptor components. Science. 1994; 263:92–95. doi: 10.1126/science.8272873. [DOI] [PubMed] [Google Scholar]
- 39.Giordano V, et al. Shc mediates IL-6 signaling by interacting with gp130 and JAK2 kinase. J Immunol. 1997; 158:4097–4103. [PubMed] [Google Scholar]
- 40.Walker CM, Moody DJ, Stites DP, Levy JA. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science. 1986; 234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- 41.Pinto LA, Blazevic V, Patterson BK, Dolan MJ, Shearer G. Alloantigen-induced anti-HIV activity occurs prior to reverse transcription and can be generated by leukocytes from HIV-infected individuals. Blood. 2000; 95:1875–1876. [PubMed] [Google Scholar]
- 42.Wetzler M, et al. Alteration in bone marrow adherent layer growth factor expression: a novel mechanism of chronic myelogenous leukemia progression. Blood. 1991; 78:2400–2406. [PubMed] [Google Scholar]
- 43.Metcalf D, Nicola NA, Gearing DP. Effects of injected leukemia inhibitory factor on hematopoietic and other tissues in mice. Blood. 1990; 76:50–56. [PubMed] [Google Scholar]
- 44.Anegon I, et al. Production of human interleukin for DA cells (HILDA) by activated monocytes. Cell Immunol. 1990; 130:50–65. doi: 10.1016/0008-8749(90)90161-j. [DOI] [PubMed] [Google Scholar]