Abstract
Tpl-2 expression is induced within 30–60 min after ConA stimulation of rat splenocytes, suggesting that it may contribute to the induction of IL-2 during T cell activation. Herein we show that wild-type and carboxyl-terminally truncated (activated) Tpl-2 activate the nuclear factor of activated T cells (NFAT) and induce interleukin 2 (IL-2) expression in EL4 cells. In Jurkat cells the truncated Tpl-2 activates NFAT and induces IL-2, whereas wild-type Tpl-2 activates NFAT only when cotransfected with NFAT expression constructs, suggesting that Tpl-2 may induce NFAT activation signals. Experiments in NIH 3T3 cells revealed that the NFATp isoform, but not the NFATc or NFATx isoform, undergoes nuclear translocation when coexpressed with wild-type Tpl-2 and confirmed this hypothesis. Activation of NFAT by anti-CD3 stimulation but not by phorbol 12-myristate 13-acetate and ionomycin in Jurkat cells was inhibited by the kinase-dead Tpl-2K167M, suggesting that Tpl-2 contributes to the transduction of NFAT activation signals originating in the T cell receptor. The Tpl-2-mediated induction of IL-2 was not observed in T cell lymphoma lines other than EL4 and Jurkat, as well as in normal T cells. NFAT activation by Tpl-2, however, was observed in several cell lines including some of nonhematopoietic origin. The activation of NFAT by Tpl-2 in different cell types defines a molecular mechanism that may contribute to its oncogenic potential.
T cell activation is induced via the interaction of antigenic peptides associated with major histocompatibility complex molecules on the surface of antigen-presenting cells with the T cell receptor (TCR)/CD3 complex on T cells, in combination with signals delivered by one or more auxiliary receptors. Alternatively, the activation of T cells can be induced by signals generated by phorbol esters (activation of protein kinase C) and ionomycin (induction of Ca2+ influx). The synergistic effect of these two signals leads to the activation of distinct signaling pathways that ultimately converge to induce interleukin 2 (IL-2) transcription within 2–5 h from the start of the stimulation (1–3).
Induction of IL-2 during T cell activation depends on the concerted action of several transcription factors, one of which, nuclear factor of activated T cells (NFAT), is obligatory for IL-2 expression (4). NFAT, which until recently was known as a DNA binding activity that is induced shortly after T cell stimulation, is a complex factor composed of AP1 and NFAT components (5). Four isoforms of NFAT identified to date, NFATp (NFAT1 or NFATc2), NFATc (NFAT2 or NFATc1), NFAT3, and NFATx (NFAT4 or NFATc3), are encoded by different genes (6).
All NFAT isoforms are phosphoproteins that are expressed in different combinations in many types of cells including cells of both hematopoietic and nonhematopoietic origin (6). Phosphorylation of NFATp at amino-terminal phosphorylation sites, mapped between residues 172–176, 178–181, and 184–188, is under the control of a kinase complex that is regulated by the phosphatidylinositol 3-kinase (7). One component of this complex, GSK3, is negatively regulated by Akt (8). Phosphorylation at these sites inactivates NFAT by promoting nuclear export and cytoplasmic localization (9). Similarly, nuclear translocation of NFATx is inhibited by phosphorylation at Ser163 and Ser165 mediated by JNK2 (10). Dephosphorylation of these residues by calcineurin, a phosphatase activated by signals that increase cytoplasmic calcium levels, induces nuclear translocation and activation of the protein. Therefore, NFAT activity depends on opposing signals that regulate its phosphorylation (11).
Signals that activate NFAT may do so both by regulating the phosphorylation of different NFAT isoforms and by regulating their expression. Thus, in unstimulated human peripheral blood T cells NFATc is not expressed, whereas NFATp and NFATx are expressed constitutively but are inactive. Ionomycin stimulation induces NFATc expression and activates NFATc and NFATp but not NFATx (12).
Earlier studies identified NFAT as a transcription factor involved in the induction of IL-2 during T cell activation. More recent studies, however, revealed that activated NFAT contributes to the expression of several additional cytokines in different cell types (6). In T cells, activation of NFAT is not only observed during stimulation but also during development. Thus, it has been shown that NFAT is not active in CD4+/CD8+ thymocytes and that it is activated as the cells undergo positive selection and progress to the single positive state. The lack of NFAT activity in double positive cells correlates with their inability to secrete cytokines (13). Finally, NFAT interacts with the cell cycle and apoptotic machineries. Thus, NFAT activation promotes G1 phase progression and, in the absence of complimentary signals, induces apoptosis (14). Bcl-2, a molecule that inhibits G1 phase progression and apoptosis, binds calcineurin and inhibits NFAT activation (14). On the other hand, Bax, a molecule that promotes G1 phase progression and apoptosis, inhibits the interaction between Bcl-2 and calcineurin and ablates the Bcl-2 inhibitory effect on NFAT activation (15).
The gene encoding the Tpl-2 kinase was cloned by provirus tagging from Moloney murine leukemia virus (MoMuLV)-induced rat thymomas and is primarily expressed in spleen, thymus, and lung. Provirus integration in Tpl-2 occurs during tumor progression in 22.5% (9 of 40 thymomas) of MoMuLV-induced thymomas (16, 17). Tpl-2 spans a 35-kb genomic DNA region and consists of eight exons (17). The provirus always integrates in the last intron of, and in the same transcriptional orientation as, Tpl-2. Provirus insertion at this site induces high levels of expression of a Tpl-2 mRNA transcript that encodes a protein with an altered carboxyl-terminal tail. In this truncated protein, the 43 carboxyl-terminal amino acids encoded by the last exon are replaced by 7 amino acids encoded by intron sequences (16). The carboxyl-terminally truncated Tpl-2 exhibits a 5- to 7-fold increase in its constitutive kinase activity and is highly oncogenic (18).
Earlier studies on the signaling role of Tpl-2 had shown that it transduces signals that activate the mitogen-activated protein kinase and stress-activated protein kinase (SAPK) pathways (18–20). In this report we present evidence that Tpl-2 activates the transcription factor NFAT and induces IL-2 expression in EL4 and Jurkat cells. NFAT activation by anti-CD3 in the same cells is blocked by the kinase-dead Tpl-2K167M mutant, suggesting that in these cells Tpl-2 is involved in the transduction of NFAT activation signals originating in the T cell receptor. Although the Tpl-2-mediated induction of IL-2 is limited to only a few T cell lymphoma lines, the Tpl-2-mediated activation of NFAT appears to be a universal feature of both hematopoietic and nonhematopoietic cells. Of the various NFAT isoforms, Tpl-2 activates NFATp but not NFATx and affects the activity of NFATc only marginally.
MATERIALS AND METHODS
Cells and Cell Culture.
Jurkat cells were maintained at 37°C and 5% CO2 in RPMI 1640 medium supplemented with 10% (vol/vol) fetal bovine serum and penicillin (50 international units/ml), streptomycin (50 μg/ml), and kanamycin (100 μg/ml) (PSK). Jurkat cells stably transfected with a simian virus 40 (SV40) large tumor antigen expression construct and an NFAT/lacZ reporter construct (TAg-Jurkat) (5) were provided by G. Crabtree (Stanford University) and they were cultured in the same medium as Jurkat cells but supplemented with G418 (400 μg/ml) and hygromycin (400 μg/ml). EL4 cells were maintained also at 37°C and 5% CO2 in DMEM supplemented with 10% horse serum and PSK. NIH 3T3 cells and SRα or SRα-Tpl-2wt-transfected NIH 3T3 cells were maintained in DMEM supplemented with 10% calf serum and PSK. To induce activation, EL4, Jurkat, and TAg-Jurkat cells were treated with phorbol 12-myristate 13-acetate (PMA; 10 nM) and ionomycin (2 nM) for 8 h. TAg-Jurkat cells were alternatively treated for 8 h with the anti-CD3 monoclonal antibody OKT3 (1 μg/ml), provided by A. Altman (La Jolla Institute of Allergy and Immunology).
Plasmids.
The reporter plasmid IL-2/CAT (15deltacxCAT) containing 326 bp of the IL-2 promoter upstream of the transcriptional initiation site (21) was provided by G. Crabtree. The reporter construct NFAT/CAT (pSPNFAT/CAT) containing three NFAT binding sites (AGAAAGGAGGAAAAACTGTTTCATACAGAAGGCGTT) upstream of the minimal SV40 promoter and the mutant pSPNFATmAP-1/CAT containing three mutant NFAT sites (AGAAACGTCCTAAAACTGTTTCATACAGAAGGCGTT) (22) were provided by C. Thompson (University of Chicago). HA-NFATc (pSH102) (23), Flag-NFATp (pSH202), and Flag-NFATx (pSH250) constructs were provided by G. Crabtree.
IL-2 Assays.
EL4 cells stably transfected with wild-type or carboxyl-terminally truncated Tpl-2 were selected with puromycin (2.5 μg/ml). Culture supernatants were harvested during logarithmic-phase growth. Supernatants of transiently transfected EL4 or Jurkat cells were harvested 36 h after transfection. Human IL-2 (Jurkat cells) and mouse IL-2 (EL4 cells) were measured by quantitative ELISA assays according to the protocols provided by the manufacturers of the IL-2 ELISA kits (Intertest-2X, Genzyme, for the murine IL-2; IL-2EASIA, Medgenics, Fleurus, Belgium for the human IL-2). The concentration of IL-2 was estimated in pg/ml.
Transient Transfections and Chloramphenicol Acetyltransferase (CAT) Assays.
Approximately 107 EL4 or Jurkat cells were transiently transfected by electroporation at 260 mV and then harvested 44 h later. Selected cultures were stimulated with PMA and ionomycin 8 h prior to harvesting. Transfections were carried out with 8 μg of IL-2/CAT or 5 μg of pSPNFAT/CAT reporter constructs in combination with 2.5 μg of each expression construct (5 μg, total) and 1 μg of a CMV-βgal expression construct. If only one expression construct was used in addition to CMV-βGal, 2.5 μg of the CMV5 empty vector was added to bring the total amount of vector DNA containing the cytomegalovirus (CMV) promoter to 6 μg per transfection. The amount of electroporated DNA was brought to a total of 20 μg by adding pBluescript. Each transfection was carried out in quadruplicate and every experiment was repeated at least twice. Transient transfections in NIH 3T3 cells were carried out by using Lipofectamine (GIBCO/BRL) according to the protocols provided by the manufacturer. Briefly, 1 μg of each expression plasmid (2 μg, total) and 500 ng of reporter plasmid were transfected transiently in 2.5 × 105 NIH 3T3 or NIH 3T3⋅Tpl-2wt plated 1 day earlier in six-well plates.
CAT assays were performed as described (24). The acetyltransferase reaction was carried out for 3 h. The acetylated and the nonacetylated forms of [14C]chloramphenicol (Amersham) were separated on TLC plates (Sigma). Chloramphenicol acetylation was quantitated by using a Fuji phosphor-imager and Fuji mac-bas V2.2 software. CAT assays in NIH 3T3 cells were performed as described (25). Cells were lyzed by three freeze–thaw cycles 48 h after transfection, normalized for transfection efficiency according to the results of a β-galactosidase (βgal) assay (26) and analyzed for CAT activity by measuring diffusion of the 3H-acetylated form in scintillation fluid (24). The relative CAT activity in each experiment was calculated by assigning the highest CAT activity in each experiment the arbitrary value of 100. Other values were calculated as percentages of the highest value.
Nuclear Extracts and Electrophoretic Mobility Shift Assays (EMSA).
Jurkat cells were transiently transfected by electroporation with the indicated constructs and then harvested 42 h later. Some of the nontransfected cultures were stimulated with PMA and ionomycin for 6 h prior to harvesting, as indicated. At harvesting, cells were lysed in a buffer containing 10 mM Hepes, 10 mM KCl, and 0.2 mM EDTA. Nuclei were isolated by centrifugation at 4,000 rpm in a microcentrifuge and nuclear proteins were extracted by using a buffer containing 20 mM Hepes, 25% glycerol, 0.4 M NaCl, 0.2 mM EDTA, 0.5 mM DTT, and complete proteinase inhibitors (Boehringer Mannheim). Protein concentration was measured by the Bradford assay (Bio-Rad). Double-stranded oligonucleotides were end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs) (24). Binding of nuclear extracts to labeled oligonucleotides was carried out by incubating approximately 7 μg of protein and 105 cpm (0.5 ng) of labeled oligonucleotides in 20 μl at room temperature (∼20°C) for 15 min. The binding buffer contained 4 mM Tris⋅HCl (pH 7.5), 80 mM NaCl, 0.5 mM ZnSO4, 5% glycerol, 0.5% DTT, and poly(dI-dC) (0.2 mg/ml). To compete the binding, a 100-fold excess of unlabeled oligonucleotide was added to the binding reaction before adding the labeled oligonucleotide. Unbound oligonucleotides were separated from DNA–protein complexes in a 6% polyacrylamide gel.
Detection of NFAT Proteins by Immunofluorescence.
NIH 3T3 cells stably transfected with an SRα-based wild-type Tpl-2 construct, or the empty SRα vector, were transiently transfected with hemagglutinin (HA)-NFATc, Flag-NFATp, or Flag-NFATx expression constructs. Transient transfections were carried out by using Lipofectamine (GIBCO/BRL). Thirty-six hours later cells were fixed in 3.5% paraformaldehyde and stained with anti-HA (HA.11 mouse monoclonal antibody, Babco, Richmond, CA) or anti-Flag (M2, Kodak) antibodies and with fluorescein isothiocyanate-conjugated anti-mouse IgG secondary antibody (Sigma). Cells were visualized by confocal microscopy.
RESULTS
Tpl-2 Induces IL-2 Expression in EL4 and Jurkat Cells.
Stimulation of T cells with a combination of signals induces expression of IL-2 and its receptor (IL-2Rα). This event marks the commitment of T cells to later activation events (27). Interaction of IL-2 with the IL-2 receptor leads to the expression of a series of cytokines that promote cell growth and differentiation (28). In addition to normal T cells, certain T cell lines retain the ability to express IL-2 either via the combined stimulation of TCR and CD28 or via stimulation with pharmacological agents. EL4 and Jurkat cells, for example, express IL-2 when stimulated with antibodies to CD3 and CD28 (29) or with PMA and ionomycin (30).
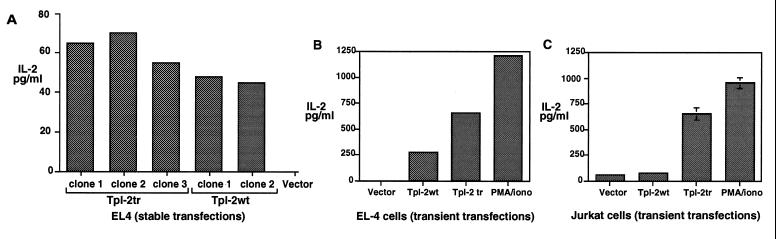
Tpl-2 expression precedes the expression of IL-2 after stimulation of rat spleen cells with Con A. Therefore, it could contribute to the induction of IL-2 in stimulated T cells (16). To test this hypothesis, EL4 cells stably transfected with Tpl-2 expression constructs were analyzed to determine whether they secrete IL-2. The results showed that cell clones transfected with both wild-type and carboxyl-terminally truncated Tpl-2 express Tpl-2, as determined by Northern blotting (data not shown) and secrete IL-2 (Fig. 1A). To determine whether Tpl-2 induces IL-2 expression in cells other than EL4, we transiently transfected both wild-type and carboxyl-terminally truncated Tpl-2 constructs in Jurkat cells. EL4 cells transiently transfected with the same Tpl-2 expression constructs were used as controls. Thirty-six hours later IL-2 secretion was measured by ELISA in the medium of the transfected cultures. The results showed that although both wild-type and truncated Tpl-2 trigger IL-2 secretion in EL4 cells, only the truncated Tpl-2 induces IL-2 in Jurkat cells (Fig. 1 B and C). The Tpl-2-mediated induction of IL-2 in different cell lines suggests that Tpl-2 may contribute to the transduction of signals that regulate the activity of the IL-2 promoter. While these data were being prepared for publication, an independent study was published also showing that carboxyl-terminally truncated Tpl-2 induces IL-2 expression in Jurkat cells (31).
Figure 1.
Overexpression of Tpl-2 induces IL-2 expression in EL4 and Jurkat cells. (A) IL-2 was measured by ELISA in culture supernatants of three clones of EL4 cells transfected stably with the carboxyl-terminally truncated Tpl-2 construct (Tpl-2tr) and two independent clones of EL4 cells transfected stably with the wild-type Tpl-2 construct (Tpl-2wt). Vector (pBabe) transfected cells secrete no IL-2. (B and C) EL4 and Jurkat cells were transiently transfected with Tpl-2 and carboxyl-terminally truncated Tpl-2 expression constructs. Thirty-six hours after transfection, empty vector-transfected control cells were stimulated with PMA plus ionomycin and cultured for an additional 8 h period. Culture supernatants were harvested and IL-2 was measured by ELISA.
Tpl-2 Activates the IL-2 Promoter: The Role of NFAT.
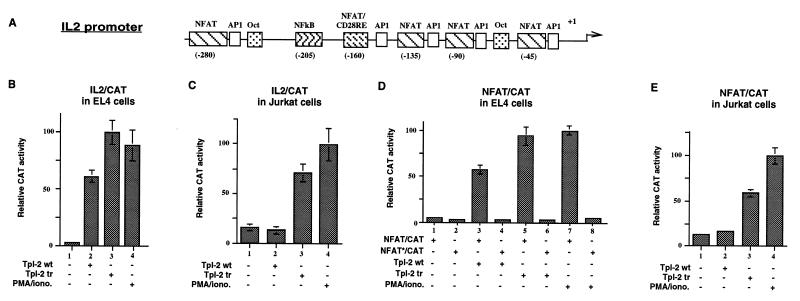
To determine whether the induction of IL-2 was due to the activation of the IL-2 promoter, we cotransfected Tpl-2 with a construct containing a 326-bp IL-2 promoter fragment linked to the CAT gene (Fig. 2A). Expression of the CAT reporter was measured enzymatically. The results showed that both wild-type and truncated Tpl-2 activate the IL-2 promoter in EL4 cells (Fig. 2B), whereas only the truncated form of the protein activates the IL-2 promoter in Jurkat cells (Fig. 2C). These results correspond to the results in Fig. 1 showing that both forms of Tpl-2 induce IL-2 expression in EL4 cells whereas only the truncated form induces IL-2 expression in Jurkat cells.
Figure 2.
Tpl-2 activates the IL-2 promoter: The role of NFAT. (A) Diagrammatic representation of the IL-2 promoter showing the position of previously mapped transcription factor binding sites. (B) Both wild-type and truncated Tpl-2 activate an IL-2 promoter/CAT reporter in EL4 cells. (C) The same reporter is activated by truncated Tpl-2 but not wild-type Tpl-2 in Jurkat cells. (D) Both wild-type and truncated Tpl-2 activate a CAT reporter construct containing a minimal SV40 promoter with three NFAT binding sites upstream, in EL4 cells. The asterisk indicates that the NFAT binding site is mutated and fails to bind NFAT. (E) The same reporter construct is activated by truncated but not wild-type Tpl-2 in Jurkat cells. Reporter constructs containing a mutant NFAT binding site are inactive in both EL4 and Jurkat cells. All transfections were done in quadruplicate and expression was normalized based on the expression of βgal from a cotransfected CMV-β-Gal construct.
The IL-2 promoter contains five NFAT binding sites, an NF-κB binding site, and two Oct-1 sites (4). It has been previously shown that, among these factors, NFAT is obligatory for the induction of IL-2 expression during T cell activation (32). We, therefore, examined the effects of Tpl-2 on the activity of a CAT reporter construct in which the reporter is under the control of a minimal SV40 promoter with three copies of the NFAT binding site placed upstream. This reporter was cotransfected with Tpl-2 into EL4 cells and the activity of the promoter was monitored by measuring CAT activity. A reporter construct with three mutant NFAT binding sites upstream of the minimal SV40 promoter was used as a control. The results showed that Tpl-2 activates the promoter flanked by the wild-type (Fig. 2D, bars 3 and 5) but not the mutant NFAT binding sites (Fig. 2D, bars 4 and 6). In agreement with the IL-2 promoter data, only the truncated form of Tpl-2 induced NFAT activation in Jurkat cells (Fig. 2E).
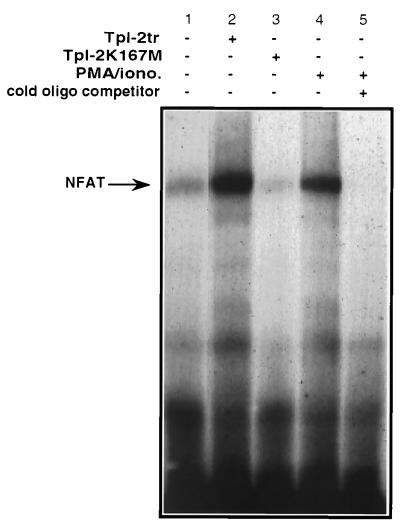
Additional experiments used EMSAs to determine whether Tpl-2 induces NFAT binding activity in nuclear extracts of transiently transfected Jurkat cells. The results showed that the carboxyl-terminally truncated Tpl-2 induces NFAT binding activity in these cells (Fig. 3, lane 2) whereas the kinase-dead Tpl-2K167M does not (Fig. 3, lane 3).
Figure 3.
Tpl-2 induces NFAT binding activity. EMSA of a 32P-labeled double-stranded oligonucleotide containing an NFAT binding site after incubation with nuclear extracts of Jurkat cells transiently transfected with the indicated constructs or stimulated with PMA and ionomycin.
Wild-Type Tpl-2 Acts Synergistically with NFATp and NFATc to Activate the IL-2 Promoter and an NFAT-Driven Promoter in Jurkat Cells.
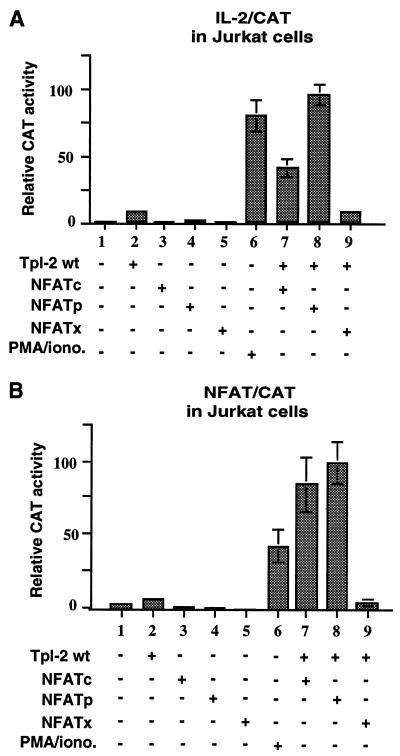
To confirm that Tpl-2 activates NFAT and to determine which NFAT isoform(s) may be activated by Tpl-2, we examined the potential synergism between wild-type Tpl-2 and the NFAT isoforms NFATc, NFATp, and NFATx. To this end, we cotransfected expression constructs of these NFAT isoforms and wild-type Tpl-2 with the reporter constructs IL-2/CAT or NFAT/CAT in Jurkat cells. The results showed that wild-type Tpl-2 does not activate either of the two promoters. However, it acts synergistically with NFATp and, to a lesser degree, with NFATc but not with NFATx to induce IL-2/CAT (Fig. 4A, bars 7–9) and NFAT/CAT activity (Fig. 4B, bars 7–9).
Figure 4.
Tpl-2 collaborates with NFATp and NFATc but not NFATx to activate IL-2/CAT and NFAT/CAT reporters. Jurkat cells were cotransfected with expression constructs of wild-type Tpl-2 and NFATp, NFATc, or NFATx in combination with IL-2/CAT (A) or NFAT/CAT (B) reporter constructs. All transfections were done in quadruplicate and expression was normalized based on the expression of βgal from a cotransfected CMV-β-Gal construct.
Tpl-2 Activates NFATp in NIH 3T3 Cells.
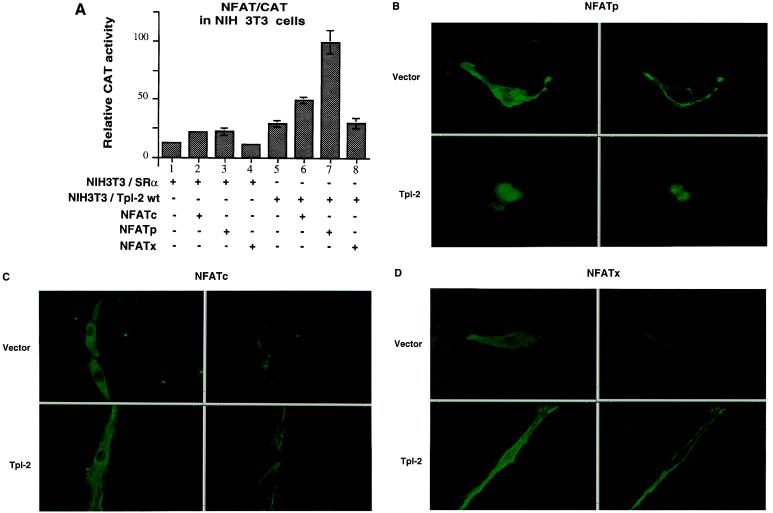
To determine whether the synergism of Tpl-2 with NFATp and perhaps NFATc occurs in cells other than the two T cell lines tested, we examined the ability of wild-type Tpl-2 to act synergistically with NFATp, NFATc, and NFATx in NIH 3T3 cells. To this end, we tested NIH 3T3 cells transfected stably with an SRα-based expression construct of the wild-type Tpl-2 tagged at its carboxyl terminus with an HA-tag (Tpl-2wt-HA) or with vector only. These cells were transiently cotransfected with SRα-based expression constructs of Flag-NFATp, HA-NFATc, or Flag-NFATx and the NFAT/CAT reporter construct. CAT activity was measured in cell lysates harvested 48 h later. The results (Fig. 5A) confirmed that wild-type Tpl-2 acts synergistically with NFATp but not with NFATx. Similar results were obtained in transient cotransfections of Tpl-2 and the same NFAT isoforms in NIH 3T3 and 293 cells (data not shown).
Figure 5.
Tpl-2 activates NFATp. (A) Relative CAT activity in freeze–thaw lysates of NIH 3T3 cells stably transfected with an SRα-based Tpl-2 construct or with SRα and cotransfected with expression constructs of NFATp, NFATc, or NFATx and the reporter construct NFAT/CAT. All transfections were performed in triplicate and transfection efficiency was monitored by using βgal. (B–D) Subcellular localization of NFATp, NFATc, and NFATx expressed from transiently transfected expression constructs in the NIH 3T3 shown in A. Localization was determined by immunofluorescence using anti-HA (HA-NFATc) or anti-Flag (Flag-NFATp or Flag-NFATx) mouse monoclonal antibodies and an anti-mouse IgG secondary antibody conjugated with fluorescein isothiocyanate. Stained cells were examined by confocal microscopy. (Left) Entire cell. (Right) Confocal image of a slice through the nucleus.
The synergism between wild-type Tpl-2 and NFATp suggests that either Tpl-2 induces signals that activate NFATp or that Tpl-2 and NFATp induce complementary signals that converge to activate the NFAT-driven promoter. To distinguish between these possibilities, we examined whether Tpl-2 alters the subcellular localization of NFATp. To this end, the stably transfected NIH 3T3 cells used in the preceding experiment (Fig. 5A) were transiently transfected with an SRα-based expression construct of Flag-NFATp. HA-NFATc and Flag-NFATx expression constructs were transfected separately as controls. Thirty-six hours later, the subcellular localization of the NFAT proteins was determined by immunofluorescence with antibodies against the HA- or Flag-epitope tags. The results showed that all NFAT isoforms were cytoplasmic in cells that did not overexpress Tpl-2 (Fig. 5 B–D). In cells that overexpressed Tpl-2, NFATp but not NFATc or NFATx was nuclear (Fig. 5 B–D). This observation confirmed the activation of NFAT by Tpl-2. Because Tpl-2 also activates NFATc although weakly (Fig. 4 and 5A), our inability to detect nuclear translocation of NFATc in Tpl-2 expressing cells is likely to reflect the fact that NFATc may translocate in response to Tpl-2 but that its translocation is minimal and quantitatively below the level of detection.
The Dominant Negative Mutant Tpl-2K167M Inhibits Anti-CD3-Induced but Not PMA- Plus Ionomycin-Induced Activation of NFAT.
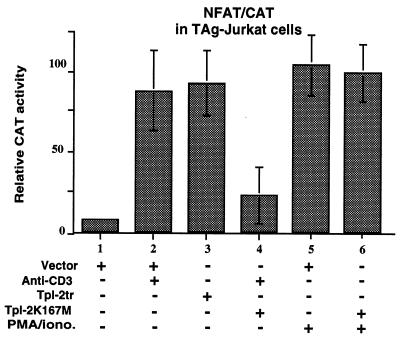
To determine the physiological relevance of the activation of NFAT by Tpl-2, we tested whether Tpl-2K167M, a dominant negative mutant of Tpl-2, can block NFAT activation by extracellular signals. In these experiments we used Jurkat cells stably transfected with an SV40 large tumor antigen expression construct (TAg-Jurkat cells) (5). These cells were chosen because they respond to stimulation by anti-CD3 (33), because of the large tumor antigen, they express transiently transfected constructs at high levels. Stimulation of the TAg-Jurkat TCR with the anti-CD3 antibody OKT3 induces activation of the NFAT/CAT reporter construct (Fig. 6, bars 1 and 2). When the kinase-dead mutant of Tpl-2 was expressed in this cell line, however, anti-CD3 could no longer activate this promoter (Fig. 6, bar 4), suggesting that signals originating in the T cell receptor are transduced via Tpl-2. The same kinase-dead mutant was unable to inhibit NFAT activation induced by PMA and ionomycin (Fig. 6, bars 5 and 6).
Figure 6.
Dominant negative mutant Tpl-2K167M inhibits the activation of NFAT by anti-CD3 but not by PMA plus ionomycin. Relative CAT activity in lysates of TAg-Jurkat cells transiently transfected with the NFAT/CAT reporter construct and carboxyl-terminally truncated or kinase-dead (K167M) Tpl-2 expression constructs is shown. Selected cultures were stimulated with the human anti-CD3 antibody OKT3 or PMA and ionomycin as indicated. All transfections were done in quadruplicate. Similar results were obtained in two independent experiments.
DISCUSSION
Evidence presented in this report shows that forced expression of Tpl-2 in T cell lines induces IL-2 gene expression and activates transiently transfected reporter constructs driven by the IL-2 promoter or an NFAT-regulated minimal SV40 promoter. The activation of NFAT by Tpl-2 suggested by these results was confirmed by EMSA. Of the various NFAT isoforms, Tpl-2 activates NFATp but not NFATx and affects the activity of NFATc minimally. This was suggested by experiments showing that Tpl-2 acts synergistically with NFATp to activate an NFAT-driven promoter in Jurkat, NIH 3T3, and 293 cells. This suggestion was confirmed by additional experiments showing that NFATp expressed from a transiently transfected expression construct in Tpl-2-expressing NIH 3T3 cells is nuclear. The kinase-dead Tpl-2K167M inhibits NFAT activation by signals originating in the TCR, suggesting that Tpl-2 plays a role in the physiological pathway of NFAT activation after TCR stimulation.
The activation of NFAT by Tpl-2 is not limited to hematopoietic cells in that it was observed in at least two additional cell lines of nonhematopoietic origin. The activation of the IL-2 promoter, on the other hand, may be unique to some transformed T cell lines. Thus, T cells from spleen and thymus of Tpl-2 transgenic mice, as well as other T cell lymphoma lines overexpressing Tpl-2, do not express IL-2 in the absence of stimulation. Moreover, spleen cells from Tpl-2 “knock-out” mice express IL-2 after stimulation with anti-CD3 and anti-CD28 antibodies (data not shown). Therefore, Tpl-2 activates NFAT in all cells tested to date but induces IL-2 expression in some T cell lymphoma lines but not in normal T cells. The activation of NFAT by Tpl-2 may, therefore, allow induction of IL-2 and other cytokines in some hematopoietic cell tumors depending on the activity of other complementary pathways. Independent of its involvement in cytokine gene expression, however, the activation of NFAT in different cell types is likely to play an important role in mediating the Tpl-2-induced oncogenic effects by altering the regulation of cell growth and apoptosis (14).
The data in this report showed that in Jurkat cells, Tpl-2 is involved in the transduction of signals that originate in the T cell receptor and activate NFAT and the IL-2 promoter. However, Tpl-2 may also be involved in the transduction of other extracellular signals. Thus, we have shown that Tpl-2 transduces tumor necrosis factor (TNF) α signals originating in TNF-R1 (C.P., C.T., and P.N.T., unpublished results). Although TNF-R1 signals may not activate NFAT, it was recently shown that NFAT is activated by TACI, a CAML-interacting molecule with similarities to TNF-R1 (34). The potential role of Tpl-2 in the transduction of NFAT activation signals originating in the TACI receptor is currently under investigation.
Nuclear localization of NFAT is controlled by phosphorylation. Thus, it has been shown that a GSK3 containing kinase complex, present in the nucleus, phosphorylates and stimulates nuclear export of NFATc (9). Because GSK3 has been reported to be negatively regulated by phosphorylation by the phosphatidylinositol 3-kinase-regulated protein kinase Akt (8), these findings suggest that nuclear export of NFAT may be inhibited by phosphatidylinositol 3-kinase and Akt. The findings reported here suggest that Tpl-2 may contribute to the nuclear translocation of NFAT perhaps by mediating signals that lead to the activation of phosphatases, which, in turn, promote NFAT dephosphorylation. Collectively, therefore, these findings suggest that Akt and Tpl-2, two oncogenes associated with the induction of T cell neoplasms (16, 35), are likely to cooperate in T cell oncogenesis by promoting NFAT activation by different mechanisms.
Tpl-2 induces nuclear translocation of NFATp in NIH 3T3 cells but is unable to induce nuclear translocation of NFATx. The failure to activate NFATx could be due to the Tpl-2-mediated activation of the SAPK/JNK pathway (18, 20). MKK7 (JNK-2) phosphorylates NFATx on Ser163 and Ser165 and blocks the nuclear translocation of NFATx in response to calcineurin (10). The potential involvement of the Tpl-2-mediated activation of the SAPK/JNK pathway in blocking the nuclear translocation of NFATx in response to Tpl-2 is currently under investigation.
Overall, the data presented in this report show that Tpl-2, a protooncogene associated with tumor progression in retrovirus-induced rodent T cell lymphomas (16) and mammary carcinomas (36), activates NFAT in addition to the mitogen-activated protein kinase and SAPK pathways (18–20). The Tpl-2-mediated activation of NFAT in both hematopoietic and nonhematopoietic cells defines a molecular mechanism that may contribute to the oncogenic potential of Tpl-2.
Acknowledgments
We thank A. Altman, G. Crabtree, and C. Thompson for providing reagents used in the course of this work. We also thank J. Caamano, A. Makris, H. L. Grimes, and the other members of the Tsichlis laboratory for stimulating discussions; J. Boyd for the confocal images; and Pat Bateman for secretarial assistance. This work was supported by Public Health Service Grant CA38047. Additional support was provided by Public Health Service Grant CA06927 and by an appropriation from the Commonwealth of Pennsylvania to the Fox Chase Cancer Center. C.P. is a special fellow of the Leukemia Society of America, Inc.
ABBREVIATIONS
- NFAT
nuclear factor of activated T cells
- IL-2
interleukin 2
- HA
hemagglutinin
- PMA
phorbol 12-myristate 13-acetate
- TCR
T cell receptor
- SAPK
stress-activated protein kinase
- CAT
chloramphenicol acetyltransferase
- EMSA
electrophoretic mobility shift assay
- SV40
simian virus 40
- βgal
β-galactosidase
References
- 1.Jain J, Loh C, Rao A. Curr Opin Immunol. 1995;7:333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 2.Crabtree G R. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- 3.Clipstone N A, Crabtree G R. Nature (London) 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 4.Rooney J W, Sun Y L, Glimcher L H, Hoey T. Mol Cell Biol. 1995;15:6299–6310. doi: 10.1128/mcb.15.11.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Northrop J P, Ullman K S, Crabtree G R. J Biol Chem. 1993;268:2917–2923. [PubMed] [Google Scholar]
- 6.Rao A, Luo C, Hogan P G. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 7.Jascur T, Gilman J, Mustelin T. J Biol Chem. 1997;272:14483–14488. doi: 10.1074/jbc.272.22.14483. [DOI] [PubMed] [Google Scholar]
- 8.Alessi D R, Caudwell F B, Andjelkovic M, Hemmings B A, Cohen P. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 9.Beals C R, Sheridan C M, Turck C W, Gardner P, Crabtree G R. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 10.Chow C W, Rincon M, Cavanagh J, Dickens M, Davis R J. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 11.Dolmetsch R E, Lewis R S, Goodnow C C, Healy J I. Nature (London) 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 12.Lyakh L, Ghosh P, Rice N R. Mol Cell Biol. 1997;17:2475–2484. doi: 10.1128/mcb.17.5.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rincon M, Flavell R A. Mol Cell Biol. 1996;16:1074–1084. doi: 10.1128/mcb.16.3.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linette G P, Li Y, Roth K, Korsmeyer S J. Proc Natl Acad Sci USA. 1996;93:9545–9552. doi: 10.1073/pnas.93.18.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibasaki F, Kondo E, Akagi T, McKeon F. Nature (London) 1997;386:728–731. doi: 10.1038/386728a0. [DOI] [PubMed] [Google Scholar]
- 16.Patriotis C, Makris A, Bear S E, Tsichlis P N. Proc Natl Acad Sci USA. 1993;90:2251–2255. doi: 10.1073/pnas.90.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makris A, Patriotis C, Bear S E, Tsichlis P N. J Virol. 1993;67:4283–4289. doi: 10.1128/jvi.67.7.4283-4289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ceci J D, Patriotis C P, Tsatsanis C, Makris A M, Kovatch R, Swing D A, Jenkins N A, Tsichlis P N, Copeland N G. Genes Dev. 1997;11:688–700. doi: 10.1101/gad.11.6.688. [DOI] [PubMed] [Google Scholar]
- 19.Patriotis C, Makris A, Chernoff J, Tsichlis P N. Proc Natl Acad Sci USA. 1994;91:9755–9759. doi: 10.1073/pnas.91.21.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salmeron A, Ahmad T B, Carlile G W, Pappin D, Narsimhan R P, Ley S C. EMBO J. 1996;15:817–826. [PMC free article] [PubMed] [Google Scholar]
- 21.Durand D B, Shaw J P, Bush M R, Replogle R E, Belagaje R, Crabtree G R. Mol Cell Biol. 1988;8:1715–1724. doi: 10.1128/mcb.8.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson C B, Wang C Y, Ho I C, Bohjanen P R, Petryniak B, June C H, Miesfeldt S, Zhang L, Nabel G J, Karpinski B, et al. Mol Cell Biol. 1992;12:1043–1053. doi: 10.1128/mcb.12.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Northrop J P, Ho S N, Chen L, Thomas D J, Timmerman L A, Nolan G P, Admon A, Crabtree G R. Nature (London) 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- 24.Zweidler M P, Grimes H L, Flubacher M M, Tsichlis P N. Mol Cell Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimes H L, Chan T O, Zweidler-McKay P A, Tong B, Tsichlis P N. Mol Cell Biol. 1996;16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bignon C, Daniel N, Djiane J. Biotechniques. 1993;15:243–246. [PubMed] [Google Scholar]
- 27.Crabtree G R, Clipstone N A. Annu Rev Biochem. 1994;63:1045–1083. doi: 10.1146/annurev.bi.63.070194.005145. [DOI] [PubMed] [Google Scholar]
- 28.Nelson B H, Lord J D, Greenberg P D. Nature (London) 1994;369:333–336. doi: 10.1038/369333a0. [DOI] [PubMed] [Google Scholar]
- 29.Boise L H, Noel P J, Thompson C B. Curr Opin Immunol. 1995;7:620–625. doi: 10.1016/0952-7915(95)80067-0. [DOI] [PubMed] [Google Scholar]
- 30.Fraser J D, Irving B A, Crabtree G R, Weiss A. Science. 1991;251:313–316. doi: 10.1126/science.1846244. [DOI] [PubMed] [Google Scholar]
- 31.Ballester A, Tobena R, Lisbona C, Calvo V, Alemany S. J Immunol. 1997;159:1613–1618. [PubMed] [Google Scholar]
- 32.Bierer B E, Mattila P S, Standaert R F, Herzenberg L A, Burakoff S J, Crabtree G, Schreiber S L. Proc Natl Acad Sci USA. 1990;87:9231–9235. doi: 10.1073/pnas.87.23.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y C, Elly C, Langdon W Y, Altman A. J Biol Chem. 1997;272:168–173. doi: 10.1074/jbc.272.1.168. [DOI] [PubMed] [Google Scholar]
- 34.Vonbulow G U, Bram R J. Science. 1997;278:138–141. doi: 10.1126/science.278.5335.138. [DOI] [PubMed] [Google Scholar]
- 35.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 36.Erny K M, Peli J, Lambert J F, Muller V, Diggelmann H. Oncogene. 1996;13:2015–2020. [PubMed] [Google Scholar]