Abstract
Approximately one-half of Caucasians with newly diagnosed insulin-dependent diabetes mellitus (IDDM) have autoantibodies to insulin, and the majority of those express the HLA-DR4 genotype [Ziegler, R., Alper, C. A., Awdeh, Z. L., Castano, L., Brink, S. J., Soeldner, J. S., Jackson, R. A. & Eisenbarth, G. S. (1991) Diabetes 40, 709–714]. However, it has been difficult to demonstrate T cell proliferative responses to human insulin in IDDM patients [Durinovic-Bello, I., Hummel, M. & Ziegler, A. G. (1996) Diabetes 45, 795–800]. We have immunized transgenic mice expressing the susceptible HLA-DR (α1*0101,β1*0401) (hereafter called DRB1*0401) and human CD4 molecules on a murine major histocompatibility complex class II null background, with human preproinsulin (PPI), proinsulin (PI), and insulin and derived large panels of T cell hybridomas to determine the immunogenic epitopes of these proteins. These results show that the prohormones PI or PPI carry the major immunogenic T cell epitope in the DRB1*0401 transgenic mice. The PPI/PI immunodominant epitope LALEGSLQK was localized at the C-peptide/A-chain junction. This T cell epitope PPI/PI LALEGSLQK is unusual because, normally, it is proteolytically destroyed during the maturation of the insulin molecule. Additionally, this T cell epitope is both processed and presented by human DRB1*0401-positive Epstein–Barr virus transformed B cells, and it can also stimulate T cells from the peripheral blood of HLA-DR4-positive patients with type 1 diabetes. These findings may partly explain why susceptibility to type 1 diabetes is associated with HLA-DR4-positive individuals and why T cell responses to the mature insulin protein are rarely detected in IDDM patients.
It has been known for more than 20 years that susceptibility to insulin-dependent diabetes mellitus (IDDM) has a very strong genetic component. Two major susceptibility regions have been identified in the human genome: the major histocompatibility complex (MHC) region on chromosome 6p21 (IDDM1) and the insulin gene region (IDDM2) on chromosome 11p15.5 (1–3). In Caucasians, susceptibility to IDDM is strongly associated with HLA-DR3, -DR4 haplotypes, whereas resistance is associated with HLA-DR2. (4, 5). In addition, certain HLA-DQB1, -DQA1, and DRB1 alleles confer susceptibility, whereas others provide resistance to disease (6, 7). Recent studies have shown that differences of a few amino acid residues in DR4 alleles are sufficient to modify the risk of developing IDDM conferred by the high-risk DQA1*03-DQB1*0302 (DQ8) haplotype (8–10). Among the HLA-DR4 alleles, DRB1*0405 confers the highest risk, DRB1*0401 has a higher risk than DRB1*0404, whereas DRB1*0403 confers a “dominant” protection even when the susceptible DRB1*0301-DQB1*0201 haplotype is present on the other chromosome (11, 12).
IDDM2 maps to a variable number of tandem repeats (VNTR) minisatellite upstream of the insulin gene (INS) (13). The short class I VNTR alleles (26–63 repeats) predispose to IDDM, whereas class III VNTR alleles (140–210 repeats) have a dominant protective effect (14, 15). Interestingly, in multiplex IDDM families, IDDM2 class I VNTR alleles are transmitted preferentially to HLA-DR4-positive diabetic offspring, suggesting that the effect of the susceptibility gene(s) in IDDM2 is dependent on the presence of HLA-DR4 (16).
The high level of insulin autoantibodies associated with expression of DR4 (17) and the early age of disease onset (18, 19) are consistent with this notion.
In the present study, using HLA-DRB1*0401 transgenic mice, we have found that the immunodominant epitopes restricted by this DR4 allele are located in the precursor forms of the hormone. These findings provide a possible explanation for how the insulin gene region-encoded susceptibility to type 1 diabetes is largely associated with HLA-DR4-positive individuals. We propose that one mechanism for susceptibility to IDDM may be that susceptible HLA-DR4 molecules present fragments of insulin and/or its prohormones to CD4+ T cells, thereby initiating an autoimmune response specifically targeted to the pancreatic β cells. The identification of the PPI/PI and Ins immunodominant T cell epitopes may provide tools for modulating or preventing diabetes in DR4-positive subjects.
METHODS
Generation of HLA-DR Transgenic Mice.
All of the animals used in this study were bred and maintained at the research animal facility at Stanford University. HLA-DRB1*0401 mice co-expressing human CD4 were generated as described previously (20). Transgenic mice expressing the DRB1*0401 allele and human CD4 on the DBA/1J background were mated with I-Aβ0/0 mice (21) (provided by Drs. D. Mathis, and C. Benoit). The offspring were screened for HLA-DR expression by flow cytometry (FACSCAN) analysis of peripheral blood lymphocytes using anti-HLA-DR antibody (L243, Becton Dickinson). The DRB1*0401, human CD4, I-Aβq/0 mice were backcrossed with I-Aβ0/0 mice, and the DR4+, I-Aβ0/0 offspring were intercrossed to generate homozygous mice (HLA-DRB1*0401+/+, human CD4+/+, and I-Aβ0/0). To distinguish between heterozygotes and homozygotes for the DR4 transgene, progeny were screened by anti-DR and anti-mouse CD4 double-staining FACSCAN analysis. CD4 T cell levels higher than 20% in peripheral blood lymphocytes were indicative of a homozygous state for the DRB1*0401 and human CD4 transgenes, as shown previously (22) (homozygous DRB1*0401 and human CD4 transgenes are necessary to permit normal levels of positive selection of CD4+ T cells in I-Aβ0/0 mice).
Expression and Purification of Recombinant Human Proinsulin and Preproinsulin.
The cDNA for human preproinsulin was a gift from Dr. D. Steiner (University of Chicago) and was coupled to a 6-histidine sequence and subcloned into pVL1393, a baculovirus vector (Invitrogen). To avoid cleavage of the leader peptide within insect cells, we have used the −1, −3 rule (23). In brief, mammalian and insect cells remove the leader peptide if the last amino acid of the peptide sequence (position −1) is small and neutral and the one in position −3 is not aromatic, charged, or large and polar. The Ala in position −3 of PPI was mutated to Asp, thereby ablating the cleavage site of the leader sequence.
Each construct was used to generate recombinant baculovirus using standard methods. PPI and PI were expressed in High Five cells (Invitrogen). Purification was achieved by Ni2+ chelate affinity chromatography of a cleared lysate obtained from guanidine hydrochloride lysed cells infected for 3 days. Preparations were separated by SDS/PAGE, and the specific band was visualized with KCl and purified to homogeneity by electroelution. Purity was checked by analytical SDS/PAGE and by Western blot analysis using anti-insulin polyclonal antibodies (Biodesign).
Generation of T Cell Hybridomas.
The DRB1*0401+/+, human CD4+/+, I-Aβ0/0 mice were immunized in the footpads and at the base of the tail with 50 μg of gel purified protein (PPI, PI) or with 50 μg of Ins (Human recombinant, Sigma) emulsified in incomplete Freund’s adjuvant (Difco). After 10 days, the draining lymph nodes were removed. Lymph node cells were resuspended at a concentration of 107 cells/ml in complete medium (RPMI 1640, Bethesda Research Laboratories) containing 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 50 μM 2-mercaptoethanol, and Hepes and restimulated in vitro for 3 days with 10 μg/ml of gel-purified human PPI, PI, or Ins, using the same protein used for immunization. At the end of the stimulation period, cells were purified by Ficoll separation, washed, and restimulated with interleukin (IL)-2 conditioned complete medium for 24 h. Cells were then washed and fused to the TCRα-β− variant of the BW5147 cell line, in 50% polyethylene glycol at a ratio of 1:3, and plated out in 96-well flat bottom plates (20). Hypoxanthine/aminopterin/thymidine (HAT) medium was added at day 2 postplating, and the plates were incubated for 10 days. Growing hybridomas were expanded in 24-well plates prior to testing.
Generation of Overlapping Peptides.
Preproinsulin peptides. A set of 10 overlapping peptides spanning the entire PPI sequence was synthesized by standard F-moc chemistry on an Applied Biosystem Peptide Synthesizer by the Protein and Nucleic Acid (PAN) Facility at Stanford University. To fine map each epitope, 33 overlapping 13-mer peptides were purchased from Research Genetics (Huntsville, AL) shifted by 3 amino acids, thus generating all possible decamers of PPI. In addition, to define the minimal core motif for the immunodominant region 70–93 of PPI more precisely, nine different N- or C-terminal truncated variant peptides were synthesized by the PAN facility at Stanford University.
The purity of all these peptides was verified by reverse phase HPLC and mass spectroscopic analysis. Before use, the peptides were resuspended in sterile PBS at a concentration of 1 mg/ml and stored at −20°C.
Assessment of Hybridoma Reactivity Using a Europium-Based IL-2 Immunoassay.
Antigen-presenting cells (APC). Transgenic mouse spleen cells or typed Epstein—Barr virus (EBV) transformed B cells were washed in HAT medium (complete medium with HAT) and irradiated (spleen cells, 3000 rads; human lymphoblastoid B cells, 12,000 rads) with a cesium source. Antigens were added to 3 × 105 irradiated transgenic APC or 1–3 × 105 EBV cells per well, in a final volume of 50 μl. Plated cells were incubated with antigen while the T cell hybridomas were prepared.
Antigen.
Recombinant antigen purified by Ni2+ columns was gel purified by electroelution of the relevant band and resuspended in PBS. Whole proteins were used at a concentration of 5–10 μg/ml.
T cell hybridomas.
T cell hybridomas were washed, and 2–5 × 104 cells in 100 μl were added per well to the APCs.
IL-2 immunoassay.
All antibody incubation and blocking steps were performed in blocking buffer (PBS/1% BSA/0.2% sodium azide) and all washes in wash buffer (PBS containing 0.05% Tween 20). ELISA plates (Corning) were coated with rat anti-mouse IL-2 monoclonal antibody (PharMingen) at a concentration of 2 μg/ml by overnight incubation at 4°C. Plates were blocked for 3 h with blocking buffer, washed once, and incubated overnight at 4°C with 100 μl of supernatant from a 48-h stimulation assay. Following five washes, biotinylated rat anti-mouse IL-2 monoclonal antibody (PharMingen) was added at a final concentration of 2 μg/ml in 100 μl. After 2 h of incubation at room temperature, the plates were washed five times, and 100 μl of 0.1 ng/ml europium-labeled streptavidin was added. After 1 h, the plates were washed five times; 100 μl of Enhancement solution (LKB-Wallac, Gaithersburg, MD) was added, and the plates were left for 5 min at room temperature. The resulting fluorescence was read using an LKB-Wallac fluorescence plate reader.
RESULTS
Details of the mice used for immunization have been described elsewhere (20, 22). In brief, DRB1*0401-restricted T cell hybridomas were generated in mice lacking murine class II genes by crossing the I-Aβ null mutation onto HLA-DR4, human CD4 transgenic mice. Using these mice, we found that PPI and PI are strongly T cell immunogenic, whereas Ins is only weakly immunogenic for CD4-positive T cells (as distinct from the B cell antibody response). Table 1 shows the DRB1*0401-restricted T cell epitopes found in transgenic mice.
Table 1.
Amino acid sequence and frequency of PPI, PI, and insulin-specific, HLA-DRB1*0401-restricted epitopes identified in DR4, human CD4, Aβ0/0 triple transgenic mice
| Peptide epitopes | T cell hybridomas, n | Protein used for immunization |
|---|---|---|
| LALLALWGPDPAAAFVp11-26 | 1 | PPI |
| PAAAFVNQHLCGSHLVp20-36 | 1 | PPI |
| GAGSLQPLALEGSLQKR↓G p73-90 | 25 | PPI, PI |
| SLQKRGIVEQCCTSICSp85-101 | 1 | Insulin |
↓ indicates the endopeptidase II cleavage site; the 9-mer MHC binding core sequence of the immunodominant HLA-DRB1*401-restricted T cell epitope is shown in boldface type; underlining indicates the tworesidues normally removed by CPH.
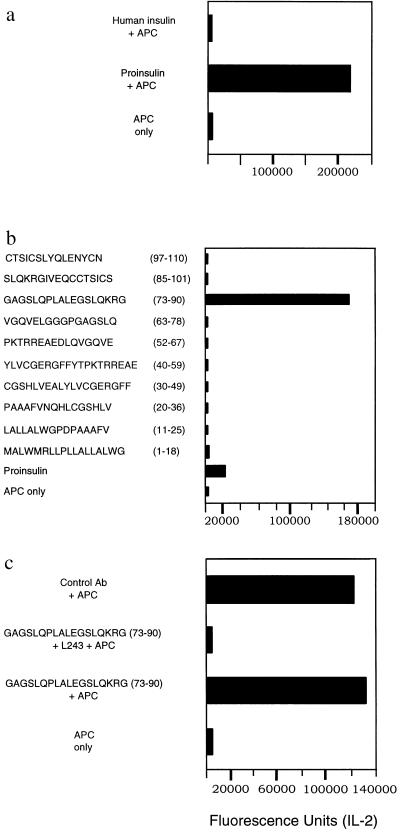
To identify specific hybridomas, T cell hybridomas were first tested using whole protein (PPI, PI, or Ins) and transgenic spleen cells as APC. Subsequently, the antigen-positive T cell hybridomas were tested using whole PPI, PI, insulin, or individual peptides and DRB1*0401-positive EBV (Preiss) cells as APCs. Figure 1 a–c shows the results of a representative T cell hybridoma, c 228. Figure 1a shows hybridoma c 228 responding to intact human insulin or human proinsulin using HLA DRB1*0401-positive transgenic spleen cells as APC. Figure 1b shows T cell hybridoma c 228 responding to a single peptide from a pool of 10 peptides using a human HLA DRB1*0401-positive EBV cell line as APC. The restriction element was determined by using anti-DR blocking antibody (L243) and DRB1*0401-expressing EBV cells (Preiss) as APC (Fig. 1c). Isotype-matched murine antibodies were used as a negative control (Fig. 1c). Spleen cells from HLA DRα1*0101, IA-β0/0 transgenic mice, in which the only functional MHC class II heterodimer is the DRα1*0101, IE-β chimeric molecule, were also used as APC to exclude participation of the chimeric class II molecule as a restriction element (see below).
Figure 1.
(a) The primary screening of T cell hybridoma c 228, using human insulin (no response), and recombinant PI as the antigen source (PI was used for immunization in generating hybridoma c 228) and transgenic spleen cells as APC. (b) The recognition of the peptide sequence p73–90, and intact PI protein by hybridoma c 228 using synthetic peptides, or recombinant PI as the antigenic stimulus and DRB1*0401-positive EBV cells as APC. (c) Selective blocking of the peptide specific response of hybridoma c 228 using a monoclonal anti-DR antibody (L243) and DRB1*0401-positive EBV cells as APC. Isotype-matched control antibodies were used as a negative control. The values correspond to IL-2 production measured in fluorescence units.
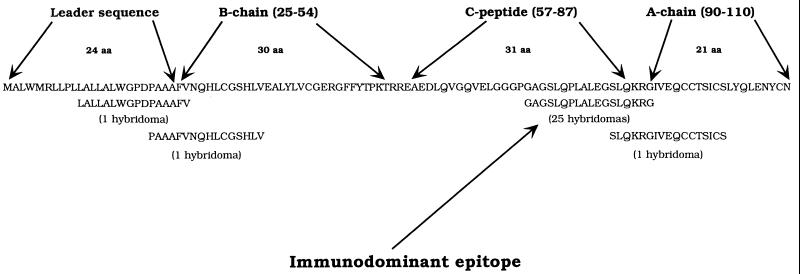
The immunodominant epitope (p73–90) is located in a region spanning the C peptide and the A chain. The C peptide is absent in the mature hormone. In addition, responses to a number of other T cell epitopes were also found at low frequency: one peptide epitope was specific for the leader sequence (p11–26), another spanning part of the leader sequence and the B chain (p20–36), and a fourth T cell epitope was recognized by a single T cell hybridoma, obtained after insulin immunization, specific for the insulin A chain (p85–101; Table 1 and Fig. 2). These results demonstrate that the major T cell epitope of Ins or its prohormones presented by DRB1*0401 is located in the precursor forms of the hormone.
Figure 2.
Amino acid sequence of human preproinsulin and a summary of the identified epitopes of PPI, PI, and Ins.
In the DRB1*0401 transgenic mice on the I-Aβ0/0 background, the IE-β gene product can form a functional heterodimer with the human HLA-DRα chain. T cell hybridomas were identified that were specific for a peptide of the leader sequence (p1–18), and for a peptide spanning the junction of the insulin B chain and the C peptide (p40–59), both of which were restricted by the DR(α1*0101), I-Eβ chimeric heterodimer (data not shown).
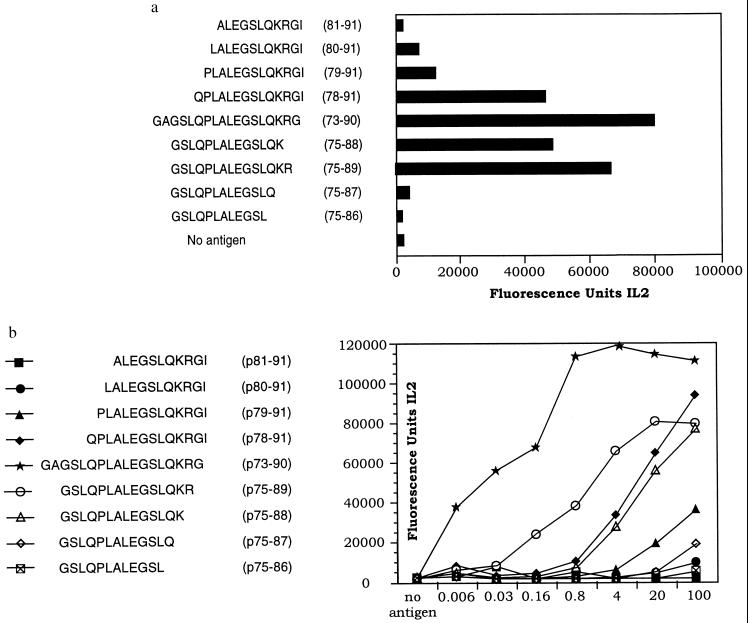
The core epitope sequence of the immunodominant p73–90 DRB1*0401 restricted peptide was identified using a set of 9 N- or C-terminal truncated peptides. The response of T cell hybridoma, c 228, to these peptides is shown in Fig. 3. The resulting minimal core epitope was found to be LALEGSLQK (Fig. 3a). This peptide sequence is compatible with the DRB1*0401 peptide binding motifs recently published (24, 25) and with the model derived from the crystal structure of an influenza virus peptide (HA 306–318) complexed with an HLA-DR1 molecule (26). According to this model, the major anchor positions of the peptide with the HLA-DR peptide complex are found at peptide positions 1, 4, 6, 7, and 9 (26). The sequence LALEGSLQK contains a Leu as a large hydrophobic anchor residue at position P1. The specificity of the MHC pocket accommodating this residue is modulated by the Gly/Val dimorphism at position 86 of the DRβ chain. The protective DRB1*0403 allele has Val at this position, as opposed to the Gly of the susceptible DRB1*0401 (the transgenic allele used here) and DRB1*0405 alleles. The presence of a Val could potentially restrict the size or change the conformation of this pocket. The presence of a Lys at position DRβ-71 encoded by the DRB1*0401 allele has been shown probably to influence electrostatic interaction with the negatively charged Glu at position P4 of the peptide, whereas the presence of a Glu at position 74 of the DRβ-chain encoded by the protective DRB1*0403 allele (also contributing to pocket P4) would disfavor such an interaction. Pocket P6 is occupied by Ser, which is also found commonly in that position in other DRB1*0401 binding epitopes (22).
Figure 3.
Fine mapping of the p73–90 9-mer core sequence in hybridoma c 228 using the peptides at a concentration of 20 μg/ml (a) and representative dose-response curves, giving peptide concentrations in μg/ml and the corresponding IL-2 production in fluorescence units, to demonstrate a similar response pattern by T cell hybridoma c 228 for variant peptides of the core sequence (b).
The dose-response curve for the individual truncated peptides using T cell hybridoma c 228 (Fig. 3b) showed a similar response pattern to the truncated peptides for different p73–90 specific, DRB1*0401-restricted T cell hybridomas (results not shown).
DISCUSSION
Using this transgenic mouse model, we have identified one major immunodominant, naturally processed, DRB1*0401-restricted T cell epitope from human PPI/PI in the region between the C peptide and the A chain. T cell responses to this peptide epitope have also been found in peripheral blood of HLA DRB1*0401-positive patients with recent onset type 1 diabetes (M.C. et al., unpublished results). Peptide p73–90 spans a region of the C peptide and the A chain of PI that is first cleaved by the endopeptidase II resulting in the split 64,65 proinsulin. Through removal of Lys and Arg (amino acid residues 64 and 65, respectively) by carboxypeptidase H, the intermediate des 64,65 proinsulin is generated. Thus, the last three amino acids of the PPI/PI region peptide GAGSLQPLALEGSLQKR↓G (the arrow indicates the endopeptidase II cleavage site, the minimal peptide sequence presented by DRB1*0401 is in boldface, and the two residues normally removed by carboxypeptidase H are underlined), removed by the cleavage of 64–65 split proinsulin, exists under normal conditions only as a short-lived intermediate during processing of the hormone in the granules of the β cells.
Another epitope, p11–26, was located in the region spanning the leader peptide. The recognition of this epitope is even more intriguing because that epitope is present only when the PPI is still anchored to the endoplasmic reticulum. The presence of these DRB1*0401-restricted T cell epitopes indicates that precursors of insulin normally confined to the endoplasmic reticulum or the cytoplasm of the β cell may be involved in targeting T cell autoreactivity to the islets.
Very recently, Pugliese and collaborators have shown that during fetal development and in childhood, mRNAs for insulin and other islet cell autoantigens (GAD, ICA69, IA-2) are expressed at low levels in the human thymus (27). Proinsulin protein was detected in the thymus at levels four times higher than insulin (27), suggesting that the possibility of a PI conversion into mature protein is unlikely. These authors and others (28) have found that VNTR class III alleles are associated with higher levels of proinsulin mRNA in the thymus, while homozygosity for class I VNTR alleles correlates with lower transcriptional activity. Under these circumstances, the higher levels of PI generated in the thymus (class III VNTR alleles) could promote negative selection of PI-specific T lymphocytes, which could play a critical role in protection from IDDM (27, 28).
It is possible that in DRB1*0401-positive patients, IDDM2 class I alleles, which express lower levels of PPI/PI proteins in the thymus, may result in a larger repertoire of autoreactive T cells specific for the DRB1*0401 immunodominant GAGSLQPLALEGSLQKR PPI/PI peptide. The same class I VNTR alleles are associated in the pancreas with increased expression of PPI and PI (16, 17). Thus, a viral infection or other exogenous damage to the β cells of the islets may release increased amounts of the immunogenic portions of the hormone, which are located in the prohormones PI or PPI. Because the HLA-DR3 genotype is negatively correlated with the presence of anti-insulin autoantibodies (2, 29). A scenario with a functional linkage between IDDM2 and a common HLA DR4 allele is an attractive possibility.
Additional evidence suggestive of a primary role for PPI and/or PI expression in generating or deleting autoreactive T cells may be seen in the prevention of insulitis and diabetes in NOD mice by expression of mouse proinsulin II under the control of the MHC class II I-Eα promoter in MHC class II positive cells (30). Moreover, it has been reported that insulitis can be induced in rats receiving T cells specific for a proinsulin peptide sequence spanning the cleavage site between the B chain and the C peptide (31). T cell reactivity against a peptide from the same proinsulin region has also recently been reported in individuals at risk for IDDM (32).
The weaker T cell immunogenicity of Ins compared with the prohormones PPI and PI is consistent with the notion that T cell reactivity to mature insulin is rare in IDDM patients. It is generally acknowledged that T cell responses to insulin in IDDM patients are very much lower in frequency than T cell reactivity against GAD 65 or the tyrosine phosphatase ICA 512 (2).
In summary these data indicate that one of the specific β cell autoantigens involved in human IDDM is PPI and/or PI. Our findings also provide a possible explanation for why the insulin gene region-encoded susceptibility to type 1 diabetes (IDDM2) is associated with HLA-DR4-positive patients. They also have important implications for the use of mature insulin in trial to prevent IDDM in prediabetics.
Acknowledgments
We are grateful to Peggy Sullivan, Mary Vadeboncoeur, and Lou Hidalgo for transgenic mice production (P.S., M.V.) and animal handling (L.H.). This work was supported in part by grants from the National Institutes of Health, the American Diabetes Association, and the Greenwall Foundation and by fellowships from the Wellcome Trust (A.P.C.), the American Diabetes Association (S.D.P.), and the Greenwall Foundation (M.C.).
ABBREVIATIONS
- APC
antigen presenting cells
- EBV
Epstein—Barr virus
- HAT
hypoxanthine/aminopterin/thymidine
- IDDM
insulin-dependent diabetes mellitus
- IDDM2
insulin gene region
- IL
interleukin
- Ins
insulin
- MHC
major histocompatibility complex
- PI
proinsulin
- PPI
preproinsulin
- VNTR
variable number of tandem repeats
References
- 1.Todd J A. Diabetic Med. 1994;11:6–16. doi: 10.1111/j.1464-5491.1994.tb00222.x. [DOI] [PubMed] [Google Scholar]
- 2.Bell G I, Horita S, Karam J H. Diabetes. 1984;33:176–183. doi: 10.2337/diab.33.2.176. [DOI] [PubMed] [Google Scholar]
- 3.Davies J L, Kawaguchi Y, Bennett S T, Copeman J B, Cordell H J, Pritchard L E, Reed P W, Gough S C, Jenkins S C, Palmer S M, et al. Nature (London) 1994;371:130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- 4.Svejgaard A, Platz P, Ryder L P. Immunol Rev. 1983;70:193–218. doi: 10.1111/j.1600-065x.1983.tb00715.x. [DOI] [PubMed] [Google Scholar]
- 5.Thomson G. Am J Hum Genet. 1984;36:1309–1317. [PMC free article] [PubMed] [Google Scholar]
- 6.Todd J A, Bell J I, McDevitt H O. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 7.Khalil I, d’Auriol L, Gobet M, Morin L, Lepage V, Deschamps I, Park M S, Degos L, Galibert F, Hors J. J Clin Invest. 1990;85:1315–1319. doi: 10.1172/JCI114569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheehy M J, Scharf S J, Rowe J R, Neme de Gimenez M H, Meske L M, Erlich H A, Nepom B S. J Clin Invest. 1989;83:830–835. doi: 10.1172/JCI113965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erlich H A, Zeidler A, Chang J, Shaw S, Raffel L J, Klitz W, Beshkov Y, Costin R, Pressman S, Bugawan TL, Rotter J I. Nat Genet. 1993;3:358–364. doi: 10.1038/ng0493-358. [DOI] [PubMed] [Google Scholar]
- 10.Cucca F, Muntoni F, Lampis R, Frau F, Argiolas L, Silvetti M, Angius E, Cao A, De Virgiliis S, Congia M. Hum Immunol. 1993;37:85–94. doi: 10.1016/0198-8859(93)90146-r. [DOI] [PubMed] [Google Scholar]
- 11.Cucca F, Lampis R, Frau F, Macis D, Angius E, Masile P, Chessa M, Frongia P, Silvetti M, Cao A, De Virgiliis S, Congia M. Hum Immunol. 1995;43:301–308. doi: 10.1016/0198-8859(95)00042-3. [DOI] [PubMed] [Google Scholar]
- 12.Undlien D E, Friede T, Rammensee H G, Joner G, Dahl-Jorgensen K, Sovik O, Akselsen H E, Knutsen I, Ronningen K S, Thorsby E. Diabetes. 1997;46:143–149. doi: 10.2337/diab.46.1.143. [DOI] [PubMed] [Google Scholar]
- 13.Lucassen A M, Julier C, Beressi J P, Boitard C, Froguel P, Lathrop M, Bell J I. Nat Genet. 1993;3:305–310. doi: 10.1038/ng0793-305. [DOI] [PubMed] [Google Scholar]
- 14.Bennett S T, Lucassen A M, Gough S C, Powell E E, Undlien D E, Pritchard L E, Merriman M E, Kawaguchi Y, Dronsfield M J, Pociot F, et al. Nat Genet. 1995;9:284–292. doi: 10.1038/ng0395-284. [DOI] [PubMed] [Google Scholar]
- 15.Bennett S T, Wilson A J, Cucca F, Nerup J, Pociot F, McKinney P A, Barnett A H, Bain S C, Todd J A. J Autoimmun. 1996;9:415–421. doi: 10.1006/jaut.1996.0057. [DOI] [PubMed] [Google Scholar]
- 16.Julier C, Hyer R N, Davies J, Merlin F, Soularue P, Briant L, Cathelineau G, Deschamps I, Rotter J I, Froguel P, Boitard C, Bell J I, Lathrop G M. Nature. 1991;354:155–159. doi: 10.1038/354155a0. [DOI] [PubMed] [Google Scholar]
- 17.Eisenbarth G S, Jackson R A, Pugliese A. J Autoimmun. 1992;5:241–246. doi: 10.1016/0896-8411(92)90039-s. [DOI] [PubMed] [Google Scholar]
- 18.Ziegler R, Alper C A, Awdeh Z L, Castano L, Brink S J, Soeldner J S, Jackson R A, Eisenbarth G S. Diabetes. 1991;40:709–714. doi: 10.2337/diab.40.6.709. [DOI] [PubMed] [Google Scholar]
- 19.Durinovic-Bello I, Hummel M, Ziegler A G. Diabetes. 1996;45:795–800. doi: 10.2337/diab.45.6.795. [DOI] [PubMed] [Google Scholar]
- 20.Fugger L, Michie S A, Rulifson I, Lock C B, McDevitt G S. Proc Natl Acad Sci USA. 1994;91:6151–6155. doi: 10.1073/pnas.91.13.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosgrove D, Gray D, Dierich A, Kaufman J, Lemeur M, Benoist C, Mathis D. Cell. 1991;66:1051–1066. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- 22.Patel S D, Cope A P, Congia M, Chen T T, Kim E, Fugger L, Wherret D, Sonderstrup-McDevitt G. Proc Natl Acad Sci USA. 1997;94:8082–8087. doi: 10.1073/pnas.94.15.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Von Heijne G. Eur J Biochem. 1983;133:17–24. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 24.Rammensee H G, Friede T, Stevanoviic S. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 25.Hammer J, Gallazzi F, Bono E, Karr R W, Guenot J, Valsasnini P, Nagy Z A, Sinigaglia F. J Exp Med. 1995;181:1847–1855. doi: 10.1084/jem.181.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stern L J, Brown J H, Jardetzky T S, Gorga J C, Urban R G, Strominger J L, Wiley D C. Nature (London) 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 27.Pugliese A, Zeller M, Fernandez A, Jr, Zalcberg L J, Bartlett R J, Ricordi C, Pietropaolo M, Eisenbarth G S, Bennett S T, Patel D D. Nat Genet. 1997;15:293–297. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- 28.Vafiadis P, Bennett S T, Todd J A, Nadeau J, Grabs R, Goodyer C G, Wickramasinghe S, Colle E, Polychronakos C. Nat Genet. 1997;15:289–292. doi: 10.1038/ng0397-289. [DOI] [PubMed] [Google Scholar]
- 29.Ziegler A G, Standl E, Albert E, Mehnert H. Diabetes. 1991;40:1146–1149. doi: 10.2337/diab.40.9.1146. [DOI] [PubMed] [Google Scholar]
- 30.French M B, Allison J, Cram D S, Thomas H E, Dempsey-Collier M, Silva A, Georgiou H M, Kay T W, Harrison L C, Lew A M. Diabetes. 1997;46:34–39. doi: 10.2337/diab.46.1.34. [DOI] [PubMed] [Google Scholar]
- 31.Griffin A C, Zhao W, Wegmann K W, Hickley W F. Am J Pathol. 1995;147:845–857. [PMC free article] [PubMed] [Google Scholar]
- 32.Rudy G, Stone N, Harrison L C, Colman P G, McNair P, Brusic V, French M B, Honeyman M C, Tait B, Lew A M. Mol Med. 1995;1:625–633. [PMC free article] [PubMed] [Google Scholar]