Abstract
To initiate infection, HIV-1 requires a primary receptor, CD4, and a secondary receptor, principally the chemokine receptor CCR5 or CXCR4. Coreceptor usage plays a critical role in HIV-1 disease progression. HIV-1 transmitted in vivo generally uses CCR5 (R5), but later CXCR4 (X4) strains may emerge; this shift heralds CD4+ cell depletion and clinical deterioration. We asked whether antiretroviral therapy can shift HIV-1 populations back to R5 viruses after X4 strains have emerged, in part because treatment has been successful in slowing disease progression without uniformly suppressing plasma viremia. We analyzed the coreceptor usage of serial primary isolates from 15 women with advanced disease who demonstrated X4 viruses. Coreceptor usage was determined by using a HOS-CD4+ cell system, biological and molecular cloning, and sequencing the envelope gene V3 region. By constructing a mathematical model to measure the proportion of virus in a specimen using each coreceptor, we demonstrated that the predominant viral population shifted from X4 at baseline to R5 strains after treatment. Multivariate analyses showed that the shift was independent of changes in plasma HIV-1 RNA level and CD4+ cell count. Hence, combination therapy may lead to a change in phenotypic character as well as in the quantity of HIV-1. Shifts in coreceptor usage may thereby contribute to the clinical efficacy of anti-HIV drugs.
Introduction
HIV-1 requires contact with two receptors to gain entry to cells and initiate infection; CD4 is the primary receptor and the chemokine receptors CCR5 and CXCR4 serve as secondary receptors (1–3). Coreceptor usage plays a critical role in viral tropism, pathogenesis, and disease progression. HIV-1 strains that are transmitted in vivo generally use CCR5 (R5 viruses) (3–6). These viruses typically infect macrophages and primary CD4+ lymphocytes and do not form syncytia in vitro (7). Years after chronic infection is established, CXCR4-using strains (X4 viruses) emerge in approximately 50% of infected individuals (3, 6, 8, 9). The X4 viruses usually coexist with R5 viruses in the viral swarm (9, 10). X4 strains not only infect primary T-lymphocytes but also replicate in T-cell lines and induce syncytia (7). More striking is their influence on pathogenesis. Although a significant fraction of patients with late stage HIV-1 infection have only R5 viruses detectable, for those individuals in whom X4 strains do emerge, the expansion in coreceptor usage usually predicts rapid depletion of CD4+ cells and acceleration of HIV-1 disease progression (3, 6, 8, 9, 10). The effect of antiretroviral therapy on coreceptor usage has not been studied quantitatively.
A recent analysis of HIV-1 coreceptor usage in infected individuals suggests a mechanism for the rapid CD4+ cell decline; X4 viruses infect an expanded spectrum of crucial target cells as compared with R5 strains (11). Thus, abundant evidence suggests that reversing the emergence of X4 strains would be desirable in the HIV-1–infected individual. Whether the predominant populations of viruses can be shifted back to R5 once the X4 strains have emerged has not been studied systematically.
To address this issue, we first quantified the proportion of viruses in primary isolates that used each coreceptor and then investigated whether combination antiretroviral therapy, particularly highly active antiretroviral therapy (HAART), influences coreceptor usage. Treatment of infected individuals with HAART has led to a dramatic decline in both HIV-1–related illness and death (12). Early clinical trials demonstrated a reduction of plasma HIV-1 RNA loads to undetectable levels in the majority of treated individuals (13, 14). Subsequent studies, however, showed more limited success in achieving and maintaining viral suppression (15, 16). Yet many patients experienced immunologic and clinical responses to HAART without sustained suppression of plasma viremia (15–17). These data led us to test the hypothesis that HAART not only reduces the quantity of virus but also affects the characteristics of HIV-1 by influencing coreceptor usage.
Methods
Study population.
We examined coreceptor usage in 22 women who participated in two prospective studies of HIV-1 infection. Nineteen were enrolled in the Bronx-Manhattan site of the Women’s Interagency HIV Study (WIHS), a NIH multicenter study of the natural history of HIV-1 infection in women. Three took part in a study of HIV-1 pathogenesis performed at the Wadsworth Center of the New York State Department of Health in Albany, New York, USA. These cohorts have been described in detail previously (18, 19). It is of note that both included individuals with a broad spectrum of HIV-1 disease. The institutional review boards at each clinical site and the New York State Department of Health approved the investigation. Each woman provided informed consent at enrollment.
To examine the effect of combination antiviral therapy on HIV-1 coreceptor usage, we sought women infected with X4 strains. After screening 22 women, most with advanced HIV-1 disease, we studied the 15 participants who met the following criteria: (a) their viral isolates displayed X4 strains either exclusively or in addition to R5 strains; (b) they were initially untreated or taking nucleoside analogues alone; and (c) antiretroviral therapy, when initiated, was clearly documented by the WIHS database, Wadsworth study questionnaires, and records of treating physicians. The 15 women who met the study criteria included 10 African Americans, 4 Latinas, and 1 Caucasian. HAART refers here to three or more antiretroviral drugs in combination, including at least one protease inhibitor.
Sample collection, preparation, and analysis.
Peripheral blood was separated into plasma and cell components (18, 19). We quantitated HIV-1 RNA in plasma by using NucliSens (Organon Teknika Corp., Durham, North Carolina, USA) with a lower limit of quantitation of approximately 80 copies/ml. The CCR5 genotype of each patient was determined as described (4).
Derivation of primary viral isolates and biological clones.
Primary isolates of HIV-1 were obtained by coculture with normal donor PBMCs (19). Viral isolates were titrated in PBMCs (19). Biological clones were derived from primary isolates by short-term limiting dilution cloning (9).
Assay for coreceptor usage.
We followed temporal changes in coreceptor usage of primary HIV-1 isolates and biological clones from participants in the study by using a HOS-CD4+ cell system. The parental HOS-CD4+ line is a human osteogenic sarcoma cell line stably expressing high levels of CD4. HOS-CD4+ cells transfected with genes encoding either CCR5 or CXCR4 in addition to CD4 (cell lines HOS-CD4.CCR5 and HOS-CD4.CXCR4, respectively) served as indicator lines for coreceptor usage (1). To determine coreceptor usage, HOS-CD4.CCR5 and HOS-CD4.CXCR4 cells were seeded onto 12-well plates and, after 24 hours, inoculated with a standard quantity of titered virus; 102 50% tissue culture infective dose (TCID50) of first-passage primary viral isolates or biological clones were assayed in duplicate. HIV JR-FL and LAV/HTLV-IIIB were inoculated in parallel as R5- and X4-specific positive control viruses, respectively, and uninoculated cells were used as negative controls. To eliminate any artifacts resulting from infection due to low levels of endogenous coreceptor expression, parental HOS-CD4+ cells were also inoculated with duplicate primary and control isolates.
Supernatants were harvested at day 10 after infection and analyzed for HIV-1 p24 antigen using a commercially available ELISA assay (NEN Life Science Products Inc., Boston, Massachusetts, USA). ELISA values were standardized so that 0 pg/ml was set at the level equal to three times the mean value of the negative controls. A culture was considered positive if the p24 antigen level was equal to or greater than 25 pg/ml. Experimental results were discarded if: (a) any parental HOS-CD4+ culture tested positive, or (b) any JR-FL– or LAV/HTLV-IIIB–positive control culture tested negative. If the variance in p24 antigen level between duplicate cultures was greater than 25%, the coreceptor usage assay for that particular viral isolate was repeated. Results of the coreceptor usage assay were then categorized in a semiquantitative manner according to p24 antigen level as follows: negative (p24 < 25 pg/ml), +/– (25–50 pg/ml), 1+ (50–250 pg/ml), 2+ (250–500 pg/ml), and 3+ (≥ 500 pg/ml).
Phenotypic characterization.
The presence of syncytium-inducing (SI) variants of HIV-1 in patient primary viral isolates was determined by infection of MT-2 cell cultures as described previously (8). A pooled stock of HIV LAV/HTLVIII was used as a positive control.
Sequence analyses of the HIV-1 V3 loop.
HIV-1 virions were isolated from plasma samples as described (20). RT-PCR amplification produced a 920-bp amplicon spanning the V3 region of the env gene. Reaction conditions were controlled rigorously to minimize recombination and other artifacts (20). Amplified products were cloned into a TOPO TA vector (Invitrogen Corp., Carlsbad, California, USA), verified by restriction digestion, and sequenced. Alignment of the sequences was done initially using the PILEUP program in the GCG Suite (Genetics Computer Group, Madison, Wisconsin, USA), then checked manually. Envelope sequences were used to predict coreceptor usage on the basis of the overall charge of the V3 loop and the presence of basic or acidic residues at positions 275 and 287 of the env gene (21, 22).
Statistical methods.
The Wilcoxon rank sum test was used to make comparisons between the magnitude of log viral level, CD4+ counts, and λ values. Data for factors relating to changes in λ values were analyzed by multivariate Poisson regression. Variables included log HIV-1 RNA levels, changes in viral levels, CD4+ cell counts, changes in CD4+ cell counts, and indicator variables for levels of antiviral therapy.
To quantitate HIV-1 coreceptor usage, we constructed a variable, λ, as the proportion of strains using CCR5: λ = 1 represents an isolate in which all strains prefer the CCR5 receptor but λ = 0 indicates that all prefer CXCR4. Values for λ were assessed by using qualitative assay data derived from primary isolates, biologic clones, and sequences of the V3 portion of the env gene. In our determination of the coreceptor usage of 525 biologic clones, none was dual tropic, suggesting that true dual-tropic viruses are rare when using our assay method. We therefore assumed for this calculation that the probability of a single virion possessing the phenotypic attributes of both coreceptors is small. Thus, for the vast majority of virions, each virion uses either CCR5 or CXCR4. This relationship can be stated as a mixture distribution:
D = λ(CCR5) + (1–λ)(CXCR4); 0 ≤ λ ≤ 1, where D is the distribution of viral phenotypes. By design, we have a binomial population.
Values for λ were constructed by relating data derived from the same patient sample by using three different analyses: biologic cloning, V3 sequencing, and qualitative assays of primary isolates. To construct λ values, we first calculated the proportion of biologic and, if available, molecular clones using R5 at each time point, then linked the proportion to the qualitative coreceptor usage score (– to 3+) of primary isolates obtained simultaneously. Data that were not available were interpolated. The data were transformed to approximate a Poisson distribution. Poisson regression analysis was then performed to determine the factors associated with changes in λ values.
Results
Patient population and response to therapy.
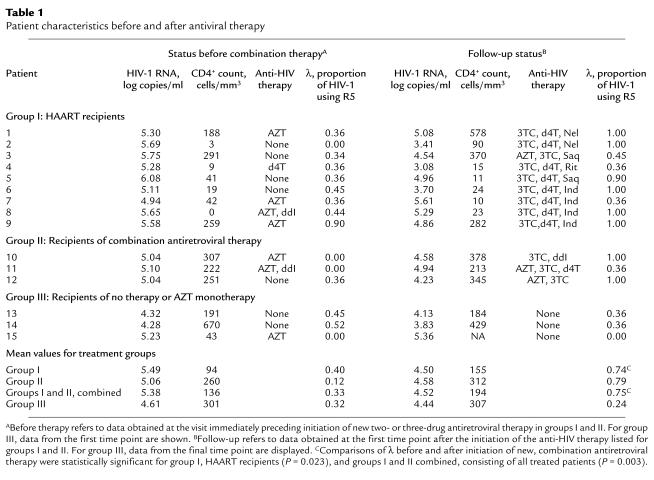
To test whether antiretroviral therapy may influence HIV-1 coreceptor usage, we analyzed serial viral isolates from 15 women who demonstrated X4 viruses. In 14 women R5 viruses were present in addition to the X4 strains. The 15 women participated in prospective studies of the natural history of HIV-1 infection during the 1990s; 14 were enrolled in the WIHS (18). All 15 women carried the homozygous wild-type genotype for the coreceptor CCR5. Initially, most displayed high-plasma HIV-1 RNA levels and CD4+ cell depletion (means of 5.22 log10 copies/ml and 147 cells/mm3, respectively). At that time, eight women were receiving antiretroviral therapy, primarily zidovudine monotherapy. While under study, however, 12 initiated new combination regimens; nine received HAART (group I), and three received two or more nucleoside analogues (group II). Three individuals, by contrast, did not initiate new therapy during the study (group III) (Table 1). For those initiating new therapy, HIV-1 RNA levels dropped by an average of 0.86 log10 copies/ml and CD4+ counts increased by an average of 58 cells/ml by the first study visit after starting the new regimens. The viral levels rebounded by 0.69 log10 copies/ml, however, by the end of the 28.5-month mean follow-up period for treated patients, at which time 11 of the 12 women continued to take antiviral therapy (six HAART, five two-drug regimens).
Table 1.
Patient characteristics before and after antiviral therapy
Antiviral therapy may preferentially suppress X4 strains.
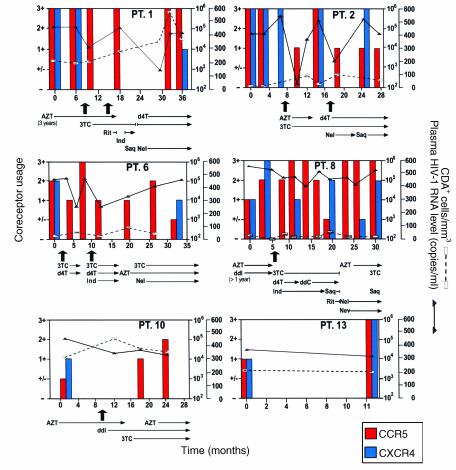
We followed temporal changes in HIV-1 coreceptor usage in the 15 participants in the study by using a HOS-CD4+ cell system, a cell line genetically engineered to indicate HIV-1 coreceptor usage in a semiquantitative manner (1). Fourteen women initially displayed viral populations composed of both R5 and X4 viruses (Figure 1), and one displayed virus that exclusively used X4. X4 viruses persisted at subsequent time points in patients who did not initiate new combination therapy, a finding exemplified in Figure 1 by patient 13, who remained untreated throughout the study, and patients 1, 2, and 8, whose virus was sampled on multiple occasions before new therapy commenced. Viruses using X4 appeared to be preferentially suppressed, however, when new regimens were initiated. Not only were X4 strains eliminated by the first time point after starting new therapy in half of the treated women (Figure 1; patients 1, 2, 6, 8, and 10), but the proportion of these viruses seemed to be diminished in most of the others. In addition, patients who experienced a rebound in HIV-1 RNA levels and X4 strains while on therapy often achieved suppression of X4 strains a second time when the antiviral regimen was changed (Figure 1; patients 2 and 8).
Figure 1.
The effect of combination antiretroviral therapy on HIV-1 coreceptor usage over time in representative study subjects. Patients 1, 2, 6, 8, and 10 received new combination therapy and patient 13 remained untreated. Arrows note the first time during the study period that a new combination of antiretroviral drugs was initiated. Two arrows appear if a patient received a two-drug regimen first, then HAART. The duration of treatment with each agent is indicated. PT., patient; AZT, zidovudine; 3TC, lamivudine; Rit, ritonavir; Ind, indinavir; Saq, saquinavir; d4T, stavudine; Nel, nelfinavir; ddI, didanosine; ddC, zalcitabine; Nev, nevirapine.
Coreceptor usage by biologically cloned viruses.
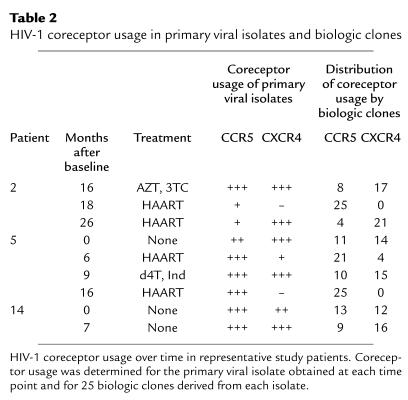
Two aspects of the pattern of HIV-1 coreceptor usage in these individuals prompted us to delineate the proportion of individual viruses using each coreceptor. First, analyses of primary viral isolates by the HOS-CD4+ system indicated coreceptor usage by both R5 and X4 viruses at many time points (Figure 1). Because primary isolates comprise a molecular mixture of viral quasispecies, we wished to determine whether usage of both coreceptors was due to dual tropic viruses or a mixture of individual viruses with R5 and X4 tropisms. In addition, to compare coreceptor usage rigorously over time, it is desirable to quantitate the proportion of virus using each coreceptor. For these reasons, we isolated biologic clones that were derived from the patients’ primary isolates by performing limiting dilution cultures. Coreceptor usage was then determined for 25 clones from each isolate by employing the HOS-CD4+ cell system. Biologic clones from these patients used either R5 or X4; no dual tropic viruses were detected among the 525 clones by using our assay system. In addition, the distribution of coreceptor usage by the clones generally confirmed the semiquantitative results obtained for primary isolates; proportions of HIV-1 using each coreceptor appeared roughly similar whether the cloned virus or primary isolates were examined (Table 2).
Table 2.
HIV-1 coreceptor usage in primary viral isolates and biologic clones
Studies of biologic clones obtained at serial time points also confirmed that the predominant viral population shifted from X4 to R5 after initiation of new combination antiretroviral therapy (Table 2). For patient 2, for example, analyses of virus obtained 16 months after baseline and 8 months after initiation of double therapy showed only eight clones that used R5 as compared with 17 that used X4. After a switch to a HAART regime that included two new drugs, however, the viral population in this patient shifted, and all 25 biologic clones used R5. A similar pattern was exhibited by biologic clones from patient 5, whose virus shifted dramatically to R5 on the two occasions that HAART was initiated. Patient 14, by contrast, remained untreated, and her viral population evolved to comprise a larger proportion of clones using X4 over time.
The MT2 assay to detect SI viruses in culture was also performed on primary isolates derived at each time point. These results confirmed the pattern of HIV-1 coreceptor usage described here. Thirteen of the fifteen patients were infected initially with SI virus. In all 11 of those who displayed SI virus and received new combination therapy, the phenotype changed, at least transiently, to non–syncytia-inducing (NSI) after treatment (data not shown).
Coreceptor usage determined by sequence analysis of molecular clones derived from HIV-1 RNA.
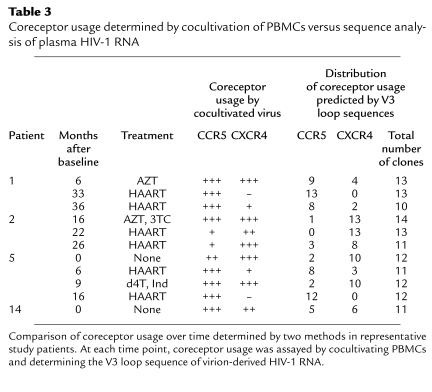
The determination of HIV-1 coreceptor usage described thus far analyzed virus cultivated from PBMCs with viral genomes derived primarily from HIV-1 DNA. Because plasma HIV-1 RNA represents replicating virus better than proviral DNA (23) and cultivation in vitro can result in selection of particular viral strains, we performed sequence analyses of molecular clones of HIV-1 to predict coreceptor usage of uncultivated, replicating virus (21, 22). We sequenced the V3 region of 221 env genes derived from uncultivated plasma virus obtained at key time points. These sequences predicted a pattern of coreceptor usage that essentially paralleled the one obtained by using viral culture (Table 3). The sequence data underscored the change in coreceptor usage seen after initiation of treatment. These experiments suggest that study of cultivated virus reflects the coreceptor usage of currently replicating virus and is likely to reveal the shifts in viral populations that occur as a result of recent antiviral therapy.
Table 3.
Coreceptor usage determined by cocultivation of PBMCs versus sequence analysis of plasma HIV-1 RNA
Quantitation of coreceptor usage by R5 and X4.
The large number of biologic and molecular clones permitted us to derive a system to quantitate the proportion of virus in a primary isolate that uses each coreceptor. In this system, λ is a continuous, nonlinear variable between 1 and 0 derived from the results presented here showing coreceptor usage by biologically and molecularly cloned virus; it describes the mixed proportion of viruses using R5 and X4. A λ value near 1 describes a population of viruses that almost all use R5; a value near 0 describes a population that almost all use X4. By applying this method, we determined the proportion of virus using each coreceptor for each patient over time.
To quantitate the effect of combination therapy on HIV-1 coreceptor usage, we compared the λ values of virus obtained at the visits before and immediately after initiating new combination therapy. This comparison demonstrated a clear, statistically significant shift of the predominant viral population from X4 to R5 (Table 1). The mean λ values for virus from all 12 patients starting combination therapy (groups I and II) changed from 0.33 to 0.75 (P = 0.003 by using the binomial proportion comparison test). For the subset of nine who initiated HAART (group I), the shift in λ extended from 0.40 to 0.74 (P = 0.023). In addition, we wished to assess separately the effect of initiating treatment with two or more nucleoside analogues and no protease inhibitor on coreceptor usage. Five of the patients who ultimately received HAART had received regimens consisting of two nucleoside analogues previously. We therefore compared the λ values of virus obtained before and after initiation of two or more nucleoside analogues in a group of eight patients (group II and patients 1, 2, 6, 7, and 9); in this group the λ values changed from 0.30 to 0.84 (P = 0.008). By contrast, in the group III patients, who did not initiate combination therapy, the mean λ value decreased from 0.32 to 0.24 during the course of this study. These numerical comparisons of coreceptor usage demonstrated a shift in the predominant viral population from X4 to R5 after initiation of a variety of combination antiviral regimens.
Long-term analysis of the effect of antiviral therapy, viral level, and CD4+ cell count on coreceptor usage.
The period of follow-up for treated women in this study averaged 28.5 months, during which their coreceptor usage, plasma HIV-1 RNA levels, and CD4+ cell count varied, sometimes in concert (Figure 1). We performed a multivariate Poisson regression to analyze the relationship of these variables to antiretroviral treatment and to each other over time. The mulitvariate regression indicated that antiretroviral therapy with two or more drugs was by far the most significant factor in determining λ, the numerical expression of the proportion of viruses using R5 (P = 0.01). Although changes in viral level and the CD4+ cell count had a significant effect on λ in univariate analysis, they lost all significance when considered in a multivariate regression analysis with antiretroviral therapy. The strength of the relationship between initiation of therapy and shift in HIV-1 coreceptor usage is reflected in the course of treated individuals such as patient 8, who maintained high plasma HIV-1 RNA levels during treatment, but demonstrated a substantial, long-term shift in viral population toward R5 (Figure 1).
Discussion
Coreceptor usage plays a major role in pathogenesis and disease progression. The emergence of X4-using HIV-1 strains occurs in approximately half of infected patients and heralds CD4+ cell depletion and accelerated disease progression. This study presents numerous lines of evidence demonstrating that in HIV-1–infected individuals with advanced disease and X4 virus, combination antiviral therapy leads to a shift in the predominant viral population from X4- to R5-using strains. This shift was seen by examining cultivated primary viral isolates, biologically cloned virus, and uncultivated virion-derived HIV-1 RNA from plasma. In addition, it was seen not only in response to HAART, but also to combinations of two or more nucleoside analogues.
The alteration in HIV-1 coreceptor usage was most dramatic at the first visit immediately after initiation of combination therapy, but Poisson regression analyses showed a significant change that persisted for the mean 28.5-month follow-up period for treated patients. Furthermore, multivariate analyses showed that the shift in viral population was dependent upon treatment, not the changes in plasma HIV-1 RNA level and CD4+ cell count that often accompany it. Because therapy was associated with changes in viral level and CD4+ cell count, λ did change with these two variables, but the regression suggested that the treatment brought about the shift and the other variables had only a secondary effect.
The data presented here suggest that antiretroviral therapy is capable of affecting not only the quantity of HIV-1 in an infected individual but the character of the virus as well. Slowing of HIV-1 disease progression and prevention of the emergence of resistance have been associated with suppression of plasma virus, making the elimination of detectable plasma HIV-1 a major goal of treatment (13, 24, 25). Recent reports, however, have shown that many individuals treated by HAART did not experience sustained suppression of plasma viremia, although some did derive immunologic and clinical benefits (15–17). The overall drop in mortality and illness due to HIV-1 in the US after the introduction of combination therapy provides evidence that the benefit may be widespread. The clinical benefits of HAART may thus stem from two effects on the virus: a suppression of plasma viremia and a shift in the viral population toward R5-using strains.
Three studies previously addressed the relationship of HIV-1 phenotype to antiviral therapy in clinical settings that differ from the one described here. One report noted a transient loss of the SI phenotype in 30% of late-stage patients taking didanosine but not zidovudine monotherapy (26). More recently, a paper addressed HIV-1 coreceptor usage in patients whose mean viral levels remained very elevated (>500,000 copies of HIV-1 RNA per milliliter) despite HAART (27). In that cross-sectional study, patients were infected with a predominance of X4 strains after therapy. A recent study of HIV-1–infected children showed that HAART was associated with a restriction in the number of coreceptors used. Although most of the children did not have X4-using virus strains before treatment, a shift in coreceptor usage from X4 to R5 was seen in a few children who had a response to HAART (28).
The analyses presented here focussed on women enrolled in natural history studies. It would be of interest to study HIV-1 coreceptor usage in individuals participating in a controlled treatment trial, where patient profile and therapy are standardized and participants are followed more frequently. Because of the controlled nature of a treatment trial, it may be possible to assess the contribution of a shift in viral population to clinical response. In addition, such a trial may provide an opportunity to determine whether coreceptor usage may predict which patients are likely to achieve a durable response to HAART. Finally, because gender, race, and host chemokine-receptor genotype can influence the HIV-1 RNA level and rate of CD4+ cell decline (18, 29), it would be worthwhile to study patients of both genders, varied racial backgrounds, and different CCR5 genotypes.
The data presented here enabled us to quantitate the proportion of virus using each coreceptor and to make numerical comparisons of coreceptor usage over time and in different individuals. As demonstrated in this and previous studies, once X4 strains evolve in an infected individual, the majority of primary isolates and plasma virus populations include both X4 and R5 viruses (Figure 1, Tables 2 and 3). To rigorously compare the effect of a therapeutic intervention or follow the natural history of HIV-1 coreceptor usage, it is necessary to quantitate the proportion of each strain. This quantitative comparison allowed us to document that antiviral therapy led to a clear, statistically significant shift in coreceptor usage (Table 1).
Studies have documented that both R5 and X4 using HIV-1 strains can be detected in advanced disease (6, 8, 10). Very few studies, however, have examined HIV-1 clones for dual tropism. Previously, a molecularly cloned, dual-tropic primary HIV-1 isolate named 89.6 was analyzed and found to use both X4 and R5, perhaps representing an important transitional phenotype in the evolution from NSI to SI viruses (2). A recent paper examined the dual-tropic 89.6 viral isolate in greater detail (30), reporting that functional clones were derived from the quasispecies comprising the 89.6 primary isolate. Even among these clones, however, 14% were not dual tropic, but instead used either R5 or X4 exclusively (30). To address the issue of coreceptor usage by individual viruses, two pioneering studies derived a small number of biologic clones, with results varying according to the indicator cell line employed. In a study using CCC/CD4 cells, three clones from a primary SI isolate were found to be dual tropic, although a biologic clone from another such isolate used X4 alone (31). In a second study that employed the HOS-CD4+ cell system to assay coreceptor preference, seven biologic clones from four individuals used either X4 or R5, but not both (9). To ascertain the proportion of virus in a primary isolate using each coreceptor, we analyzed 525 biologic and 221 molecular clones but found none that were dual tropic. The analyses reported here add a substantial body of data to show that dual-tropic cloned viruses are unusual when assayed by the HOS-CD4+ system. Further studies are needed to determine how frequently dual tropic clones of HIV-1 can be isolated.
The reason for the preferential suppression of X4 strains and shift in HIV-1 coreceptor usage is not clear, but several mechanisms are possible. Most depend on the concept that combination therapy reduces the HIV-1 burden greatly, leaving a viral reservoir consisting of a larger proportion of R5 than X4 strains. X4 and R5 viruses infect different cell populations, with R5 viruses targeting relatively long-lived CD4+ memory cells and macrophages that may serve as reservoirs of HIV-1 (10, 32). Other cells infected by R5 virus may be inaccessible to antiviral therapy because of their location or drug permeability. Immune activation may periodically result in the stimulation of HIV-1 replication and release of R5 virus from these cells (33, 34).
HAART has been found to suppress a large fraction of viral replication, resulting in recovered viruses that are derived in part from a latent reservoir of HIV-1 (33, 34). Analyses of these strains show that latent viruses vary, but there is evidence that at least a portion of them may represent ancestral species; these latent viruses may include R5 strains. There are also data showing that HAART leads to a reduction in expression of coreceptors in lymphoid tissue (35). This reduction may change the balance of different coreceptors and result in differential replication of R5 strains after therapy.
Finally, an immune mechanism may contribute to the selective suppression of X4 strains over time. A study of viral phenotypes during primary infection showed that soon after infection SI strains are detected, but selectively eliminated, perhaps by immune surveillance (36). After an immunologic response to HAART, a newly reconstituted immune response might help to suppress X4 strains in a similar manner.
Although this study reports that the viral population in treated patients shifted toward R5 after initiation of therapy, the X4 virus eventually rebounded in most individuals, usually to a limited extent (Figure 1). It is not known whether these X4 strains reflected the reemergence of virus present before therapy or the evolution of new, drug-resistant strains. The source of virus may also have been a reservoir other than blood. Understanding coreceptor usage during potent antiretroviral therapy is relevant to HIV-1 dynamics and the maintenance of viral suppression and clinical response. Coreceptor usage studies may have a role in virologic monitoring of patients on antiviral therapy, particularly as drugs are developed that target R5 or X4 strains of HIV-1. Additional studies of viral variation, drug resistance, and coreceptor usage in well-characterized, treated patients are needed.
Acknowledgments
We thank R. Collman for scientific discussion; T. Moran and the Wadsworth Center Molecular Genetics Core Laboratory for oligonucleotide synthesis and DNA sequence analysis; D. Bopst, M. Bradley, M. Dupuis, L. Hirsch, and L. Townsend for technical help and specimen shipment; S. Back, R. Castro, M. Vasco, and W. Gao for obtaining and processing patient data; M. Wren for graphics; A. Klugo for preparing the manuscript; and the study patients for their participation. The NIH AIDS Research and Reference Reagent Program provided the HOS-CD4+ cell lines and the clones of HIV LAV/HTLVIII and JR-FL. This work was supported in part by grant U01A135004 from the National Institute for Allergy and Infectious Diseases. S. Philpott is the recipient of a National Research Service Award (1F32HD08478-01) from the National Institute of Child Health and Human Development. This paper is dedicated to the memory of C.O., a devoted WIHS participant.
References
- 1.Deng H, et al. Identification of a major coreceptor for primary isolates for HIV-1. Nature. 1996; 381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 2.Doranz BJ, et al. A dual-tropic, primary HIV-1 isolate that uses fusin and the B-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusin cofactors. Cell. 1996; 86:1149–1159. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 3.Berger EA, et al. A new classification for HIV-1. Nature. 1998; 391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 4.Samson M, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of CCR-5 chemokine receptor gene. Nature. 1996; 382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 5.Shankarappa R, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999; 73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scarlatti G, et al. In vivo evolution of HIV-1 coreceptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997; 3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 7.Bjorndal A, et al. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997; 71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koot M, et al. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993; 118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 9.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997; 185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuitemaker H, et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell tropic virus populations. J Virol. 1992; 67:1772–1778. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaak H, et al. In vivo HIV-1 infection of CD45RA+CD4+ T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4+ cell decline. Proc Natl Acad Sci USA. 2000; 97:1269–1274. doi: 10.1073/pnas.97.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palella FJ, Jr, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med. 1998; 338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 13.Hammer SM, et al. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997; 337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 14.Autran B, et al. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science. 1997; 277:112–116. doi: 10.1126/science.277.5322.112. [DOI] [PubMed] [Google Scholar]
- 15.Deeks SE, Barbour JD, Martin JN, Swanson MD, Grant RM. Sustained CD4+ T cell response after virologic failure of protease inhibitor-based regimens in patients with human immunodeficiency virus infection. J Infect Dis. 2000; 181:946–953. doi: 10.1086/315334. [DOI] [PubMed] [Google Scholar]
- 16.Mezzaroma I, et al. Clinical and immunologic response without decrease in virus load in patients with AIDS after 24 months of highly active antiretroviral therapy. Clin Infect Dis. 1999; 29:1423–1430. doi: 10.1086/313520. [DOI] [PubMed] [Google Scholar]
- 17.Grabar S, et al. Clinical outcome of patients with HIV-1 infection according to immunologic and virologic response after 6 months of highly active antiretroviral therapy. Ann Intern Med. 2000; 133:401–410. doi: 10.7326/0003-4819-133-6-200009190-00007. [DOI] [PubMed] [Google Scholar]
- 18.Anastos K, et al. Association of race and gender with HIV-1 RNA levels and immunologic progression. J Acquir Immune Defic Syndr Hum Retrovirol. 2000; 24:218–226. doi: 10.1097/00126334-200007010-00004. [DOI] [PubMed] [Google Scholar]
- 19.Fang G, et al. Maternal plasma human immunodeficiency virus type 1 RNA level: a determinant and projected threshold for mother-to-child transmission. Proc Natl Acad Sci USA. 1995; 92:12110–12114. doi: 10.1073/pnas.92.26.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang G, Weiser B, Visosky L, Townsend L, Burger H. Molecular cloning of full-length HIV-1 genomes directly from plasma viral RNA. J Acquir Immune Defic Syndr Hum Retroviral. 1996; 12:352–357. doi: 10.1097/00042560-199608010-00004. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharyya D, Brooks BR, Callahan L. Positioning of positively charged residues in the V3 loop correlates with HIV type 1 syncytium-inducing phenotype. AIDS Res Hum Retroviruses. 1996; 12:83–90. doi: 10.1089/aid.1996.12.83. [DOI] [PubMed] [Google Scholar]
- 22.Hung C-S, Vander Heyden N, Ratner L. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1gp120 involved in CCR5 utilization. J Virol. 1999; 73:8216–8226. doi: 10.1128/jvi.73.10.8216-8226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995; 267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 24.Gunthard HF, et al. Human immunodeficiency virus replication and genotypic resistance in blood and lymph nodes after a year of potent antiretroviral therapy. J Virol. 1998; 72:2422–2428. doi: 10.1128/jvi.72.3.2422-2428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Brien WA, Hartigan PM, Daar ES, Simberkoff MS, Hamilton JD. Changes in plasma HIV RNA levels and CD4+ lymphocyte counts predict both response to antiretroviral therapy and therapeutic failure. Ann Intern Med. 1997; 126:939–945. doi: 10.7326/0003-4819-126-12-199706150-00002. [DOI] [PubMed] [Google Scholar]
- 26.Delforge ML, et al. In vivo inhibition of syncytium-inducing variants of HIV in patients treated with didanosine. AIDS. 1995; 9:89–90. [PubMed] [Google Scholar]
- 27.Holtkamp N, et al. Unexpected coreceptor usage of primary human immunodeficiency virus type 1 isolates from viremic patients under highly active antiretroviral therapy. J Infect Dis. 2000; 181:513–521. doi: 10.1086/315240. [DOI] [PubMed] [Google Scholar]
- 28.Equils O, et al. Recovery of replication-competent virus from CD4 T cell reservoirs and change in coreceptor use in human immunodeficiency virus type 1-infected children responding to highly active antiretroviral therapy. J Infect Dis. 2000; 182:751–757. doi: 10.1086/315758. [DOI] [PubMed] [Google Scholar]
- 29.Michael NL, et al. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997; 3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 30.Singh A, Collman RG. Heterogeneous spectrum of coreceptor usage among variants within dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies. J Virol. 2000; 74:10229–10235. doi: 10.1128/jvi.74.21.10229-10235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons G, et al. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996; 70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perelson AS, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997; 387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 33.Finzi D, et al. Latent infection of CD4+ cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999; 5:512–516. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 34.Chun T-W, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997; 94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersson J, et al. Early reduction of immune activation in lymphoid tissue following highly active HIV therapy. AIDS. 1998; 12:F123–F129. doi: 10.1097/00002030-199811000-00004. [DOI] [PubMed] [Google Scholar]
- 36.Cornelisson M, et al. Syncytium-inducing (SI) phenotype suppression at seroconversion after intramuscular inoculation of a non-syncytium-inducing/SI phenotypically mixed human immunodeficiency virus population. J Virol. 1995; 69:1810–1818. doi: 10.1128/jvi.69.3.1810-1818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]