Abstract
To determine the effect of aging on IFN-γ–induced MHC class II antigen expression, we produced bone marrow–derived macrophages in vitro. In these conditions, we analyzed the effect of aging on the genomic expression of macrophages without the influence of other cell types that may be affected by aging. Although macrophages from young and aged mice showed an identical degree of differentiation, after incubation with IFN-γ, the expression at the cell surface of the IA complex and the levels of IAβ protein and mRNA were lower in aged macrophages. Moreover, the transcription of the IAβ gene was impaired in aged macrophages. The amount of transcription factors that bound to the W and X, but not to the Y, boxes of the IAβ promoter gene was lower in aged macrophages. Similar levels of CIITA mRNA were found after IFN-γ treatment of both young and aged macrophages. This shows that neither the initial cascade that starts after the interaction of IFN-γ with the receptor nor the second signals involved in the expression of CIITA are impaired in aged macrophages. These data indicate that aging is associated with low levels of MHC class II gene induction by IFN-γ because of impaired transcription.
Introduction
In aged populations, infectious diseases are generally more serious and cause higher mortality. The responses to the influenza virus and to vaccination against this virus have been extensively studied, in an attempt to overcome the increased risk of lethal infection in elderly people (1). This has been attributed to a decline in the functional activity of the immune system. Many factors have been implicated, e.g., thymus involution and progressive impairment of the tissues and cells involved in the generation of an immune response (2).
MHC class II proteins play a key role in the development and maintenance of the immune system (3). They participate in the generation of the T-cell repertoire in the thymus and are required for antigen presentation to T lymphocytes (4). Aberrant expression of class II proteins has been linked to immune dysfunction. Lack of class II expression in humans (5) and in animal models (6) leads to severe combined immunodeficiency, and abnormal expression is probably linked to the development of autoimmune diseases (7). The level of class II antigen expression has been correlated with the intensity of the immune response in physiological conditions (8). Class II proteins are normally expressed on a limited number of cell types, including B, thymic epithelial, dendritic, and glial cells, as well as activated macrophages (9). Their expression is regulated by cytokines, mainly IFN-γ (10), primarily through transcriptional activation. We have recently reported that IFN-γ also regulates the translational process (11).
In this study, we used primary nontransformed macrophages grown in vitro from aged and control mice. We analyzed the effect of aging on the genomic expression of macrophages without the interactions of other cell types that may be affected by aging. We assessed the effect of aging on the IFN-γ–induced expression of the MHC-II IAβ gene in macrophages. Our results showed that macrophages from aged mice had lower expression of class II molecules. This is due to decreased DNA-binding activity in the promoter of the IAβ gene.
Methods
Reagents.
Recombinant M-CSF was from R&D Systems Inc. (Minneapolis, Minnesota, USA). IFN-γ was kindly donated by Genentech (San Francisco, California, USA). The products used were of the best grade available and were purchased from Sigma Chemical Co. (St. Louis, Missouri, USA). Deionized water further purified with a Millipore Milli-Q system (Millipore Corp., Bedford, Massachusetts, USA) was used.
Cell culture.
Bone marrow–derived macrophages were isolated from 6-week– and 20-month–old C57/BL6 mice (Charles River Laboratories Inc., Wilmington, Massachusetts, USA) as described elsewhere (12). The cells were cultured in plastic tissue culture dishes (150 mm) in 40 ml DMEM containing 20% FBS and 30% L-cell conditioned media as a source of M-CSF. The cells were incubated at 37°C in a humidified 5% CO2 atmosphere. After 7 days of culture, a homogeneous population of adherent macrophages was obtained.
Protein cell-surface expression.
Surface expression of the IAβ was analyzed with monoclonal anti-mouse IAd,b antibodies (06281D; PharMingen, San Diego, California, USA) as described previously (11). The cells were activated with saturating amounts of IFN-γ (300 U/ml) (13) at different times and then harvested and washed in ice-cold PBS. After fixing with 2% paraformaldehyde for 30 minutes at 4°C, they were resuspended in 50 μl PBS containing 5% FBS and then incubated at 4°C for 15 minutes with 1 μg/106 cells of anti-CD16/CD32 mAb (PharMingen) to block Fc receptors. They were then incubated for 1 hour at room temperature with murine IAd,b specific antibody, washed by centrifugation through a FBS cushion, and finally incubated with FITC-conjugated anti-mouse IgG antibody for 1 hour at 4°C. The levels of Mac1 were determined using an anti-CD11b as primary antibody (PharMingen). Stained cell suspensions were analyzed using an Epics XL flow cytometer (Coulter Corp., Hialeah, Florida, USA). FITC excitation was obtained using a 488-nm argon laser lamp, and its fluorescence was collected with a 525-nm band-pass filter. The parameters used to select cell populations for analysis were forward and side light scatter. As a control for specificity, we used a irrelevant antibody of the same isotype.
Analysis of DNA content with DAPI.
A total of 106 cells previously subjected to a specific treatment were resuspended and fixed in ice-cold 70% ethanol (14). They were then washed in PBS; resuspended in 0.2 ml of a solution containing 150 mM NaCl, 80 mM HCl, and 0.1% Triton X-100; and incubated at 4°C for 10 minutes. Thereafter, 1 ml of a solution containing 180 mM Na2HPO4, 90 mM citric acid, and 2 mg/ml DAPI (pH 7.4) was added to each sample. After incubating the cells at 4°C for 24 hours, their fluorescence was measured with an Epics Elite flow cytometer (Coulter Corp.). For this analysis, we used an ultraviolet laser with a 25 mW excitation beam at 333–364 nm, and fluorescence was collected with a 525-nm band-pass filter. Cell doublets were gated out by comparing the pulse area versus the pulse width. Twelve thousand cells were counted for each histogram, and cell cycle distributions were analyzed with the Multicycle program (Phoenix Flow Systems Inc., San Diego, California, USA).
Plasmids and constructs.
The cDNA probes for IAβ and IEβ used in Northern blotting were a kind gift from P. Cosson (Basel Institute for Immunology, Basel, Switzerland) (15). An 18S ribosomal RNA probe was used as a control for the amount of loaded RNA (16). For TAP1 determination, we used a 199-bp probe obtained by PCR using the following primers: AACCCTGTCTCCTGGCGAAG and GCCGCATCACTGACTGGATT. The probe for the lmp2 determination had a length of 560 bp and was also cloned by PCR, using the following primers: CATGGCAGTGGAGTTTGACG and CCAGGATGACTCGATGGTCC. The fragments were cloned and sequenced, showing 100% homology with the reported sequence (17). The cDNA probe for CIITA was a gift from R. Flavell (18).
Northern blot analysis.
Total cellular RNA was extracted by the acid guanidinium thiocyanate-phenol-chloroform method, as described previously (19). RNA was separated by using 1% agarose with 5 mM MOPS (3-[N-morpholino]propanesulfonic acid) (pH 7.0) / 1 M formaldehyde buffer. The RNA was transferred overnight to a GeneScreen nitrocellulose membrane (Life Science Products, Boston, Massachusetts, USA) and fixed by ultraviolet irradiation (150 mJ). To check for differences in RNA loading, we analyzed the expression of 18S ribosomal RNA. All probes were labeled with γ-32P-dCTP (ICN Pharmaceuticals, Costa Mesa, California, USA) following the oligolabeling kit method (Pharmacia Biotech AB, Uppsala, Sweden). After incubating the membranes for 18 hours at 65°C in hybridization solution (20% formamide, 5× Denhart’s, 5× SSC, 10 mM EDTA, 1% SDS, 25 mM Na2HPO4, 25 mM NaH2PO4, 0.2 mg/ml salmon sperm DNA, and 106 cpm/ml of 32P-labeled probe), they were exposed to Kodak X-AR films (Kodak Co., Rochester, New York, USA). The bands of interest were quantified with a Molecular Imaging System (Bio-Rad Laboratories Inc., Richmond, California, USA).
In vitro nuclear transcription assay.
In vitro nuclear transcription run-on assay was performed as reported elsewhere (20). Briefly, individual plates of cells were cultured until the cell density reached approximately 5 × 105 cells/ml, after which the cells were treated with IFN-γ for the indicated times. All subsequent steps and solutions were at 4°C. The nuclei were isolated and resuspended in 100 μl of storage buffer containing 50 mM Tris-HCl (pH 8.2), 5 mM MgCl2, 40% glycerol and 0.1 mM EDTA and frozen at –90°C.
For the labeling reaction, an equal number of nuclei per time point were thawed on ice and stained with 0.3 U/μl of RQ1 DNase (Promega Corp., Madison, Wisconsin, USA). The labeled RNA was denaturated by incubation for 10 minutes at 65°C in 100 μl final volume of hybridization solution (50% formamide, 5× SSC, 5× Denhardt’s and 100 μg/ml denaturated salmon sperm DNA; 1× SSC = 0.15 M sodium chloride, 0.015 M sodium citrate). Filters containing bound plasmid were hybridized with an equal number of labeled RNA counts in a final volume of 1 ml for 4–5 days at 42°C. The filters were washed twice, each time for 15 minutes at room temperature in 2× SSC, 1% SDS, then in 0.5× SSC, 0.1% SDS for 120 minutes at 60°C. The filters were exposed to x-ray film and quantitated by densitometry.
Nuclear extract preparation and DNA-binding assay.
Nuclear extracts were prepared as described previously (21). In brief, cells were washed in PBS and lysed in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 M EDTA, 0.1 mM EGTA and 1 mM DTT). The crude nuclei were extracted at 4°C in buffer B (20 mM HEPES [pH 7.9], 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1 mM DTT) with constant stirring, followed by centrifugation at 12,000 g for 15 minutes. The supernatant (protein concentration of 1–5 mg/ml) was frozen in aliquots and stored at –80°C.
Oligonucleotides (33-pb) containing the X, Y, and W boxes of the IAβ gene promoter were used for the DNA-binding assay (22). The oligonucleotide of the W box was the following: 5′-GATCATGCCTTGCATAGAGAGCCTTTGTAAACA-3′; for the W box mutant, the underlined sequence (W box) was replaced by TCTAACG. The sequence for the X box was: 5′-GATCGTTTACCCAGAGACAGATGACAAGCTTCG-3′; for the mutated X box, the underlined sequence (X box) was replaced by AGTCGTCTGACTC. The sequence for the Y box was the following: 5′-GATCCAATGCTGATTGGTTCCTCACTTGGGACG-3′, and for the Y box mutant the last two Gs in the underlined sequence were replaced by two Ts. DNA (10,000 cpm of 32P-labeled DNA, ∼0.1 ng) and nuclear extracts (4 μg) were mixed in a total volume of 20 μl with a buffer containing 12 mM HEPES (pH 7.9), 60 mM KCl, 5 mM MgCl2, 0.12 mM EDTA, 0.3 mM phenyl-methylsulfonyl fluoride, 0.3 mM dithiothreitol, and 12% glycerol. A total of 100 ng of poly (dI-dC) (Pharmacia Biotech AB) was also added to the reaction mix. The samples were incubated at room temperature for 30 minutes and then loaded in a 6% polyacrylamide gel (acrylamide/bisacrylamide 30:1), TBE 0.25×, and electrophoresis was carried out at 4°C and 200 V for 2 hours. Thereafter, the gels were dried and exposed to Kodak x-ray film. Radioactivity was quantified using a radioanalytic imaging system (Molecular Analyst System; Bio-Rad Laboratories Inc.).
Protein extraction and Western blot analysis.
The cells were washed twice in ice-cold PBS and lysed on ice with lysis solution (1% Triton X-100, 10% glycerol, 50 mM HEPES [pH 7.5], 150 mM NaCl, protease inhibitors) (19). The protein concentration of the samples was determined with the Bio-Rad protein assay. The proteins from the cell lysates (100 μg) were boiled at 95°C in Laemmli SDS loading buffer, separated in a 8% SDS-PAGE gel, and electrotransferred to nitrocellulose membranes (Hybond-ECL; Amersham Corp., Arlington Heights, Illinois, USA). The membranes were blocked for at least 1 hour at room temperature in Tris buffer saline-0.1% Tween-20 (TBS-T) containing 5% non-fat dry milk and then incubated with TBS-T containing the primary antibody (FF282-4 antiserum, kindly provided by R.N. Germain [NIH, Bethesda, Maryland, USA]). Incubation was performed for 1 hour at room temperature. After three 5-minute washes in TBS-T, the membranes were incubated with peroxidase-conjugated anti-rabbit antibody (Cappel, Turnhout, Belgium) for 1 hour. After three 5-minute-washes with TBS-T, ECL detection was performed (Amersham Life Science Ltd., London, United Kingdom), and the membranes were exposed to x-ray films (Amersham Life Science Ltd.). Quantification of the blots was carried out by densitometric analysis.
Statistical analysis.
The Wilcoxon test, nonparametric for paired differences, was used for all calculations (23).
Results
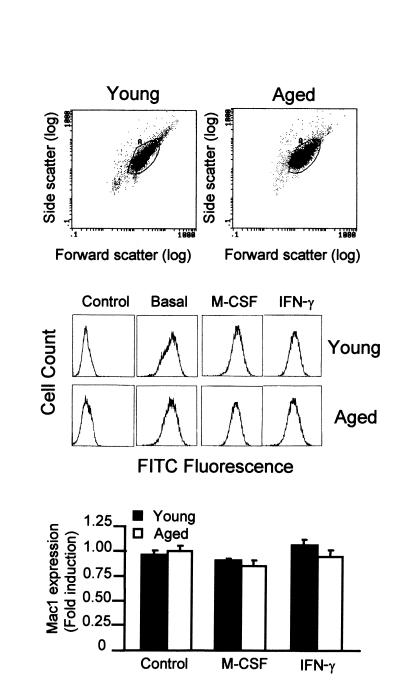
To determine whether the IFN-γ–induced expression of MHC class II molecules is affected by aging, we used bone marrow–derived macrophages from young (6-week-old) and aged (19- to 24-month-old) mice. For each experiment, bone marrow macrophages were obtained from pools of bone marrow from five young or five old mice. Macrophages are produced in vitro in media containing M-CSF as growth factor and constitute a homogeneous population of nontransformed quiescent cells. Besides, the production of macrophages in vitro avoids the presence of exogenous factors in both groups of mice that could modulate the expression of MHC class II molecules. In these conditions, we analyzed the effect of aging on the genomic expression of macrophages without the interactions of other cell types that may be affected by aging. The number, size, and DNA content of macrophages obtained from young and aged mice were similar (Figure 1). Finally, the determination of cell-surface markers that are expressed during macrophage maturation, like Mac1 (24), was similar in macrophages from aged and young mice (Figure 1). The treatment of macrophages with M-CSF or IFN-γ did not significantly modify the levels of Mac1 expression in macrophages. Therefore, this procedure enabled us to compare only the effects of aging.
Figure 1.
Similar size and antigen Mac1 expression in macrophages from aged and young mice. In each case, macrophages were prepared from the bone marrow of pools of five young (6-week-old) or five aged (24-month-old) mice. The size of macrophages was determined by cytometry. Surface expression of Mac1 (CD11b) was assessed by cytometry using specific antibodies after blocking the Fc receptors. In some experiments, macrophages were treated for 24 hours with M-CSF (1200 U/ml) or IFN-γ (300 U/ml). The first two panels are representative of six independent experiments. In the third panel, data are represented as the mean ± SD of six experiments. No significant differences were found between young and aged macrophages (P > 0.05).
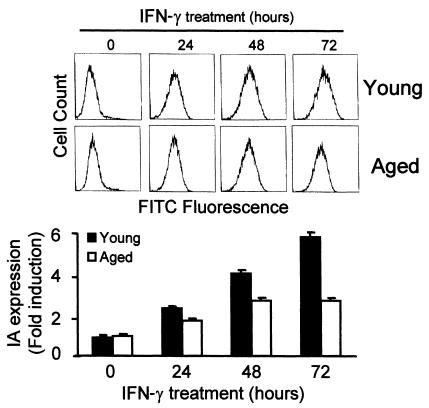
Macrophages from young and aged mice were incubated with saturating amounts of IFN-γ (300 U/ml) (13) for different periods. The cell-surface expression of class II antigens was analyzed by flow cytometry using specific purified mAb’s against IAb molecules. Macrophages from young and old mice showed a time-dependent increase in IA expression (Figure 2). However, after 72 hours of treatment, macrophages from young mice showed a sixfold increase in IA expression, whereas those from old mice showed only a threefold increase (Figure 2). This difference was significant (P < 0.01). The number of cells expressing IA on the cell surface in both types of macrophages was similar (data not shown). This suggests that the defect of IA expression is not associated with a specific population of aged macrophages.
Figure 2.
Decreased IFN-γ–induced cell-surface expression of MHC class II IA in macrophages from aged mice. Bone marrow macrophages from young and old mice were incubated with saturating amounts of IFN-γ (300 U/ml) for the indicated times. Surface expression of MHC class II was then assessed by cytometry using specific antibodies after blocking the Fc receptors. The background level of expression in young and older macrophages was similar. The upper panel is representative of six independent experiments. In the lower panel, data are represented as the mean ± SD of six experiments. After 24, 48, and 72 hours of IFN-γ treatment, there was a significant difference (P < 0.01) between young and aged macrophages.
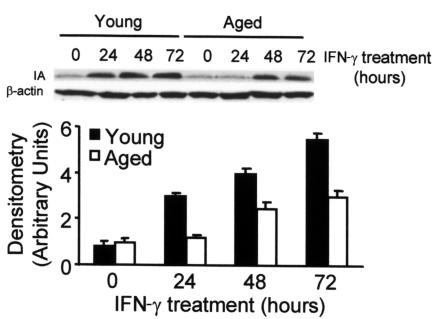
The lower expression of class II antigens in macrophages from aged mice could be caused by a defect in the transport of these molecules from the cytosol to the cell surface. To rule out this possibility, we measured the amount of IA protein by Western blotting. An antibody against β-actin, whose levels of expression do not change after treating the cells with IFN-γ, was included as a control for each time point. Again, the concentration of IA protein increased in both types of macrophages after treatment with IFN-γ (Figure 3). However, young macrophages showed a 5.8-fold increase after 72 hours of treatment with IFN-γ, whereas older macrophages showed only a threefold increase.
Figure 3.
Decreased IA protein level in bone marrow macrophages of aged mice after IFN-γ treatment. The cells were treated or not treated with 300 U/ml of IFN-γ for the indicated times and then lysed to obtain total protein extracts. After Western blotting, IA was detected with specific antibodies. β-Actin was used as a control that is not affected by the treatment to normalize the loaded samples. The upper panel is representative of six independent experiments, all of which showed the same level of basal expression in young and older macrophages. In the lower panel, data are represented as the mean ± SD of six experiments. After 24, 48, and 72 hours of IFN-γ treatment, there was a significant difference (P < 0.01) between young and aged macrophages.
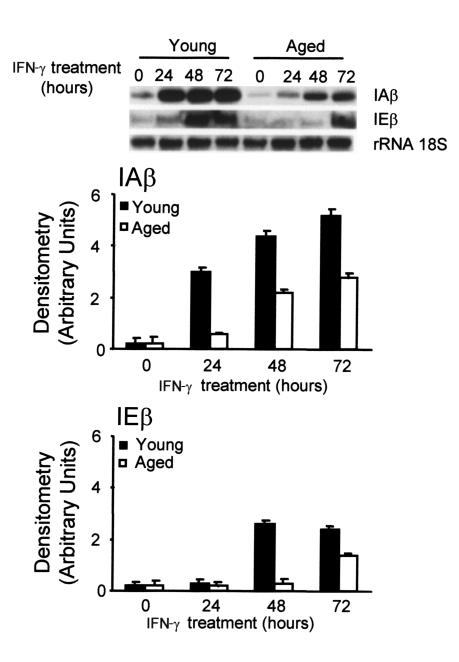
The steady-state levels of IAβ and IEβ RNA in macrophages were monitored during the treatment of the cells with IFN-γ (Figure 4). As a control, we used a probe for the 18S RNA. A low level of IAβ and IEβ mRNA was detected in untreated cells from both types of macrophages. In macrophages from young mice, there was a steady increase in IAβ and IEβ mRNA levels after treating the cells with IFN-γ, reaching a plateau after 48 hours of treatment; the final increase over background levels was 26- and 18-fold, respectively. Macrophages from aged mice also showed a steady increase in IAβ and IEβ mRNA levels, but a plateau was not reached after 48 hours of treatment and the estimated increase over background was only of 15- and 8-fold, respectively, after 72 hours of treatment. These differences were significant (P < 0.01). As determined by Northern blotting, the amount of RNA present in the whole cell was similar to the amount of cytoplasmic RNA, thus suggesting that the distribution of the mRNA between the nucleus and the cytoplasm is not different in young and old mice (data not shown).
Figure 4.
Reduced IAβ and IEβ mRNA expression in bone marrow macrophages from aged mice after IFN-γ stimulation. Macrophages from aged and young mice were treated with IFN-γ (300 U/ml) for the indicated times. The 18S RNA was used as an internal control of RNA load and was not affected by the treatment. The upper panel is representative of six independent experiments, all of which showed the same level of basal expression in young and older macrophages. In the lower panels, data are represented as the mean ± SD of six experiments. For IAβ, after 24, 48, and 72 hours, and for IEβ, after 48 and 72 hours of IFN-γ treatment, there was a significant difference (P < 0.01) between young and aged macrophages.
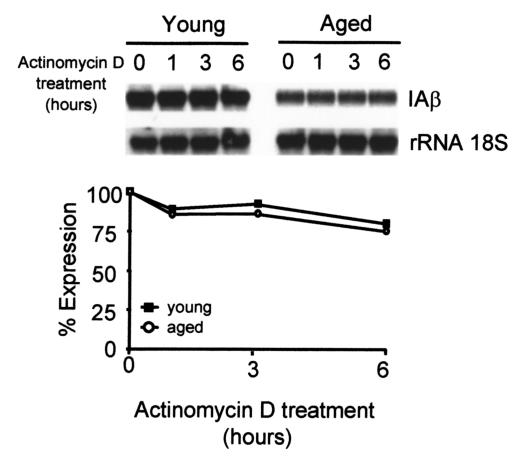
The differences in the IAβ protein expression between young and aged macrophages after treatment with IFN-γ were linked to variation in levels of IAβ mRNA. To determine whether this variation was caused by a decrease in mRNA synthesis or by an increase in mRNA degradation, we measured the rate of mRNA degradation after treatment with IFN-γ for 48 hours. Actinomycin D was then added to a concentration (5 μg/ml) sufficient to block all further RNA synthesis, as determined by [3H]UTP incorporation (25), and the RNA was isolated from aliquots of cells at various intervals thereafter. In macrophages from both aged and young mice, IAβ mRNA was stable (Figure 5).
Figure 5.
Similar half-life of IAβ mRNA in IFN-γ–treated macrophages from young and aged mice. Cells were treated with IFN-γ for 48 hours, transcription was stopped by the addition of actinomycin D (5 μg/ml), and RNA was measured after 1, 3, and 6 hours of incubation. Cell viability was greater than 95% for all culture conditions. Densitometry data are expressed as a percentage of the level determined before the addition of actinomycin D. The upper figure is representative of five independent experiments. No significant differences were found between young and aged macrophages (P > 0.05).
The similar half-life of IAβ mRNA indicated that the difference in mRNA levels between young and aged macrophages was due to different rates of transcription. To determine the levels of transcription induced by IFN-γ in macrophages, we performed nuclear run-on transcription assays for the IAβ gene. The nuclei for this series of experiments were isolated from bone marrow macrophages that had been treated for the indicated times with IFN-γ. We repeatedly observed some transcription of the IAβ gene in nuclei isolated from untreated cells (Figure 6). A 1.6- and 2.9-fold increase in the rate of transcription (average of five experiments) was observed after 24 hours of IFN-γ treatment, in aged and young macrophages, respectively (P < 0.01). No change in the level of transcription of the ribosomal L32 gene was observed throughout the experiments. These results indicate that an increase in the transcription of IAβ gene occurred after the cells were treated with IFN-γ and that the difference in transcription between young and aged macrophages is significant.
Figure 6.
Reduced IAβ gene transcription in bone marrow macrophages from aged mice after IFN-γ stimulation. For the nuclear run-on assays, bone marrow macrophages were treated with IFN-γ (300 U/ml) for 24 hours, and the nuclei were prepared and used for the transcription run-on assay. The ribosomal L32 gene was used as a control that is not affected by the conditions of the study. The autoradiographs depicted are representative of five experiments. In the lower panel, data are represented as the mean ± SD of five experiments. There are significant differences for the IAβ gene transcription between young and aged macrophages after IFN-γ treatment (P < 0.01).
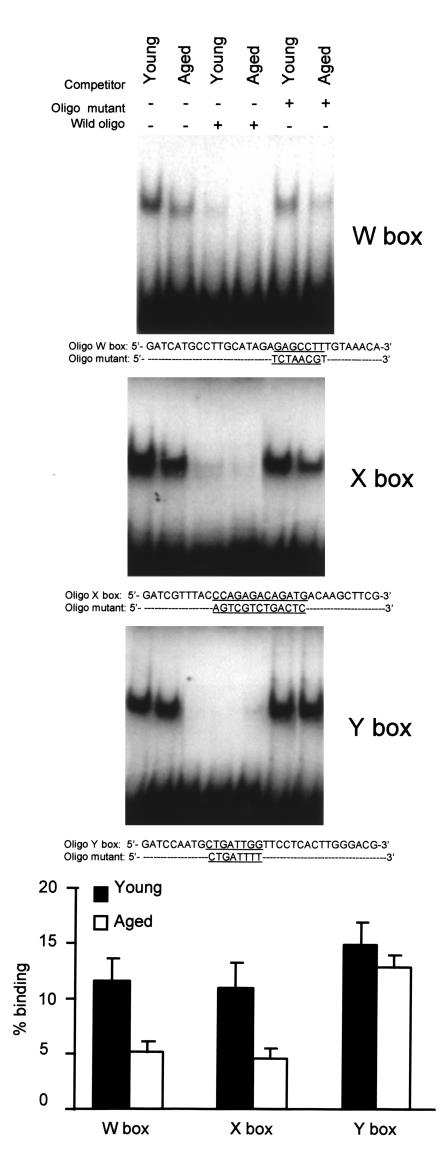
The transcriptional regulation of expression of the class II genes involves at least three cis-acting elements located upstream of the transcription initiation site (26, 27). These elements are known as W, X, and Y boxes, and nuclear factors bind to each element. Using footprinting and competition assays, we previously showed that specific nuclear factors bind to the Y box (21, 22, 28, 29), the X box (22, 30), and the W box (22, 31) of the IAβ gene. A gel electrophoresis DNA binding assay was performed using nuclear extracts prepared from macrophages of young and aged mice and oligonucleotide probes covering the W, X, and Y boxes (Figure 7). Each probe formed a specific complex with protein from macrophages of young and older mice, which appeared as a slower migrating band in the assay. The addition of cold double-stranded oligonucleotide competed for the binding of nuclear factors to the respective labeled oligonucleotide, whereas oligonucleotides containing mutations within the W, X, or Y boxes did not compete. The amount of transcription factors bound to the W and X boxes was significantly lower in macrophages from aged mice (P < 0.01). However, no significant differences were observed for the Y box (P > 0.05).
Figure 7.
Decreased binding of nuclear extracts from aged macrophages to the W and X boxes of the IAβ promoter. Nuclear extracts (4 μg) were used for gel retardation assays with probes for W, X, and Y boxes. A 50-fold excess of cold DNA was used in the competition experiments. The amount of DNA free or bound to protein was quantitated, and the figure represents the mean ± SD of five independent experiments. There are significant differences between young and aged macrophages in the binding capacity to the W and X boxes (P < 0.01), but not to the Y box (P > 0.05).
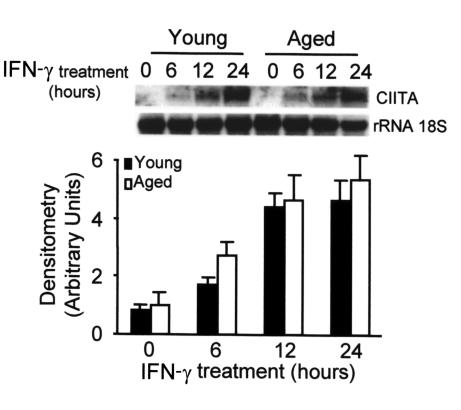
Thus, the quantitative differences in the expression of these transcription factors may account for the decreased transcription of the IAβ gene. However, other factors may be involved. CIITA is a tissue-specific and IFN-γ–inducible transactivator required for the transcriptional activation of MHC II genes (32). To study whether the expression of CIITA was affected by aging, we treated macrophages from young and aged mice with saturating amounts of IFN-γ and we determined the mRNA levels of CIITA. The expression of CIITA started after 6 hours of treatment with IFN-γ and reached a plateau after 24 hours (Figure 8). No differences were found between the levels of mRNA in macrophages from young and aged mice (P > 0.05). This shows that neither the initial cascade of events that starts after the interaction of IFN-γ with the receptor nor the signals involved in the expression of CIITA are impaired in aged macrophages.
Figure 8.
Similar kinetics of CIITA mRNA expression in macrophages from aged and young mice after IFN-γ stimulation. Bone marrow macrophages from aged and young mice were treated with IFN-γ (300 U/ml) for the indicated times. The 18S RNA was used as an internal control of RNA load, and it was not affected by the treatment. The upper panel is representative of six independent experiments, all of which showed the same level of basal expression in young and older macrophages. In the lower panel, data are represented as the mean ± SD of six experiments. At each time point, no significant differences were found between young and aged macrophages (P > 0.05).
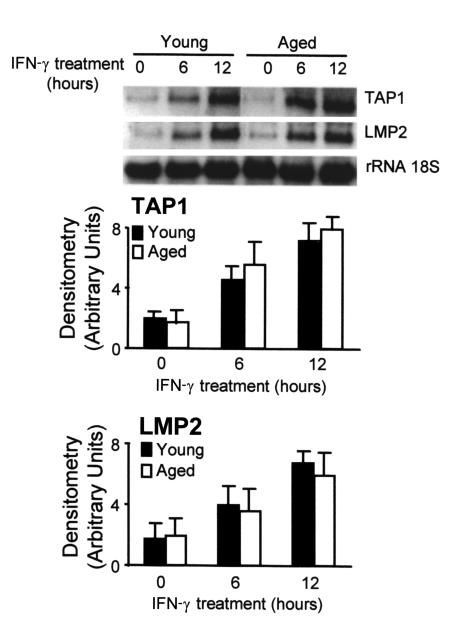
To determine whether the effect of aging on MCH class II expression was a general feature of the genes induced by IFN-γ in macrophages, we measured the IFN-γ–induced expression of TAP1 and lmp2. These two genes are located in the MHC class II region and share the same promoter but are transcribed in opposite orientations (17). The promoter of these genes does not contain the W, X, or Y boxes and thus, the transcription factors involved are different (33). The Northern blot analysis of these mRNAs showed that the expression of both genes was induced after 6 hours of treatment with IFN-γ (Figure 9). No differences were found between macrophages from young and aged mice (P > 0.05). Therefore, the effect of aging on the expression of class II antigens was specific and it was not due to a general impairment of IFN-γ–induced macrophage activation.
Figure 9.
The kinetics of TAP1 and lmp2 mRNA expression in macrophages from aged and young mice after IFN-γ stimulation are not different. Bone marrow macrophages from aged and young mice were treated with IFN-γ (300 U/ml) for the indicated times. The 18S RNA was used as an internal control of RNA load, and it was not affected by the treatment. The upper panel is representative of six independent experiments, all of which showed the same level of basal expression in young and older macrophages. In the lower panel, data are represented as the mean ± SD of six experiments. At each time point, no significant differences were found between young and aged macrophages (P > 0.05).
Discussion
Our data show, for the first time to our knowledge, that the IFN-γ–induced expression of the MHC class II IAβ gene in macrophages in aged mice is lower than in young mice. The experiments were carried out using bone marrow–derived macrophages produced in vitro, which are homogeneous populations of primary quiescent nontransformed macrophages. Besides, this method enables us to compare only the effects of aging on macrophages.
We found similar levels of IAβ and IEβ mRNA and IA protein in nonstimulated macrophages from young and aged mice. This is in accordance with previous models, which detected residual levels of class II expression despite a lack of response to IFN-γ. This is the case of the targeted gene inactivation of CIITA (18), RFX5 (34), and STAT-1 genes (35). After IFN-γ stimulation, the cell-surface expression of IA and the total amount of IA protein increased in both young and aged macrophages. However, there is a quantitative difference: both cell-surface expression and IA protein levels were lower in older macrophages.
We have previously reported that IAβ expression is regulated at the levels of transcription (20) and translation (11). The low levels of protein in older macrophages correlate with decreased levels of mRNA, and the mRNA half-life is not affected by age, suggesting that transcription is impaired in older macrophages. As measured by run-on assays, IFN-γ increased the rate of transcription of the IAβ gene, but a significant difference was found between young and aged macrophages. The low amounts of transcription factors that bind to the W and X boxes are probably related to the low levels of transcription. The same transcription factors are involved in the regulation of all MHC class II genes (9), which may account for the low levels of IEβ mRNA in aged macrophages. Therefore, the impaired expression of MHC IA molecules associated with aging may involve all the MHC class II genes. The X box plays a central role in chromatin remodeling and transcription from class II promoters. It consists of the core X and downstream X2 boxes. The former binds to the heterotrimeric regulatory factor that binds to the X box (RFX) and that is formed by three elements: RFX5, RFXAP, and RFXANK (36, 37). The interaction between RFX and the X box is enhanced by NF-Y, which binds to the Y box (27). The W box (also called S) is required for IFN-γ inducibility and optimal expression of class II genes (31). This is probably due to the recruitment of the CIITA trans-activator to the class II promoters (38). There are similarities between the sequences of the X and W boxes, and RFX binds to the W box both in vitro and in vivo (39). Therefore, a decrease in the amount of the group of RFX factors could explain the defective transcription. Unfortunately, no reagents are currently available to identify which of the murine RFX proteins are expressed at low levels. The defect in older macrophages is probably not due to a complete lack of one of the RFX complex components, as this could induce a complete block of transcription, as shown in humans (5). The levels of the CIITA trans-activator in macrophages after IFN-γ treatment are similar in macrophages from young and aged mice. This suggests that the signal transduction that follows the interaction of the IFN-γ receptor with the ligand and induces CIITA expression is not impaired in aged macrophages. This is supported by the levels of expression of the two IFN-γ–inducible MHC class II genes, TAP1 and lmp2, with a promoter that does not contain the W, X, or Y boxes (33). These results also suggest that the effect on the expression of MHC II molecules is not caused by variations in the cell-surface expression of IFN-γ receptors during aging or by a defect in the signal transduction pathway activated after the interaction of the IFN-γ receptors with the ligand that induces the expression of CIITA. As the expression of MHC class II molecules correlates with the intensity of the immune response (8), our observations could be clinically relevant. The low levels of MHC class II expressed in macrophages could partly explain the immunodeficiency associated with aging.
Our data are supported by several reports on immunosenescence. These reports suggest that the age-related decline in the functions of the immune system results from the decreased activity of several genes controlling these functions, such as those encoding IL-2 (40), IL-2–receptor (41), γ-IFN (42), and IL-3 (43). A defect in the levels of transcription factors such as Sp1 has also been associated with aging (44).
The loss of functional activity of genes is the basis of aging. The life span of an organism is the sum of deleterious changes and counteracting repair and maintenance mechanisms (45). Various factors of genome instability have been described in aging. The WRN gene encodes a DNA helicase of the RecQ family, the inactivation of which induces the Werner syndrome characterized by shortened life span and premature onset of phenotypic changes that are reminiscent of aging (46). The Werner syndrome is characterized by chromosome instability, but also enhanced frequencies of deletion and rearrangement (47). Telomerases are also implicated. Several findings relate telomere shortening to aging in vivo (48). Telomerase-knockout mice show age-dependent telomere shortening, with genetic instability and shorter life span (49). In aging, impaired repairing of damaged DNA could be the ultimate mechanism accounting for the altered expression of several transcription factors that may be crucial for the immune system.
Acknowledgments
This study was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (SAF98/0102 and PM98/0200) to A. Celada. We acknowledge the help received from Jaume Comas and Rosario Gonzalez, in charge of the flow cytometry facility of the Serveis Científico Tècnics, Universitat de Barcelona. We also acknowledge the assistance of Pierre Fabre Iberica and in particular Amadeo Perez. We thank P. Cosson for the plasmids containing the IAβ and IEβ cDNA. We thank R.N. Germain for the anti-IA antibody. We also thank Martin Cullell-Young for the revision of the manuscript.
References
- 1.Couch RB, et al. Influenza: its control in persons and populations. J Infect Dis. 1986; 153:431–440. doi: 10.1093/infdis/153.3.431. [DOI] [PubMed] [Google Scholar]
- 2.Miller, R.A. 1999. Aging and immune function. In Fundamental immunology. 4th edition. W.E. Paul, editor. Lippincott-Raven Publishers. Philadelphia, Pennsylvania, USA. 947–966.
- 3.Germain RN. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994; 76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 4.von Boehmer H. Positive selection of lymphocytes. Cell. 1994; 76:219–228. doi: 10.1016/0092-8674(94)90330-1. [DOI] [PubMed] [Google Scholar]
- 5.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996; 14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 6.Cosgrove D, Gray D, Dierich A. Mice lacking MHC class II molecules. Cell. 1991; 66:1051–1066. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- 7.Bottazzo GF, Pujol-Borrell R, Hanafusa T, Feldmann M. Role of aberrant HLA-DR expression and antigen presentation in induction of endocrine autoimmunity. Lancet. 1983; 2:1115–1119. doi: 10.1016/s0140-6736(83)90629-3. [DOI] [PubMed] [Google Scholar]
- 8.Otten LA, Steimle V, Bontron S, Mach B. Quantitative control of MHC expression by the transactivator CIITA. Eur J Immunol. 1998; 28:473–478. doi: 10.1002/(SICI)1521-4141(199802)28:02<473::AID-IMMU473>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 9.Glimcher LH, Kara CJ. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992; 10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 10.King DP, Jones PP. Induction of Ia and H-2 antigens on a macrophage cell line by immune interferon. J Immunol. 1983; 13:315–318. [PubMed] [Google Scholar]
- 11.Goñalons E, Barrachina M, Garcia-Sanz JA, Celada A. Translational control of MHC class II I-A molecules by IFN-gamma. J Immunol. 1998; 161:1837–1843. [PubMed] [Google Scholar]
- 12.Xaus J, et al. Interferon γ induces the expression of p21waf-1 and arrests macrophage cell cycle, preventing induction of apoptosis. Immunity. 1999; 11:103–113. doi: 10.1016/s1074-7613(00)80085-0. [DOI] [PubMed] [Google Scholar]
- 13.Celada A, Gray PW, Rinderknecht E, Schreiber RD. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J Exp Med. 1984; 160:55–74. doi: 10.1084/jem.160.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xaus J, et al. LPS induces apoptosis in macrophages mostly through the autocrine production of TNF-α. Blood. 2000; 95:3823–3831. [PubMed] [Google Scholar]
- 15.Cosson P, Bonifacino PS. Role of transmembrane domain interactions in the assembly of class II MHC molecules. Science. 1992; 258:659–662. doi: 10.1126/science.1329208. [DOI] [PubMed] [Google Scholar]
- 16.Torczynski P, Bollon AP, Fuke M. The complete nucleotide sequence of the rat 18S ribosomal RNA gene and comparison with the respective yeast and frog genes. Nucleic Acids Res. 1983; 11:4879–4890. doi: 10.1093/nar/11.14.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kishi F, Suminami Y, Monaco JJ. Genomic organization of the mouse Lmp-2 gene and characteristic structure of its promoter. Gene. 1993; 133:243–248. [PubMed] [Google Scholar]
- 18.Chang CH, Guerder S, Hong SC, van Ewijk W, Flavell RA. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 1996; 4:167–178. doi: 10.1016/s1074-7613(00)80681-0. [DOI] [PubMed] [Google Scholar]
- 19.Xaus J, et al. IFN-γ up-regulates the A2B adenosine receptor expression in macrophages: a mechanism of macrophage deactivation. J Immunol. 1999; 162:3607–3614. [PubMed] [Google Scholar]
- 20.Celada A, Klemsz MJ, Maki RA. Interferon gamma activates multiple pathways to regulate the expression of the genes for class II major histocompatibility I-A β, tumor necrosis factor and complement component C3 in mouse macrophages. Eur J Immunol. 1989; 19:1103–1109. doi: 10.1002/eji.1830190621. [DOI] [PubMed] [Google Scholar]
- 21.Celada A, Shiga M, Imagawa M, Kop J, Maki RA. Identification of a nuclear factor that binds to a conserved sequence of the I-A beta gene. J Immunol. 1988; 140:3995–4002. [PubMed] [Google Scholar]
- 22.Celada A, McKercher S, Maki RA. Repression of major histocompatibility complex IA expression by glucocorticoids: the glucocorticoid receptor inhibits the DNA binding of the X box DNA binding protein. J Exp Med. 1993; 177:691–698. doi: 10.1084/jem.177.3.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snedecor, G.W., and Cochran, W.G. 1967. Statistical methods. 6th edition. Iowa State University Press. Ames, Iowa, USA. 125–131.
- 24.Springer TA. Adhesion receptors of the immune system. Nature. 1990; 346:425–434. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- 25.Barrachina M, Goñalons E, Celada A. LPS upregulates MHC class II I-A expression in B lymphocytes at transcriptional and at translational level. Tissue Antigens. 1999; 54:461–470. doi: 10.1034/j.1399-0039.1999.540503.x. [DOI] [PubMed] [Google Scholar]
- 26.Benoist C, Mathis D. Regulation of major histocompatibility complex class-II genes: X, Y and other letters of the alphabet. Annu Rev Immunol. 1990; 8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- 27.Glimcher LH, Kara CJ. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992; 10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 28.Celada A, Maki RA. DNA binding of the mouse class II major histocompatibility complex CCAAT factor depends on two components. Mol Cell Biol. 1989; 9:3097–3100. doi: 10.1128/mcb.9.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Celada A, McKercher SR, Maki RA. Identification of the transcription factors NF-YA and NF-YB as factors A and B that bound to the promoter of the major histocompatibility complex class II gene I-Aβ. Biochem J. 1996; 317:771–778. doi: 10.1042/bj3170771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Celada A, Maki RA. Evidence for multiple major histocompatibility class II X-box binding proteins. Mol Cell Biol. 1989; 9:5219–5222. doi: 10.1128/mcb.9.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celada A, Gil P, McKercher SR, Maki RA. Identification of a transcription factor that binds to the S box of the I-Aβ gene of the major histocompatibility complex. Biochem J. 1996; 313:737–744. doi: 10.1042/bj3130737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993; 75:135–146. [PubMed] [Google Scholar]
- 33.Wright KL, et al. Coordinate expression of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J Exp Med. 1995; 181:1459–1471. doi: 10.1084/jem.181.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clausen BE, et al. Residual MHC class II expression on mature dendritic cells and activated B cells in RFX5-deficient mice. Immunity. 1998; 8:143–155. doi: 10.1016/s1074-7613(00)80467-7. [DOI] [PubMed] [Google Scholar]
- 35.Meraz MA, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996; 84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 36.Masternak K, et al. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998; 20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 37.Nagarajan UM, et al. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999; 10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- 38.Masternak K, et al. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000; 14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 39.Jabrane-Ferrat N, Fontes JD, Boss JM, Peterlin BM. Complex architecture of major histocompatibility complex class II promoters: reiterated motifs and conserved protein-protein interactions. Mol Cell Biol. 1996; 16:4683–4690. doi: 10.1128/mcb.16.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu WT, Pahlavani M, Cheung HT, Richardson A. The effect of aging on the expression of interleukin 2 messenger ribonucleic acid. Cell Immunol. 1986; 100:224–231. doi: 10.1016/0008-8749(86)90021-3. [DOI] [PubMed] [Google Scholar]
- 41.Nagel JE, et al. Decreased proliferation, interleukin 2 synthesis, and interleukin 2 receptor expression are accompanied by decreased mRNA expression in phytohemagglutinin-stimulated cells from elderly donors. J Clin Invest. 1988; 81:1096–1102. doi: 10.1172/JCI113422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gauchat JF, Walker C, De Weck AL, Stadler BM. Stimulation-dependent lymphokine mRNA levels on human mononuclear cells. Eur J Immunol. 1988; 18:1441–1446. doi: 10.1002/eji.1830180921. [DOI] [PubMed] [Google Scholar]
- 43.Li DD, Chien YK, Gu MZ, Richardson A, Cheung HT. The age-related decline in interleukin-3 expression in mice. Life Sci. 1988; 43:1215–1222. doi: 10.1016/0024-3205(88)90211-1. [DOI] [PubMed] [Google Scholar]
- 44.Ammendola R, Mesuraca M, Russo T, Cimino F. Sp1 DNA binding efficiency is highly reduced in nuclear extracts from aged rat tissues. J Biol Chem. 1992; 267:17944–17948. [PubMed] [Google Scholar]
- 45.Johnson FB, Sinclair DA, Guarente L. Molecular biology of aging. Cell. 1999; 96:291–302. doi: 10.1016/s0092-8674(00)80567-x. [DOI] [PubMed] [Google Scholar]
- 46.Yu CE, et al. Positional cloning of the Werner’s syndrome gene. Science. 1996; 272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 47.Ellis NA. DNA helicases in inherited human disorders. Curr Opin Genet Dev. 1997; 7:354–363. doi: 10.1016/s0959-437x(97)80149-9. [DOI] [PubMed] [Google Scholar]
- 48.Chang E, Harley CB. Telomere length and replicative aging in human vascular tissues. Proc Natl Acad Sci USA. 1995; 92:11190–11194. doi: 10.1073/pnas.92.24.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudolph KL, et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999; 96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]