Group A Streptococcus (GAS) is a Gram-positive bacterium that is responsible for a wide range of human infections, including pharyngitis and tonsillitis, skin infections (impetigo, erysipelas, and cellulitis), sepsis, osteomyelitis, a toxic-shock syndrome, and necrotizing fasciitis (1). The organism is also responsible for nonsuppurative sequelae such as acute rheumatic fever and acute glomerulonephritis. Globally, GAS causes extensive human morbidity and mortality (1). In the US, approximately 10,000 cases of severe invasive GAS disease occur each year. Direct costs associated with pharyngitis in the US have been estimated to be $1 billion annually. Although antibiotic treatment has largely eliminated rheumatic fever and subsequent rheumatic heart disease in North America and Western Europe, these crippling forms of heart disease are still very common in the developing world and in certain indigenous populations in developed countries.
The resurgence of GAS as a cause of serious human infections in the US, Europe, and elsewhere in the 1980s and into the 1990s has been thoroughly documented and has heightened public awareness about this organism (1). Disease resurgence, coupled with the lack of a licensed GAS vaccine and ongoing concern about acquisition of penicillin resistance, has stimulated renewed interest in the molecular pathogenesis of GAS disease. The return of GAS as a cause of serious infections was a strong reminder that biomedical science knows relatively little about the evolutionary and other forces that drive temporal variation in bacterial disease frequency and severity. GAS’s revival has also taught the importance of understanding the molecular basis of pathogen variation, not merely as a means to discriminate between clinically relevant strains, but to provide data relevant to pathogenesis, host adaptation, and the origin of new pathogenic forms. In this Perspective, we highlight certain aspects of the genetic variation present in GAS, and we stress the biomedical relevance of this diversity.
Extensive allelic variation in GAS structural genes
Until 1989, most information pertaining to variation in GAS was based on serologic differences in M protein, an antiphagocytic surface molecule that is a critical virulence factor in many GAS (2). Although approximately 80 M types were recognized, and knowledge of variation in this protein was quite useful for certain kinds of epidemiological studies, serologic classification alone failed to reveal the special characteristics of GAS that were associated with certain infection types, such as streptococcal toxic shock syndrome (STSS). The convention of classifying isolates on the basis of one surface antigen was not satisfactory for the study of genomic relationships because serologic methods do not reflect the complexity of the genetic structure of GAS. Hence, there was little basis for understanding the genetic variations in population structure occurring in connection with temporal changes in the frequency or character of streptococcal diseases, such as invasive infections or rheumatic fever outbreaks. In essence, protein serotyping led to the conventional but incorrect notion that strains assigned to the same antigenic class were genetically identical or nearly so.
The first realization that allelic variation in GAS structural genes is extensive was provided by a study examining the genetic diversity and clonal relationships among 108 GAS isolated from patients with invasive infections (3). These GAS isolates were studied by multilocus enzyme electrophoresis, long a workhorse for indexing levels of allelic variation in structural genes and estimating levels of chromosomal variation among eukaryotic organisms and bacterial strains (4, 5). The study found that allelic variation was extensive in GAS, and, importantly, that more than one-half of the strains were members of clonally related lineages designated as multilocus electrophoretic type-1 and -2 (ET1 and ET2), marked by serotypes M1 and M3, respectively. Moreover, the analysis identified a strong statistical relationship between the presence of the gene (speA) encoding the superantigen streptococcal pyrogenic exotoxin A (scarlet fever toxin) and STSS. Hence, analysis of allelic variation in GAS recovered from clinically relevant human infections provided new insight into host-pathogen relationships occurring in nature. Moreover, the findings stimulated investigations resulting in the discovery of three new SpeA variants (see below).
Knowledge of allelic variation in GAS genes, especially those encoding putative or proven virulence factors, expanded rapidly when automated DNA sequencing methods were formulated (Table 1). One common theme revealed by large-scale analysis of virulence gene variation is that several mechanisms have contributed to generating allelic diversity in natural populations of GAS, including point mutations, slipped-strand mispairing, and assortive recombination (6–19).
Table 1.
Allelic polymorphism in GAS genes
The results of one recent study illustrate the extensive allelic diversity that can be present in GAS. Lukomski et al. (18) identified an extracellular protein with collagen-like characteristics that is present in all GAS. Identification of an extracellular collagen-like protein made by GAS is potentially of considerable interest, given the association of the pathogen with various sequelae presumed to have an autoimmune component. The protein has a cell-wall anchor motif at the carboxy-terminus, variable numbers of contiguous collagen-like Gly-X-X triplet amino acid residue repeats, and a variable amino-terminus. This streptococcal collagen-like (Scl) protein participates in the adherence of GAS to human cells and the pathogenesis of soft tissue infection in a mouse model of human disease. The scl gene was sequenced in 50 strains representing 21 distinct M protein serotypes. The number of contiguous Gly-X-X motifs ranged from 12 in serotype M6 isolates to 62 in a serotype M42 organism. The amino acid sequence also varied in the Gly-X-X regions, with 50 distinct Gly-X-X sequences identified. Interestingly, M1 and M18 organisms had the identical allele which indicates very recent horizontal gene transfer.
The observation that Scl is an extracellular protein that is potentially subjected to selective pressure in the host suggested that molecular evolutionary genetic analysis would provide insight into the processes shaping variation at the scl locus. The level of selective constraint, a measure of the putative functional importance of a genetic region, was determined for different domains of this locus. The analysis showed that the locus is responding to selective pressure in such a way as to increase the genetic distance between scl alleles while maintaining the Gly-X-X repeat motif. The discovery of a collagen-like gene that encodes a highly variable extracellular protein provides a framework for analysis of the role of structural variation in autoimmune sequelae.
Single amino acid polymorphisms and GAS-human interactions
Allelic variation is common among all eukaryotic and prokaryotic organisms, as well as viruses, and is a well-recognized source of individual differences in genetic composition and behavior. Allelic variation in human genes that results in structural variation in encoded proteins can have a profound effect on phenotype and overall fitness. For example, variation in the CFTR and SS genes is responsible for cystic fibrosis and sickle cell anemia, respectively. In principle, allelic variation in bacterial virulence genes could have important biomedical consequences for host-pathogen interactions. There has been little investigation of the potential ramifications of naturally occurring single amino acid polymorphisms for interactions of the host with bacterial pathogens, including GAS.
An Arg-Gly-Asp (RGD) loop embellishment in an extracellular cysteine protease virulence factor.
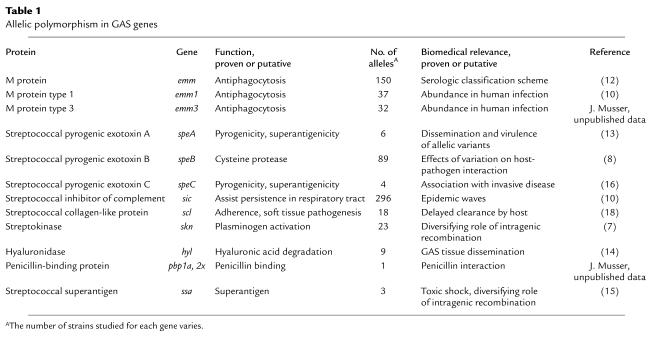
Stockbauer et al. (8) identified one example illustrating how a single naturally occurring amino acid replacement in a GAS virulence factor alters the character of interaction with human cells. GAS isolates produce a highly conserved extracellular cysteine protease known as streptococcal pyrogenic exotoxin B (SpeB) that is a critical virulence factor (20–22). The enzyme detrimentally affects many important host molecules, such as fibronectin, vitronectin, and H-kininogen (20). Motivated by the observation that immunization with SpeB confers protective immunity to mice (22), Stockbauer et al. assessed the level of allelic variation in the speB gene by sequence analysis of the gene from 200 GAS isolates (8). Three main mature SpeB variants were identified that differed from one another by only one or two amino acids. One of these variants (arbitrarily designated mSpeB2) contains an Arg-Gly-Asp (RGD) sequence, a tripeptide motif that is recognized by a variety of host integrins (Figure 1). Importantly, mSpeB2 is made by all isolates of the unusually abundant serotype M1 and several other geographically widespread clones that frequently cause invasive infections. Only the mSpeB2 variant bound specifically via the RGD motif region to host cells expressing integrin αvβ3 (also known as the vitronectin receptor) or αIIbβ3 (platelet glycoprotein IIb-IIIa). In essence, analysis of speB allelic variation resulted in (a) identification of the first Gram-positive virulence factor that directly binds integrins, (b) identification of naturally occurring variants of a documented Gram-positive virulence factor with biomedically relevant differences in their interactions with human cells, and (c) enhanced appreciation that subtle natural variation in the structure of a microbial virulence factor alters the character of host-pathogen interactions. These results stimulated Kagawa et al. (23) to solve the SpeB crystal structure, which revealed the surface location of this integrin-binding RGD motif (Figure 1).
Figure 1.
Ribbon diagram of the three-dimensional structure of SpeB, an extracellular cysteine protease virulence factor. The protease domain is shown in blue, the prosegment in brown, the catalytic site cysteine in green, and the RGD loop embellishment in red. Dotted lines indicate flexible regions. The RGD loop is magnified to reveal the exposed Asp-164 residue required for binding of αvβ3 and αIIbβ3 human integrins. Serotype M1 strains that are abundant causes of human disease contain the RGD motif in the integrin-binding loop, whereas other strains possess an RSD sequence that does not bind integrins (8). N, amino-terminus; C, carboxy-terminus. Modified from ref. 23with permission.
SpeA and the resurgence of invasive GAS disease.
SpeA is one of numerous pyrogenic toxins produced by GAS (24). Many of these toxins are superantigens that stimulate proliferation of T-cell subsets bearing specific T-cell receptor (TCR) variable domains, a process that can lead to a cytokine storm, shock, and death (24).
The identification of STSS, and the significant association of SpeA production with organisms causing this syndrome, led to an interest in allelic variation in speA. Population genetic analysis predicted the likelihood of new variants of speA in part on the basis of the observation that this gene was distributed among strains representing the breadth of genomic diversity in GAS (3). Comparative sequencing of the speA gene in isolates representing a range of ETs identified four alleles, designated speA1 through speA4 (9). SpeA2 and SpeA3 each differed from the presumed ancestral variant (SpeA1) by single amino acid replacements at positions 80 (Gly→Ser) and 76 (Val→Ile), respectively. Importantly, all contemporary M1/ET1 and M3/ET2 strains had the speA2 and speA3 alleles, respectively, whereas strains of these same ETs recovered before 1950 had the ancestral speA1 allele (25, 26).
The association of two variant speA alleles with two widely disseminated GAS clones that have caused the majority of invasive disease episodes in the last two decades suggests that the mutants differ functionally. In this regard, Kline and Collins reported (27) that the new SpeA3 toxin variant was more mitogenic and had tenfold higher affinity for the class II MHC molecule than the old SpeA1 variant. Crystal structure analysis revealed that the two amino acid replacements, Gly80Ser in SpeA2 and Val76Ile in SpeA3, are located close to a zinc-binding site in SpeA1, a region postulated to be involved in class II MHC recognition (28, 29). Taken together, the results suggest that clones expressing the mutant toxins might have more severe effects in vivo. Although the exact role of SpeA in GAS infections and STSS remains unresolved, it is clear that allelic variants of speA are molecular markers of very successful clones of this human pathogen.
Structural variation in the streptococcal inhibitor of complement (Sic) protein and GAS epidemic waves
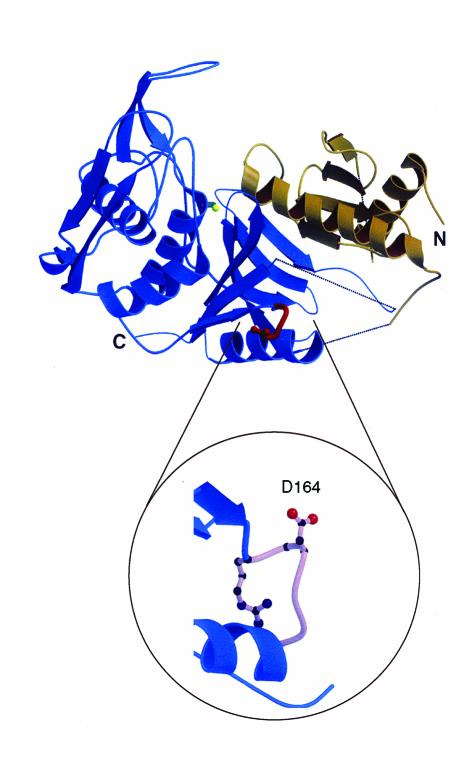
In addition to being the most common serotype recovered from invasive disease episodes, M1 strains display epidemic wave behavior (1). The molecular mechanisms responsible for the abundance of M1 organisms in invasive disease and the factors contributing to the emergence and perpetuation of epidemic waves are now being revealed. Conventional wisdom and published data led to the concept that most M1 epidemic waves are mono- or oligoclonal. Recent analysis of allelic variation in the sic virulence gene has proven otherwise (6, 10, 11) (Figure 2).
Figure 2.
Summary of variation identified in the Sic protein among 1,673 serotype M1 GAS strains. Left: Positions of the polymorphic nucleotides and amino acids. The single-letter amino acid abbreviations are used. Far right: Domains of Sic located in the proximal half of the protein. SRR, short repeat region; R1–R3, repeats R1, R2, and R3. Modified from ref. 11with permission.
The sic gene encodes an extracellular protein known as streptococcal inhibitor of complement that inhibits the normal cytolytic function of the complement C5b-C9 membrane attack complex in vitro (30). This gene is present in M1 strains but occurs very rarely in other GAS organisms. Analysis of allelic variation in the sic gene in 1,673 M1 GAS isolates obtained from comprehensive, population-based surveillance studies of invasive disease discovered a strikingly high level of diversity among organisms that are otherwise very closely related genetically (Figure 2) (6, 10, 11). Hence, M1 epidemic waves are composed of an array of subclones characterized by Sic variants that are rapidly selected in human populations. Remarkably, virtually all polymorphisms result in structural variation in the Sic protein, indicating that strong positive (Darwinian) selection acts on sic. Several lines of evidence suggest that Sic variants are selected on the mucosal surface by a mechanism involving host antibody (10). Recently, to gain further understanding of the molecular mechanisms of M1 epidemic waves, we have sequenced the sic gene in GAS isolates causing pharyngitis and invasive disease in metropolitan Helsinki, Finland, recovered during a 37-month population-based surveillance study (31). Our results indicate that the majority of invasive organisms are represented in the pool of organisms causing pharyngitis in the same geographic area. More significantly, we discovered that new Sic variants occur among pharyngitis isolates an average of 9.8 months prior to their recovery from invasive episodes. These data unambiguously demonstrate mucosal selection of Sic variants. The significant time lapse that occurs between the appearance of an M1 GAS clone first in noninvasive and subsequently in invasive disease may have implications for the development of a practical strategy to predict epidemics of invasive GAS disease. The mechanism contributing to the abundance of M1 in human infections involves a process whereby Sic significantly enhances persistence in the mammalian upper respiratory tract (32). Hence, analysis of allelic variation in the sic gene has provided critical new insight into the molecular population genetics of serotype M1 GAS epidemic waves (10, 31). Surprisingly, analysis of allelic variation in the gene (emm1) encoding M1 protein ruled out the idea that diversity at this locus contributes to these epidemic waves. Similarly, recent results indicate that structural variation in the M18 protein does not account for variation in the frequency of occurrence of acute rheumatic fever episodes in the region of Salt Lake City, Utah (J.M. Musser, unpublished data).
Future considerations: genome-scale investigations
Currently, GAS genome sequencing projects are nearing completion at the University of Oklahoma (Norman, Oklahoma) (serotype M1), the Sanger Centre (Cambridge, United Kingdom) (serotype M5), and the Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories (multiple strains). The volume of resulting data will be extensive, but will the information move us closer to understanding the complexities of GAS pathogenesis and to formulating new therapeutics? Analysis of the available GAS sequence data (commonly known as data mining) has already produced new discoveries (18, 19, 33), and, undoubtedly, more will rapidly accrue. One of the unexpected insights obtained from population genetic analyses of GAS is that alleles of many genes are reassorting more or less at random over loci, as opposed to the clonal nature of many other bacterial pathogens (34). The resultant new combinations of virulence genes are rapidly expressed, and the overall rate of evolution in the species is accelerated compared with that due to simple nucleotide substitutions (1). Comparison of GAS genomes by DNA microarray technology (35) has already begun to reveal sequences that are common or unique to distinct strains (Figure 3). Genome comparisons with DNA microarrays are being conducted on a large scale in our laboratory. Use of global genomic and proteomic analysis methods will provide substrates to generate additional testable hypotheses regarding host-GAS interactions, an especially important issue because long-described associations of M types with certain clinical conditions such as rheumatic fever defy adequate molecular explanation. In addition, because most or all GAS strains have one or more bacteriophages integrated into their genomes (36, 37), it will be critical to assess the extent to which transduction events have mediated interstrain differences in virulence gene complement. We are certain that investigation of GAS allelic variation, population genetics, and host interactions on a genome scale will provide many new insights pertaining to human disease caused by this pathogen.
Figure 3.
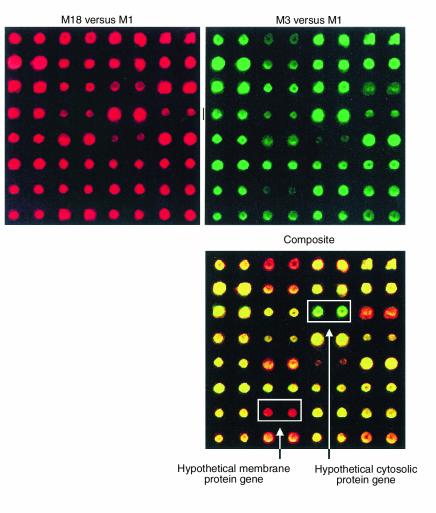
Comparative genome analysis of three GAS strains with a DNA microarray. The array was constructed with PCR products formulated on the basis of a genome sequence available for a serotype M1 strain (38). Approximately 99% of the M1 open reading frames were represented in duplicate on the array. Shown for illustrative purposes are the hybridization results obtained from only one sector of the entire genome array. Sheared genomic DNA from an M18 strain and an M3 strain were labeled with fluorescent dyes Cy5 and Cy3, respectively, and hybridized to the M1 microarray. The hybridization results were analyzed with a ScanArray 5000 laser scanner and QuantArray software package (Packard Biosciences, Meriden, Connecticut, USA). The composite image (bottom) shows that a genomic segment encoding a hypothetical cytosolic protein (green) is unique to this M3 strain, whereas a genomic segment encoding a hypothetical membrane protein (red) is unique to this M18 strain. The yellow spots represent DNA that is present in these M1, M3, and M18 strains.
Acknowledgments
J.M. Musser wishes to thank R.K. Selander for encouraging and supporting research into the molecular population genetics of human bacterial pathogens. We thank T. Schwan and J. Portis for review of the manuscript and T. Kindt, R.M. Krause, and A. Fauci for their critical contributions to the GAS research effort at Rocky Mountain Laboratories. Some of the research on GAS conducted by J.M. Musser and described herein was supported by Public Health Services grant AI-33119. We apologize to colleagues whose important work could not be cited due to space limitations.
References
- 1.Musser, J.M., and Krause, R.M. 1998. The revival of group A streptococcal diseases, with a commentary on staphylococcal toxic shock syndrome. In Emerging Infections. R.M. Krause, editor. Academic Press. New York, New York, USA. 185–218.
- 2.Fischetti, V.A. 2000. Surface proteins on Gram-positive bacteria. In Gram-positive pathogens. V.A. Fischetti, R.P. Novick, J.J. Ferretti, D.A. Portnoy, and J.I. Rood, editors. American Society for Microbiology Press. Washington, DC, USA. 11–24.
- 3.Musser JM, et al. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc Natl Acad Sci USA. 1991; 88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musser JM. Molecular population genetic analysis of emerged bacterial pathogens: selected insights. Emerg Infect Dis. 1996; 2:1–17. doi: 10.3201/eid0201.960101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selander, R.K., and Musser, J.M. 1990. The population genetics of bacterial pathogenesis. In Molecular basis of bacterial pathogenesis. B.H. Iglewski and V.L. Clark, editors. Academic Press Inc. Orlando, Florida, USA. 11–36.
- 6.Hoe N, et al. Rapid molecular genetic subtyping of serotype M1 group A Streptococcus strains. Emerg Infect Dis. 1999; 5:254–263. doi: 10.3201/eid0502.990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapur V, et al. Molecular population genetic analysis of the streptokinase gene of Streptococcus pyogenes: mosaic alleles generated by recombination. Mol Microbiol. 1995; 16:509–519. doi: 10.1111/j.1365-2958.1995.tb02415.x. [DOI] [PubMed] [Google Scholar]
- 8.Stockbauer KE, et al. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins αvβ3 and αIIbβ3. Proc Natl Acad Sci USA. 1999; 96:242–247. doi: 10.1073/pnas.96.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson K, Schlievert PM, Selander RK, Musser JM. Characterization and clonal distribution of four alleles of the speA gene encoding pyrogenic exotoxin A (scarlet fever toxin) in Streptococcus pyogenes. J Exp Med. 1991; 174:1271–1274. doi: 10.1084/jem.174.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoe NP, et al. Rapid selection of complement-inhibiting protein variants in group A Streptococcus epidemic waves. Nat Med. 1999; 5:924–929. doi: 10.1038/11369. [DOI] [PubMed] [Google Scholar]
- 11.Stockbauer KE, et al. Hypervariability generated by natural selection in an extracellular complement-inhibiting protein of serotype M1 strains of group A Streptococcus. Proc Natl Acad Sci USA. 1998; 95:3128–3133. doi: 10.1073/pnas.95.6.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Facklam R, et al. emm typing and validation of provisional M types for group A streptococci. Emerg Infect Dis. 1999; 5:247–253. doi: 10.3201/eid0502.990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bessen DE, et al. Genetic linkage of exotoxin alleles and emm gene markers for tissue tropism in group A streptococci. J Infect Dis. 1999; 179:627–636. doi: 10.1086/314631. [DOI] [PubMed] [Google Scholar]
- 14.Marciel AM, Kapur V, Musser JM. Molecular population genetic analysis of a Streptococcus pyogenes bacteriophage-encoded hyaluronidase gene: recombination contributes to allelic variation. Microb Pathog. 1997; 22:209–217. doi: 10.1006/mpat.1996.9999. [DOI] [PubMed] [Google Scholar]
- 15.Reda KB, et al. Phylogenetic distribution of streptococcal superantigen SSA allelic variants provides evidence for horizontal transfer of ssa within Streptococcus pyogenes. Infect Immun. 1996; 64:1161–1165. doi: 10.1128/iai.64.4.1161-1165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norrby-Teglund A, Holm SE, Norgren M. Detection and nucleotide sequence analysis of the speC gene in Swedish clinical group A streptococcal isolates. J Clin Microbiol. 1994; 32:705–709. doi: 10.1128/jcm.32.3.705-709.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapur V, Nelson K, Schlievert PM, Selander RK, Musser JM. Molecular population genetic evidence of horizontal spread of two alleles of the pyrogenic exotoxin C gene (speC) among pathogenic clones of Streptococcus pyogenes. Infect Immun. 1992; 60:3513–3517. doi: 10.1128/iai.60.9.3513-3517.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukomski S, et al. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect Immun. 2000; 68:6542–6553. doi: 10.1128/iai.68.12.6542-6553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukomski, S., et al. 2001. Identification and characterization of a second extracellular collagen-like protein made by Group A Streptococcus: control of production at the level of translation. Infect. Immun. In press. [DOI] [PMC free article] [PubMed]
- 20.Musser JM. Streptococcal superantigen, mitogenic factor, and pyrogenic exotoxin B expressed by Streptococcus pyogenes. Structure and function. Prep Biochem Biotechnol. 1997; 27:143–172. doi: 10.1080/10826069708000074. [DOI] [PubMed] [Google Scholar]
- 21.Lukomski S, et al. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Invest. 1997; 99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapur V, et al. Vaccination with streptococcal extracellular cysteine protease (interleukin-1 beta convertase) protects mice against challenge with heterologous group A streptococci. Microb Pathog. 1994; 16:443–450. doi: 10.1006/mpat.1994.1044. [DOI] [PubMed] [Google Scholar]
- 23.Kagawa TF, et al. Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: an integrin-binding cysteine protease. Proc Natl Acad Sci USA. 2000; 97:2235–2240. doi: 10.1073/pnas.040549997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlievert PM. Role of superantigens in human disease. J Infect Dis. 1993; 167:997–1002. doi: 10.1093/infdis/167.5.997. [DOI] [PubMed] [Google Scholar]
- 25.Musser JM, et al. Geographic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (scarlet fever toxin) J Infect Dis. 1993; 167:337–346. doi: 10.1093/infdis/167.2.337. [DOI] [PubMed] [Google Scholar]
- 26.Musser JM, et al. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995; 63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kline JB, Collins CM. Analysis of the superantigenic activity of mutant and allelic forms of streptococcal pyrogenic exotoxin A. Infect Immun. 1996; 64:861–869. doi: 10.1128/iai.64.3.861-869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papageorgiou AC, et al. Structural basis for the recognition of superantigen streptococcal pyrogenic exotoxin A (SpeA1) by MHC class II molecules and T-cell receptors. EMBO J. 1999; 18:9–21. doi: 10.1093/emboj/18.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Earhart C, Vath G, Roggiani M, Schlievert P, Ohlendorf D. Structure of streptococcal pyrogenic exotoxin A reveals a novel metal cluster. Protein Sci. 2000; 9:1847–1851. doi: 10.1110/ps.9.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Åkesson P, Sjöholm AG, Björck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996; 271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 31.Hoe NP, et al. Distribution of streptococcal inhibitor of complement variants in pharyngitis and invasive isolates in an epidemic of serotype M1 group A Streptococcus infection. J Infect Dis. 2001; 183:633–639. doi: 10.1086/318543. [DOI] [PubMed] [Google Scholar]
- 32.Lukomski S, et al. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000; 68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen M, Müller H-P, Björck L. Protein GRAB of Streptococcus pyogenes regulates proteolysis at the bacterial cell surface by binding α2-macroglobulin. J Biol Chem. 1999; 274:15336–15344. doi: 10.1074/jbc.274.22.15336. [DOI] [PubMed] [Google Scholar]
- 34.Kehoe MA, Kapur V, Whatmore AM, Musser JM. Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol. 1996; 4:436–443. doi: 10.1016/0966-842x(96)10058-5. [DOI] [PubMed] [Google Scholar]
- 35.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995; 270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 36.Ohlsen, K., et al. 1999. Mobile elements, phages, and genomic islands of staphylococci and streptococci. In Pathogenicity islands and other mobile virulence elements. J.B. Kaper and J. Hacker, editors. American Society for Microbiology Press. Washington, DC, USA. 265–287.
- 37.Krause RM. Studies on bacteriophages of hemolytic streptococci. J Exp Med. 1957; 106:365–384. doi: 10.1084/jem.106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Streptococcus pyogenes Genome Sequencing Project. http://www.genome.ou.edu/strep.html.