Abstract
Females are natural mosaics for X chromosome-linked genes. As X chromosome inactivation occurs randomly, the ratio of parental phenotypes among blood cells is approximately 1:1. Recently, however, ratios of greater than 3:1 have been observed in 38–56% of women over age 60. This could result from a depletion of hematopoietic stem cells (HSCs) with aging (and the maintenance of hematopoiesis by a few residual clones) or from myelodysplasia (the dominance of a neoplastic clone). Each possibility has major implications for chemotherapy and for transplantation in elderly patients. We report similar findings in longitudinal studies of female Safari cats and demonstrate that the excessive skewing that develops with aging results from a third mechanism that has no pathologic consequence, hemizygous selection. We show that there is a competitive advantage for all HSCs with a specific X chromosome phenotype and, thus, demonstrate that an X chromosome gene (or genes) regulates HSC replication, differentiation, and/or survival.
Keywords: hematopoiesis, aging
Hematopoietic stem cells (HSCs) are primitive and generally quiescent cells that are able to support multilineage hematopoiesis. Large numbers of HSCs and redundancy in cytokine function and signal transduction mechanisms assure a stable production of blood cells throughout life. The behavior of HSCs is difficult to study, especially in large animals and humans, because they are infrequent (<1 in 106 nucleated marrow cells) and cannot be isolated by physical or immunologic characteristics (1–3). It is unclear whether the behavior of a stem cell is predominately defined by its genetic constitution or by its interactions with the cytokines and cells present in the marrow microenvironment.
The fate of HSCs with aging is also unclear. To study this, others (4–6) have analyzed clonal contributions to hematopoiesis in normal females who are heterozygous for X chromosome-linked genes. Approximately 50% of blood cells from neonates and females <40 years old express maternal X chromosome genes, and 50% express paternal X chromosome genes. Excessive skewing, defined as parental phenotype ratios greater than 3:1, is rarely seen. However, excessive skewing occurs in 38–56% of normal females over 60 years old (4–6). This has been attributed to the depletion of HSCs (and the support of blood cell production by the few remaining clones), the development of myelodysplasia (the neoplastic expansion and dominance of a single HSC clone), or hemizygous selection. Hemizygous selection (a competitive advantage to all cells that express one parental phenotype) cannot be excluded in studies in an outbred (human) population.
Safari cats are the F1 offspring of Geoffroy (G) and domestic (d) cat parents. Because of X chromosome inactivation early in embryogenesis, each somatic cell in female Safari cats, including each stem cell, expresses d or G glucose-6-phosphate dehydrogenase (G6PD) but not both (7). Each progenitor and differentiated cell expresses the G6PD phenotype of the HSC from which it derived. In previous investigations of female Safari cats in whom hematopoiesis was perturbed by the administration of chemotherapy (busulfan) or by the autologous transplantation of limited numbers of HSCs, we observed fluctuations in the percent of progenitor cells with d G6PD (2, 8–11). In this report, we analyze data obtained from all female Safari cats before their entry into experimental protocols.
MATERIALS AND METHODS
Animal Care.
Female Safari cats (the F1 offspring of G and d cat parents) were bred at Washington State University, Pullman, WA. Marrow was aspirated from the humerus or femur after anesthesia with ketamine and acepromazine (0.11 mg/kg and 22 mg/kg, respectively). Blood was obtained by jugular venipuncture. Animals were housed at the University of Washington, Washington State University, or the Pacific Northwest Research Foundation (for autologous marrow transplantation only). All studies were approved by Institutional Animal Care Committees.
Progenitor Cell Assays and G6PD Analysis.
For marrow culture studies, heparinized marrow cells were layered over percoll (1.070 g/ml; Sigma) and centrifuged at 400 × g for 20 min, and interface cells were collected and washed twice. Mononuclear cells (1 × 105) were plated in 1 ml of IMDM (GIBCO/BRL) supplemented with 1.2% methylcellulose (Kodak), 30% heat-inactivated fetal calf serum (Summit Biotech, Fort Collins, CO), 1% BSA (Intergen, Rochester, NY), 10−4 M 2-mercaptoethanol (Sigma), 1% penicillin/streptomycin/fungizone (PSF; GIBCO/BRL), 1 unit of human recombinant erythropoietin (Amgen Biologicals), and 5 ng of recombinant canine stem cell factor (Amgen). Plates were incubated at 37°C in a high-humidity environment containing 5% CO2/95% air. After 10 days, erythroid bursts and granulocyte–macrophage (GM) colonies were individually harvested with micropipets under direct vision and freeze/thaw lysates of the colonies were placed on thin-layer polyacrylamide gels, subjected to isoelectric focusing (4 W per gel for 2.5 hr), and stained to detect G6PD activity (10). In all determinations in all animals, the percentage of erythroid burst-forming units with d G6PD was similar to the percentage of GM colony-forming units with d G6PD. Therefore, total numbers of progenitors (erythroid burst-forming units plus GM colony-forming units) with d G6PD were recorded. If multiple data points were available from an animal during the time frame listed (e.g., 2–3 months), only the earliest value was included in the analyses.
G6PD Analysis of Blood Cells and Marrow Fibroblasts.
Red cells (>99% purity by morphological assessment) and granulocytes (>96% purity) were isolated as described (7). T cells (>95% purity by flow cytometry analysis with monoclonal antibodies to CD5 (from Peter Moore, University of California, Davis, CA) were prepared by isolating peripheral blood mononuclear cells and incubating cells at 106 cells per ml in RPMI 1640 medium (GIBCO/BRL), 10% fetal calf serum, 1% PSF, 2 mM glutamine (GIBCO/BRL), 10−4 M 2-mercaptoethanol, and Con A (5 μg/ml; Boehringer Mannheim) for 3 days. The cells were then expanded for 5 days in the same medium, without Con A but containing recombinant human interleukin 2 (100 units/ml; R & D Systems). Marrow fibroblasts were prepared by culturing nucleated marrow cells in RPMI 1640 medium containing 10% fetal calf serum, 1% PSF, 2 mM glutamine, and recombinant human fibroblast growth factor (4 ng/ml; Collaborative Biomedical Products, Bedford, MA). Confluent adherent cells were passaged several times to eradicate any residual macrophages. For isoelectric focusing, 2 μl of red cells were mixed in 100 μl of 1.2% methylcellulose. Granulocytes (4.0 × 105), T cells (4.8 × 106), or fibroblasts (4.8 × 106) were suspended in 100 μl of IMDM and then mixed with an equal volume of 2.4% methylcellulose. Next 5 μl of freeze/thaw lysates of the cell preparations were placed on the polyacrylamide gel. After enzymatic staining, the percentage of d versus G G6PD activities was estimated by the comparison of band intensities with those on a concurrently processed gel containing mixtures of the specific G and d blood cell type (or fibroblasts) at known ratios (7). Samples from the same cat obtained at different times were placed on a single polyacrylamide gel, so that G6PD band intensities could be compared directly.
Transplantation Protocol.
Autologous marrow transplantation was performed as described (10, 11). After marrow harvest, each cat received 920 cGy of total-body irradiation (7 cGy/min from opposing cobalt-60 sources). Marrow cells (2 × 107 nucleated cells per kg) were infused intravenously within 4 hr of collection and were not manipulated in vitro. Infection prophylaxis included neomycin (5 mg/kg) and polymyxin (5,000 units/kg) administered orally every 8 hr beginning 5 days before transplantation and continuing until hematologic recovery. After transplantation, the animals also received broad spectrum antibiotics subcutaneously (ampicillin at 6.6 mg/kg every 8 hr and gentomycin at 4 mg/kg every 12 hr for 1 day and then once every day). Transfusions of irradiated blood were given as needed for platelet support. Recovery was monitored by complete blood counts, white blood cell differentials, and platelet counts.
RESULTS
Skewing Toward the Geoffroy X Chromosome Phenotype Develops with Aging.
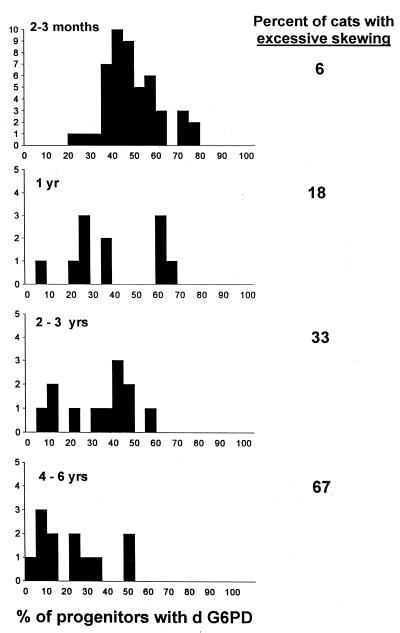
Fig. 1 shows the G6PD phenotype of erythroid burst-forming units and GM colony-forming units from 48 animals that were 2–3 months old. The percent of progenitors with d G6PD was 49 ± 13% (mean ± SD) and the median was 47%. Similar binomial distributions were obtained in analyses of red cells, granulocytes, and lymphocytes (data not shown), confirming that feline X chromosome inactivation was a random event. In three animals (6%), there was excessive skewing (<25% or >75% of progenitors had d G6PD). Twenty-two, 76, and 77% of progenitors contained d G6PD in these cats.
Figure 1.
Distribution of G6PD phenotype among progenitor cells from female Safari cats at various ages. The data at 2–3 months represent studies in 48 female Safari cats. Because most animals were entered into studies of retroviral physiology or hematopoiesis after baseline evaluations, the numbers of normal (unmanipulated) animals in the subsequent studies are smaller. Excessive skewing is defined as a distribution of G6PD phenotype in which <25% of progenitors contained d G6PD or >75% of progenitors contained d G6PD. The percent of animals with excessive skewing increased with age. This skewing was toward the G G6PD phenotype (resulting in a smaller percentage of progenitors with d G6PD). A significant change from a binomial distribution is seen in the 2- to 3-year and 4- to 6-year data when assessed by Q–Q plots (12) (analysis not shown). The X chromosome inactivation pattern observed at 2–3 months is similar to that reported in human studies (13, 14). By using the approach of these studies, the data can be further analyzed. If X chromosome inactivation took place after k divisions, 2k cells would be present at that time. Given binomial variability, the distribution would have a standard deviation of 2−(1 + k/2) × 100%, because the sample size is 2k and the probability of d G6PD is 1/2. When we equate this description of a binomial distribution with the observed data and solve for k, we estimate that k equals 4. Thus, approximately 16 cells were present at the time of X chromosome inactivation, or alternatively 16 cells gave rise to the hematopoietic system during development, similar to calculations in the referenced human studies.
In cats that were ages 4–6 months and 6–8 months, percents of progenitors with d G6PD were 49 ± 13% (mean ± SD; n = 24) and 52 ± 14% (n = 16). There was no excessive skewing. However, at 1 year (Fig. 1), this value was 39 ± 19%. Two of 11 animals (18%) had excessive skewing toward G G6PD. The percent of animals with excessive skewing increased to 33% in 2- to 3-year-old cats and 67% in 4- to 6-year-old cats (Fig. 1).
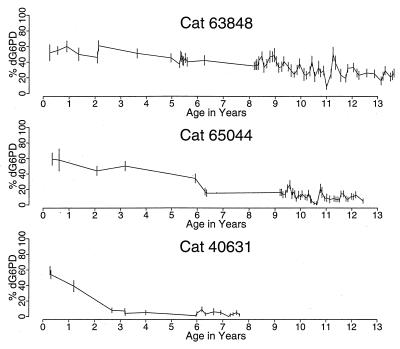
Fig. 2 shows longitudinal studies in three representative animals. The rate of change in the G6PD phenotype of progenitors varied extensively. There was an equivalent skew in the G6PD phenotype of granulocytes and red cells in each animal but not of T lymphocytes (Fig. 2).
Figure 2.
Longitudinal studies of progenitor cells from three representative female Safari cats. In cat 63848, a slow change toward a dominance of progenitor cells with a G G6PD phenotype was seen. Significant deviation from the mean value was first observed at age 8.4 years, when the data are analyzed by sequential χ2 analyses. In cat 65044, a significant change in the percent of progenitors with d G6PD was first noted at age 6.3 years, and a change in the phenotype of cells from cat 40631 was evident by year 2.7. Although a similar drift occurred among red cells and granulocytes (data not shown), the G6PD phenotype of T cells remained unchanged and specifically was 60% (year 13) in cat 63848, 60% (year 12) in cat 65044, and 60% (year 7) in cat 40631.
Sequential data from birth to >3 years of age were available in 11 animals. In 7, there was a significant change in the phenotype of progenitors, granulocytes, and red cells during the time of observation (when assessed with sequential χ2 analyses). Most importantly, in each animal, the change was toward a single parental (G) G6PD phenotype. Four additional animals were not studied until ages 4–6 years. Their progenitors, granulocytes, and red cells had excessive skewing toward the G G6PD phenotype at the time of the initial observations, and in all analyses over the next year, while T cells had a balanced distribution of G6PD phenotype. Thus, excessive skewing does not result from HSC depletion or neoplasia (where skew toward d and G phenotypes should have equal incidence). Rather, the data imply that cells that express G X chromosome genes, and thus G G6PD, have a selective growth advantage relative to cells containing d G6PD. As all hematopoietic cells in a female Safari cat are genetically equivalent and differ only in which parental X chromosome is active, an X chromosome gene (or genes) must impact hematopoietic cell kinetics in vivo. The genetic effect is direct (intrinsic to hematopoietic cells) and not due to changes in the marrow microenvironment.
Studies of T Cell Reconstitution Demonstrate Hemizygous Selection Within the HSC Compartment.
There are two possibilities why the distribution of the G6PD phenotype among T cells did not change from baseline determinations with aging. (i) Hemizygous selection may exclusively impact differentiating myeloid cells and not stem cells. (ii) Because T cells are long-lived cells that turn over slowly during adult life, the T cells studied at a time of observation could represent those made many years before the dominance of the stem cell compartment by cells containing G G6PD. To distinguish these possibilities, autologous marrow transplantations were performed. The results of these studies are shown in Table 1. At 3 years of age, 50–60% of progenitors, granulocytes, T cells, and marrow fibroblasts (a nonhematopoietic control cell) from cat 65044 contained d G6PD. These values were stable until year 6 (Fig. 2). By 12 years of age, 7% of progenitors and 15% of granulocytes contained d G6PD. However, baseline values persisted among T cells and marrow fibroblasts. After marrow harvest, this cat received 920 cGy of total-body irradiation to eliminate its hematopoietic cells (including T cells) and then 2 × 107 nucleated marrow cells per kg were infused. Three months later, the percent of progenitors, granulocytes, and T cells with d G6PD were 7, 10, and 10%, whereas 50% of marrow fibroblasts had a d G6PD phenotype. Fifteen months after transplantation, the percent of progenitors, granulocytes, and T cells with d G6PD were similar and were 3, 10, and 5%, respectively. Equivalent data were obtained in a second animal (41008, Table 1) that demonstrate that cells able to reconstitute and maintain both myelopoiesis and lymphopoiesis (i.e., HSCs) contained predominately G G6PD. Thus hemizygous selection occurs within the HSC compartment. Despite the low numbers of nucleated marrow cells infused (2 × 107 cells per kg vs. 2 × 108 cells per kg generally used in human and canine transplantation), each animal quickly reconstituted hematopoiesis, granulocyte engraftment (>200 cells per μl) was seen at day 18, and platelet engraftment (>50,000 cells per μl) was seen at day 23, which further supports the concept that HSCs are not depleted with aging.
Table 1.
Autologous marrow transplantation (AMT) studies in elderly female Safari cats
| Cat | Age, years | Time after AMT, months | % of cells with d G6PD
|
|||
|---|---|---|---|---|---|---|
| Progenitors | Granulocytes | T cells | Marrow fibroblasts | |||
| 65044 | 3 | 60 | 50 | 50 | 60 | |
| 12 | 7 | 15 | 60 | 50 | ||
| 3 | 7 | 10 | 10 | 50 | ||
| 15 | 3 | 10 | 5 | 50 | ||
| 41008 | 5 | 7 | 7 | 45 | 55 | |
| 3 | 10 | 10 | 10 | 50 | ||
| 20 | 13 | 10 | 10 | 50 | ||
Longitudinal data from cat 65044 are presented in Fig. 2. Cat 41008, used in breeding studies of F2 animals, did not have the G6PD phenotype of her progenitors measured until age 5 years. Values remained unchanged in four determinations over 9 months before transplantation and the percent of progenitors with d G6PD ranged from 2 to 7%, granulocytes from 5 to 10%, and T cells from 40 to 50%. After transplantation, the distributin of G6PD phenotype in T cells became equivalent to that among progenitors and granulocytes, and a balanced distribution of G6PD phenotype persisted among marrow fibroblasts, a nonhematopoietic (control) cell population.
DISCUSSION
In this study we analyzed the G6PD phenotype of blood and marrow progenitor cells from normal female Safari (F1 d × G) cats over time and observed skewing in the percent of cells that contained d G6PD. In population studies, a binomial distribution was no longer present by age 2–3 years (Fig. 1), and a significant change from baseline levels was observed in data from 7 of the 11 individual cats studied longitudinally for more than 3 years (Fig. 2). In each circumstance, the change resulted in the dominance of cells with a G G6PD phenotype. These data resemble observations in human studies, but because cells in all female Safari cats have one d-cat- and one G-cat-derived X chromosome, we were able to demonstrate that the acquired skewing was due to the hemizygous expansion of HSCs in which the G X chromosome was active. We suspect that a comparable mechanism is operative in human females.
There are many examples of hemizygous selection of differentiating blood cells. In women heterozygous for both the X chromosome-linked disorder Wiscott–Aldrich Syndrome (WAS) and G6PD, all platelets and T cells express a single parental G6PD phenotype (A or B), whereas both isoenzymes A and B are present among red cells and granulocytes, implying the defective (WAS) platelets and T cells fail to mature in heterozygous (clinically normal) women (15, 16). Similarly, there is a selective growth advantage for normal B cells (but not T cells or granulocytes) in mothers of boys with agammaglobulinemia (17, 18) and normal B and T cells in carriers of the X chromosome-linked severe combined immunodeficiency syndrome (19). Studies of mothers of boys with Lesch–Nyhan disorder [hypoxantine guanine phosphoribosyltransferase (HGPRT) deficiency] report a skewed distribution of G6PD phenotype among red cells, granulocytes, and T cells (but not fibroblasts) (20). To explain these findings, the authors suggested that HGPRT-positive cells preferentially formed the hematopoietic system during development, but it is also possible that the hematopoietic system was formed by cells of both phenotypes and that HGPRT-positive HSCs had a preferential growth advantage. A study by Luzzatto et al. (21) of a Nigerian family heterozygous for the Ilesha variant of G6PD and with no coexistent X chromosome-linked clinical disorder raised the question of hemizygous expansion of HSCs. As only red cells and granulocytes were studied, however, their observations are also compatible with a preferential (nonrandom) inactivation of the X chromosome (21, 22). Because female Safari cats are balanced heterozygotes early in life and a skewed G6PD phenotype is acquired with aging, nonrandom X chromosome inactivation and a nonrandom origin for the hematopoietic system during embryonic development are not relevant considerations.
The identity of the gene or genes responsible for the selective growth advantage of HSCs with G G6PD is unknown. Because G cats (South American origin) and domestic cats (Eurasian origin) have evolved independently for 12 million years (23), it is likely that F1 Safari cats will be heterozygous at many genetic loci. Candidate genes include the cell cycle relevant determinants of Xq28 (24) and the Inhibitor-of-Apoptosis gene of Xq25 (25, 26). However, in the studies of Luzzatto et al. (21), recombination between the loci for G6PD (which also maps to Xq28) and for hemizygous selection occurred in three of five individuals, suggesting that these genes cannot be too closely linked. Another attractive candidate is the PIG-A gene (Xp22.1), which is responsible for paroxysmal nocturnal hemoglobinuria (PNH; for review, see ref. 27). There is preferential expansion of the PNH clone in patients with myelodysplasia or aplastic anemia. Recent studies in mice suggest that pig-a −/− cells do not have an intrinsic growth advantage (28) and imply that PNH HSCs may preferentially survive only in circumstances where there is an altered marrow environment. Other data suggest that the protein product of the PIG-A gene may decrease HSC apoptosis (29). Of interest, a skew in progenitor G6PD phenotype was seen after busulfan administration in three of six cats previously studied, and in each instance, the change was toward the G G6PD phenotype (8). A skew toward G G6PD developed in a fourth animal after the repeated administration of busulfan (9). Because busulfan depletes and damages hematopoietic stem cells to induce aplastic anemia, a similar mechanism could be responsible for the relative growth advantage of cells containing G G6PD.
Van Zant and colleagues described a gene (called Stk), for stem cell kinetics that influences HSC behavior in allophenic (DBA/2 × C57BL/6) mice (30–32) and have recently localized the gene to chromosome 18 (33). Several features of this gene are similar to those that we observed in feline studies (e.g., with aging, C57BL/6 type HSCs dominated, although HSCs of both phenotypes were present in more equal proportions in young animals) (31), although others differ (32). It is possible that many genes influence HSC kinetics, a concept consistent with stochastic differentiation.
With computer simulation, as described (2), it appears that a 4–5% increased probability of HSC replication, a 6–8% decreased probability of HSC differentiation, or a 20–25% decreased probability of HSC apoptosis could lead to the dominance of the stem cell compartment by cells with the G G6PD over a 1- to 12-year period (34). These competitive interactions leading to the slow domination by one of two populations that share a geographic niche are conceptually similar to those observed by population geneticists (35, 36). If the difference in behavior of cells with G versus d G6PD is subtle, the identification of the relevant gene(s) will be extremely difficult. Studying the genetic factors that may influence HSC behavior in large animals or humans could be very complex and require repeated damage to the HSC compartment (i.e., with busulfan exposure) to allow phenotypic detection. Linkage studies in a large pedigree of human females may provide an alternative approach.
Thus, our data prove that a gene on the X chromosome influences the in vivo behavior of normal HSCs and that the genetic composition of this cell, and not just microenvironmental factors, influences its replication, differentiation, and/or apoptosis decisions. We confirm that the dominance of a single parental X chromosome phenotype among blood cells is a common phenomenon with aging and demonstrate that this results from hemizygous selection. Excessive skewing is not due to HSC depletion or to HSC neoplasia. A reanalysis of previous studies (8, 9) suggests that excessive skewing may also develop after busulfan administration. Therefore, caution is required when using X chromosome inactivation patterns to diagnose myelodysplasia in elderly individuals or in patients who have received repeated chemotherapy (6, 37, 38). Because T cells are long-lived, yet sensitive to radiation and chemotherapy, their use as an internal control for clonality studies in individual patients is also problematic.
Acknowledgments
We thank Stan Gartler and Thalia Papayannopoulou for insightful reviews of these data, Michael Linenberger for advice on the culture of feline T cells and marrow fibroblasts, and Allan Dimaunahan for preparation of the manuscript. These studies were supported by Grant R01 HL46598 from the National Institutes of Health. J.L.A. is the recipient of a Faculty Research Award from the American Cancer Society.
ABBREVIATIONS
- HSC
hematopoietic stem cell
- G6PD
glucose-6-phosphate dehydrogenase
- G
Geoffroy
- d
domestic
References
- 1.Orlic D, Bodine D M. Blood. 1994;84:3991–3994. [PubMed] [Google Scholar]
- 2.Abkowitz J L, Catlin S N, Guttorp P. Nat Med. 1996;2:190–197. doi: 10.1038/nm0296-190. [DOI] [PubMed] [Google Scholar]
- 3.Wang J C Y, Doedens M, Dick J E. Blood. 1997;89:3919–3924. [PubMed] [Google Scholar]
- 4.Busque L, Mio R, Mattioli J, Brais E, Blais N, Lalonde Y, Maragh M, Gilliland D G. Blood. 1996;88:59–65. [PubMed] [Google Scholar]
- 5.Gale R E, Fielding A K, Harrison C N, Linch D C. Br J Haematol. 1997;98:512–519. doi: 10.1046/j.1365-2141.1997.2573078.x. [DOI] [PubMed] [Google Scholar]
- 6.Champion K M, Gilbert J G R, Asimakopoulos F A, Hinshelwood S, Green A R. Br J Haematol. 1997;97:920–926. doi: 10.1046/j.1365-2141.1997.1933010.x. [DOI] [PubMed] [Google Scholar]
- 7.Abkowitz J L, Ott R L, Nakamura J M, Steinmann L, Fialkow P J, Adamson J W. J Clin Invest. 1985;75:133–140. doi: 10.1172/JCI111665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abkowitz J L, Ott R M, Holly R D, Adamson J W. Blood. 1988;71:1687–1692. [PubMed] [Google Scholar]
- 9.Abkowitz J L, Linenberger M L, Persik M, Newton M A, Guttorp P. Blood. 1993;82:2096–2103. [PubMed] [Google Scholar]
- 10.Abkowitz J L, Linenberger M L, Newton M F, Ott R L, Guttorp P. Proc Natl Acad Sci USA. 1990;87:9062–9066. doi: 10.1073/pnas.87.22.9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abkowitz J L, Persik M T, Shelton G H, Ott R L, Kiklevich J V, Catlin S N, Guttorp P. Proc Natl Acad Sci USA. 1995;92:2031–2035. doi: 10.1073/pnas.92.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers J M, Cleveland W S, Kleiner B, Tukey P A. Graphical Methods for Data Analysis. Belmont, CA: Wadsworth; 1983. pp. 9–16. [Google Scholar]
- 13.Fialkow J. Ann Hum Genet. 1973;37:39–48. doi: 10.1111/j.1469-1809.1973.tb01813.x. [DOI] [PubMed] [Google Scholar]
- 14.Buescher E S, Alling D W, Gallin J I. J Clin Invest. 1985;76:1581–1584. doi: 10.1172/JCI112140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gealy W J, Dwyer J M, Harley J B. Lancet. 1980;1:63–65. doi: 10.1016/s0140-6736(80)90492-4. [DOI] [PubMed] [Google Scholar]
- 16.Prchal J T, Carroll A J, Prchal J F, Crist W M, Skalka H W, Gealy W J, Harley J, Malluh A. Blood. 1980;56:1048–1054. [PubMed] [Google Scholar]
- 17.Conley M E, Brown P, Pickard A R, Buckley R H, Miller D S, Raskind W H, Singer J W, Fialkow P J. N Engl J Med. 1986;315:564–567. doi: 10.1056/NEJM198608283150907. [DOI] [PubMed] [Google Scholar]
- 18.Fearon E R, Winkelstein J A, Civin C I, Pardoll D M, Vogelstein B. N Engl J Med. 1987;316:427–431. doi: 10.1056/NEJM198702193160802. [DOI] [PubMed] [Google Scholar]
- 19.Conley M E, Lavoie A, Briggs C, Brown P, Guerra C, Puck J M. Proc Natl Acad Sci USA. 1988;85:3090–3094. doi: 10.1073/pnas.85.9.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nyhan W L, Bakay B, Connor J D, Marks J F, Keeble D K. Proc Natl Acad Sci USA. 1970;65:214–218. doi: 10.1073/pnas.65.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luzzatto L, Usanga E A, Bienzle U, Esan G F, Fasuan F A. Science. 1979;205:1418–1420. doi: 10.1126/science.472761. [DOI] [PubMed] [Google Scholar]
- 22.Zakian S M, Kulbakina N A, Meyer M N, Semenova L A, Bochkarev M N, Radjabli S I, Serov O L. Genet Res. 1987;50:23–27. doi: 10.1017/s0016672300023296. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien S J. Trends Genet. 1986;2:137–142. [Google Scholar]
- 24.Tribioli C, Droetto S, Bione S, Cesareni G, Torrisi M R, Lotti L V, Lanfrancone L, Toniolo D, Pelicci P. Proc Natl Acad Sci USA. 1996;93:695–699. doi: 10.1073/pnas.93.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajcan-Separovic E, Liston P, Lefebvre C, Korneluk R G. Genomics. 1996;37:404–406. doi: 10.1006/geno.1996.0579. [DOI] [PubMed] [Google Scholar]
- 26.Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. Nature (London) 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 27.Luzzatto L, Bessler M, Rotoli B. Cell. 1997;88:1–4. doi: 10.1016/s0092-8674(00)81850-4. [DOI] [PubMed] [Google Scholar]
- 28.Rosti V, Tremml G, Soares V, Pandolfi P P, Luzzatto L, Bessler M. J Clin Invest. 1997;100:1028–1036. doi: 10.1172/JCI119613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brodsky R A, Vala M S, Barber J P, Medof M E, Jones R J. Proc Natl Acad Sci USA. 1997;94:8756–8760. doi: 10.1073/pnas.94.16.8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Zant G, Eldridge P W, Behringer R R, Dewey M J. Cell. 1983;35:639–645. doi: 10.1016/0092-8674(83)90096-x. [DOI] [PubMed] [Google Scholar]
- 31.Van Zant G, Holland B P, Eldridge P W, Chen J-J. J Exp Med. 1990;171:1547–1565. doi: 10.1084/jem.171.5.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Zant G, Scott-Micus K, Thompson B P, Fleischman R A, Perkins S. Exp Hematol. 1992;20:470–475. [PubMed] [Google Scholar]
- 33.de Haan G, Van Zant G. J Exp Med. 1997;186:529–536. doi: 10.1084/jem.186.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catlin, S. N., Guttorp, P. & Abkowitz, J. L. (1997) Blood 90 Suppl. 1, 143b. [PubMed]
- 35.Gause G F. The Struggle for Existence. Baltimore: Williams & Wilkins; 1934. [Google Scholar]
- 36.Debach P. Annu Rev Entomol. 1966;11:183–212. [Google Scholar]
- 37.Fialkow P J, Singer J W, Raskind W H, Adamson J W, Jacobson R J, Bernstein I D, Dow L W, Najfeld V, Veith R. N Engl J Med. 1987;317:468–473. doi: 10.1056/NEJM198708203170802. [DOI] [PubMed] [Google Scholar]
- 38.Gale R E, Bunch C, Moir D J, Patterson K G, Goldstone A H, Linch D C. Br J Haematol. 1996;93:53–58. doi: 10.1046/j.1365-2141.1996.4751014.x. [DOI] [PubMed] [Google Scholar]