Abstract
The LIM-finger protein Lmo2, which is activated in T cell leukemias by chromosomal translocations, is required for yolk sac erythropoiesis. Because Lmo2 null mutant mice die at embryonic day 9–10, it prevents an assessment of a role in other stages of hematopoiesis. We have now studied the hematopoietic contribution of homozygous mutant Lmo2 −/− mouse embryonic stem cells and found that Lmo2 −/− cells do not contribute to any hematopoietic lineage in adult chimeric mice, but reintroduction of an Lmo2-expression vector rescues the ability of Lmo2 null embryonic stem cells to contribute to all lineages tested. This disruption of hematopoiesis probably occurs because interaction of Lmo2 protein with factors such as Tal1/Scl is precluded. Thus, Lmo2 is necessary for early stages of hematopoiesis, and the Lmo2 master gene encodes a protein that has a central and crucial role in the hematopoietic development.
The mechanism of development and cell differentiation is one of the main themes of biology. The process of hematopoiesis is one of the most well studied because hematopoietic stem cells in the bone marrow (BM) have a capacity of self-renewal and differentiation into all kinds of blood cells. The process of commitment is controlled by both transcriptional regulation (nuclear factors) and humoral factors. Several important transcription factors involved in hematopoiesis have been identified from the analysis of chromosome translocation associated with human tumors (1). One of these genes, LMO2 (previously called RBTN2 or TTG2), is associated with the translocations t(11;14)(p13;q11) or t(7;11)(q35;p13), in childhood T cell acute leukemia (2, 3). LMO2 encodes a LIM domain protein (in which two zinc-binding LIM domains occur) and is activated after chromosomal translocation by association with either the T cell receptor α/δ gene at chromosome 14, band q11, or the β gene at chromosome 7, band q35. As a result of the chromosomal translocations, LMO2 protein is made in the specific T cells that have the translocation, and the consequence of this aberrant expression is thought to be an alteration in the differentiation of the T cell, ultimately resulting in overt leukemia (4, 5).
The evidence of physiological role of Lmo2 protein in hematopoiesis came from gene target experiments in which null mutations were introduced into the Lmo2 gene (6). Null mutant embryonic mice die in utero due to a failure of yolk sac erythropoiesis, showing that Lmo2 is necessary for yolk sac erythropoiesis to take place. Although there is no direct evidence that Lmo2 itself has a DNA binding capacity, in normal expression sites, such as erythroid cells, Lmo2 protein directly interacts with a basic-loop–helix protein Tal1/Scl (7–9) and the GATA DNA-binding protein Gata-1 (9, 10). Furthermore, in vitro binding site selection (CASTing assays; cyclic amplification and selection of targets) led to the identification of a complex involving Lmo2 and also including Tal1/Scl, E47, Gata-1, and Ldb1 (10). This erythroid complex binds to a bipartite DNA motif consisting of E box and GATA consensus sequences in which the Tal1/Scl-E47 component binds to the E box and Gata-1 binds to the GATA site. These data strongly support the idea that Lmo2 acts as a bridging molecule bringing together the different DNA binding factors in this erythroid complex (9, 10). Because Lmo2 can bind also to Gata-2 protein (9), it is possible that a complex of Lmo2, Gata-2, and other proteins, analogous to that seen in erythroid cells, might occur at earlier times of hematopoiesis when Gata-1 is not expressed.
The possibility of a broader role of Lmo2 protein in adult hematopoiesis has been investigated by using embryonic stem (ES) cells in which both Lmo2 alleles have been inactivated. Because the early lethality of the Lmo2 null mutant mice occurs at about embryonic day 9.75, before the onset of definitive (adult type) hematopoiesis, the mutant mice could not be readily used to investigate this issue. We have exploited the use of Lmo2 null mutant (−/−) ES cells, principally by using chimeric mice, to ascertain a role in definitive hematopoiesis for this transcriptional regulator. The results of these studies show that definitive hematopoiesis does not occur in the absence of Lmo2. Thus the Lmo2 gene is necessary for the earliest point of adult hematopoietic development, perhaps even before the development of the BM stem cell.
MATERIALS AND METHODS
ES Cell Growth and Differentiation.
ES cells with null mutation of both Lmo2 alleles have been described (6) and two karyotypically normal clones (clones 53 and 71) were selected for study. For in vitro culture, ES cells from a single well of a 24-well plate were trypsinized and transferred to a fresh well for 60 min (nongelatinized and no feeder layer) in a volume of 1 ml, to allow the removal of fibroblasts by adherence. Approximately 1 × 103 ES cells were plated in a final volume of 1 ml of 0.9% α methylcellulose. The following growth factors were included: erythropoietin at 2 units/ml, interleukin 1α at 1 ng/ml, murine interleukin 3 at 100 units/ml, and murine stem cell factor at 25 ng/ml. Monothioglycerol was added to a final concentration of 150 mM. The plates were placed in a fully humidified 5% CO2/95% air incubator at 37°C for 10 days. Total RNA was isolated from embryoid bodies at day 0, 4, and 10 by use of the guanidine/phenol method and oligo(dT)-primed cDNA synthesized from about 20% of the total RNA. A fraction (5%) of the synthesized cDNA was used for PCR with internal control (actin). Each set of primers is designed to span at least one intron (11). Primers for actin were TAGGAATCCATGGCCACTGCCGCATCCTCTTCC and CACGATGGAGGGGCCGGACTCATC. Products were visualized after separation on 2% agarose by ethidium bromide staining.
Transfection of Lmo2 Expression Vector.
The Lmo2 expression segment from pEF-BOS-Lmo2 (10) was subcloned into a vector harboring a puromycin N-acetyltransferase gene (pBabePuro) (25). Linealized vector (pBabePuro EF-BOS-Lmo2) was transfected into Lmo2 −/− clone 71 by electroporation. Cells were selected with puromycin (2 μg/ml). Each independent clone was tested for Lmo2 expression by reverse transcription-coupled PCR and two (BR3 and BR12) were selected for study.
Production and Analysis of Chimeric Mice.
ES cells were microinjected into C57BL/6 blastocysts and transferred to CBA/C57BL/6 recipients. The resulting pups were assessed initially for chimerism based on the coat coloration. The following analysis was done between 2 months and 3 months after birth.
Glucose phosphate isomerase (GPI) isozyme analysis was performed on blood and tissue samples as described (12). Lymphocytes were purified from thymus and spleen by using Histopaque (Sigma) according to the manufacturers’ instruction. Peritoneal macrophages were taken by peritoneal wash and cultured for 24 h before typing. BM cell suspensions were prepare by flushing femurs with PBS and washing the eluted cells once in PBS before use.
Blood samples for globin analysis were obtained as heparinized blood after cardiac puncture. Packed red cells were lysed and modified by cystamine before electrophoresis on cellulose acetate plates as described (13).
Flow cytometry analysis was performed on pure lymphocytes; single cell suspensions were made from spleen or thymus and pure white cells prepared by centrifugation on Isopaque. Surface antigens were labeled with monoclonal antibody binding to the 129 allotypic lymphocyte marker Ly9.1 labeled with fluorescein isothiocyanate fluorchrome (PharMingen) and either phycoerythrin-conjugated antibodies binding B220 for splenic lymphocytes or phycoerythrin-conjugated CD4 for thymic lymphocytes. Analyses were conducted on a FACscan, with subtraction for fluorescence due to background binding determined with isotype controls (PharMingen).
PCR Analysis of Mouse Tissues.
For microsatellite DNA analysis, PCR amplifications were performed with genomic DNA isolated from tissues of chimeric mice and the microsatellite primer, D10Mit180 (Research Genetics, Huntsville, AL) as described (14–16). PCR products (from 30 cycles) were analyzed on 4% Nusieve agarose.
For Y chromosome analysis, PCR amplification was performed with genomic DNA isolated from tissues of female chimeric mice and specific primers for a Y chromosome gene, Uty (17). Reaction conditions were 30 cycles of 94°C for 30 s, 55°C for 40 s, and 72°C for 60 s. PCR products were analyzed on 4% Nusieve agarose.
RESULTS
Lmo2 −/− Mutant ES Cells Cannot Undergo Hematopoiesis.
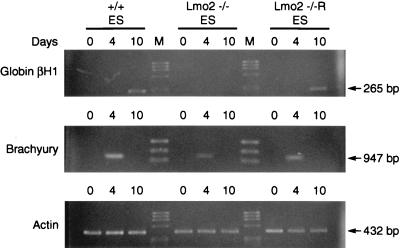
Previous studies of Lmo2 ES cells in which both alleles were rendered inactive by gene targeting (Lmo2 −/− ES cells) showed their inability to undergo erythropoiesis in vitro (6). In vitro differentiation of Lmo2 −/− ES cells was compared with that of normal ES cells and of Lmo2 −/− cells in which an Lmo2 expression vector had been introduced. Gene activation in these cultures was assayed by extracting mRNA at various time points (at 4 and 10 days after induction) and performing reverse transcription-coupled PCR. Although brachyury mRNA, a mesodermal marker (11), was observed in all cell lines at 4 days after induction, embryonic globin βH1 induction (a yolk sac hematopoietic marker) was only found at 10 days in +/+ ES cells and not in Lmo2 −/− cells. However, when Lmo2 expression was restored via transfection into the Lmo2 −/− cells (Fig. 1; −/−R), globin gene induction is also restored. These results are consistent with an essential role of Lmo2 protein in hematopoietic differentiation and demonstrate that the Lmo2 −/− cells can be reconstituted in the erythroid lineage by reexpression of Lmo2 (see also below). These Lmo2 null ES cells were, therefore, considered to have a cell autonomous defect in erythropoiesis due only to loss of the Lmo2 gene because reexpression of Lmo2 in these cells rescued the developmental defect. These cells were therefore considered appropriate for in vivo studies.
Figure 1.
Requirement for Lmo2 expression in hematopoiesis in vitro. ES cells were differentiated in the presence of interleukin 1α, stem cell factor, interleukin 3, and erythropoietin, and PCR amplifications were performed with cDNA synthesized from RNA isolated from embryoid bodies at day 0 and at 4 and 10 days after induction, with gene-specific primers for the indicated transcripts. Actin was used as a quality control for the RNA prepared from ES cells. ES cells examined were wild-type (+/+), Lmo2 −/−, and Lmo2 −/− into which an Lmo2 expression vector had been transfected (Lmo2 −/−R). Sizes of the transcripts are indicated.
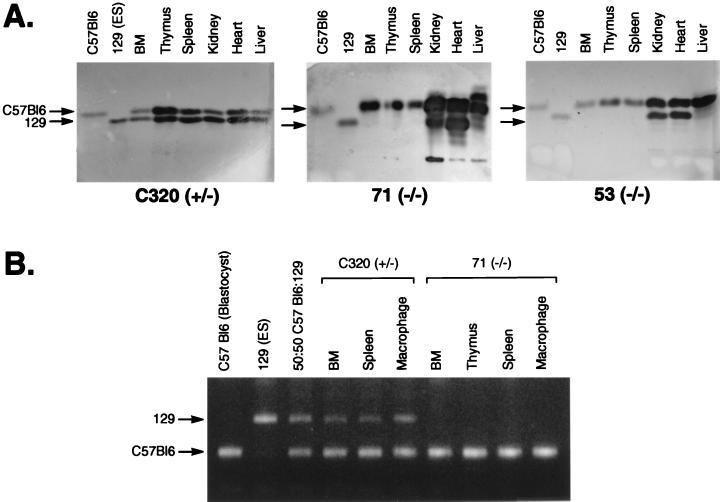
Potential in vivo effects of Lmo2 null mutation on definitive hematopoiesis was studied in chimeric mice (about 3 months old) generated by injecting Lmo2 −/− ES cells into C57BL/6 blastocysts. The ability of Lmo2-deficient ES cells to contribute to development in vivo was studied in chimeric mice made with two independent clones of Lmo2 −/− cells (clones 53 and 71) and compared with an Lmo2 +/− clone (C320). ES cell contribution was initially tested by GPI isotype difference (Fig. 2A) (12). There was substantial ES-derived component in hematopoietic organs in all Lmo2 +/− derived chimeras as well as all peripheral tissues tested (Fig. 2A, C320). Analysis of tissues from chimeras made with two Lmo2 −/− ES clones revealed no detectable contribution in any hematopoietic organ (Fig. 2A, 71 and 53 −/−), whereas the other tissues examined showed substantial contribution. This indicates that the Lmo2 gene is needed for hematopoiesis to occur in adult mice.
Figure 2.
Lmo2 −/− ES cells cannot differentiate into hematopoietic tissues in vivo. (A) Tissue contribution of Lmo2 +/− and −/− ES cells in chimeric mice. Tissue extracts from chimeric mice were prepared and the relative proportion of GPI isozymes 1B (C57BL/6 blastocysts) or 1A (129 ES cells) was assayed. Chimeric mice were generated by injecting Lmo2 +/− ES cells (C320) or two Lmo2 −/− ES cell clones, 71(−/−) and 53(−/−). (B) PCR amplifications were performed with the polymorphic microsatellite marker D10Mit180 (15) and DNA isolated from tissues of chimeric mice. PCR products differ between C57BL/6 (blastocysts) and 129 (ES cells), as seen in products from a 50:50 mixture of C57BL/6 and 129 DNA.
This defect in hematopoietic development was confirmed by using a microsatellite DNA PCR assay (14–16) to estimate ES cell contribution in the chimeras, based on sequence differences between C57BL/6 (blastocyst) and 129 (ES cells). In this assay, although the ES cell contribution was observed in BM, spleen, and macrophages of a chimeric mouse derived from Lmo2 +/− cells (Fig. 2B, C320), no signal was detectable from these tissues (as well as in thymus) of a representative high-level chimera made from the Lmo2 −/− clone 71 (Fig. 2B, 71 −/−). These data substantiate the observation that neither myelopoiesis or lymphopoiesis is taking place from the ES cells in the chimeras made with the Lmo2 −/− ES cells. This lack of lymphopoiesis from ES-cell derivatives in the chimeras was confirmed by assessing the presence of the Ly9.1 antigen on lymphocytes made from chimeric spleen and thymus (Ly9.1 is an allotype present on lymphocytes from 129 mouse cells and, therefore, also ES cells). Two-color flow cytometry analysis was performed on mononuclear cells isolated from spleen and thymus. Splenocytes were stained with antibodies binding to Ly9.1 and B220 and thymocytes with antibodies to Ly9.1 and CD4, revealing double fluorescent cells in the control 129 mouse spleen and thymus but not in high-level chimeras made with Lmo2 −/− ES cells nor in the C57BL/6 mouse lymphoid tissues (data not shown).
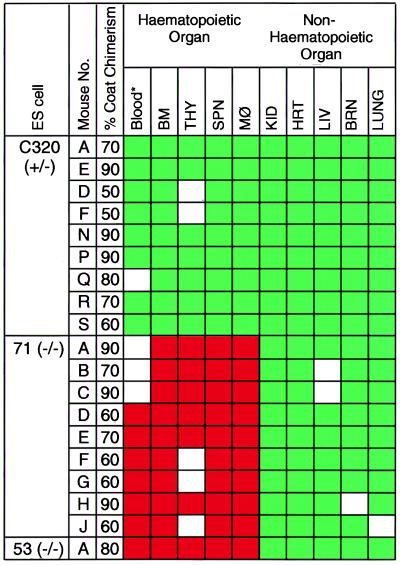
Because these experiments were carried out with individual chimeras, only those with greater than 50% coat color chimerism (total 19 chimeric mice of 28 chimeras generated) were assayed and included in the experimental group. Fig. 3 shows a compilation of data obtained with GPI and microsatellite analysis of nine chimeras obtained from Lmo2 +/− cells (C320), nine from Lmo2 −/− clone 71, and one from Lmo2 −/− clone 53. In every Lmo2 +/− chimera, all tissues analyzed (including BM, spleen, and thymus) had substantial ES cell contribution (Fig. 3), whereas only nonhematopoietic tissues of chimeras from either of the Lmo2 −/− clones showed ES cell contribution. It is noteworthy that chimeras made with Lmo2 +/− cells always had ES contribution in BM (Fig. 3), but none of the 10 mice made with Lmo2 −/− cells had any detectable contribution (either assessed by GPI or microsatellite PCR). Approximately 60–70% of cells in BM are neutrophils and myelocytes, therefore, indicating lack of Lmo2 −/− contribution to myelopoiesis, and no lymphocyte contribution was found in any of the Lmo2 −/− chimeras.
Figure 3.
Contribution of the Lmo2 +/− (C320) and −/− (clones 71 and 53) ES cells to tissues in chimeric mice. Individual mice are indicated and the approximate coat color chimerism is given. The mouse tissue was assayed with combinations of techniques. In each case, GPI analysis was first performed and these data were confirmed by the microsatellite PCR method (complete concordance was obtained). For C320 thymus sample E, no GPI data is available, the results being obtained from microsatellite and Y chromosome PCR only. Blood (∗) analysis was carried out by the modified globin analysis (13) and was only performed on C320 chimeras A, E, D, F, N, P, R, and S; clone 71 chimeras D, E, F, G, H, and J; and clone 53 chimera A. Boxes: green, ES contribution was detectable; red, no ES contribution was detectable; white, no information. Data from those with more than 50% coat color chimerism are shown. THY, thymic cells; SPN, splenic cells; MØ, peritoneal macrophage; KID, kidney; HRT, heart; LIV, liver; BRN, brain.
Rescue of Hematopoietic Development from Lmo2 −/− ES Cells.
An additional critical appraisal of the sensitivity of the assays employed to judge the contribution in vivo by the Lmo2 −/− ES cells was accomplished by repeating the in vivo chimerism experiments with Lmo2 −/− ES cells in which a Lmo2-expression vector had been introduced (Lmo2 −/−R cells). These experiments were also carried out to ascertain whether the inability of the Lmo2 −/− cells to contribute hematopoietic lineages might reflect unrecognized genetic damage, perhaps incurred during culture, rather than due to a defect of the Lmo2 mutation. Genetic damage seems unlikely, however, because two independent Lmo2 −/− clones exhibit the same behavior (i.e., Lmo2 −/− clones 53 and 71) in chimera analysis and the reexpression of Lmo2 in the −/− ES clones facilitates in vitro erythroid differentiation (Fig. 1). These points, however, were assessed by assaying hematopoiesis in chimeras made with Lmo2 −/− ES clone 71 carrying the Lmo2 vector (Lmo2 −/−R).
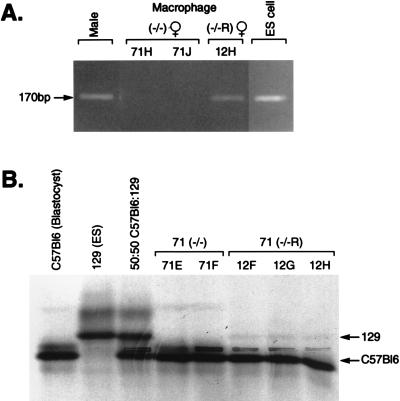
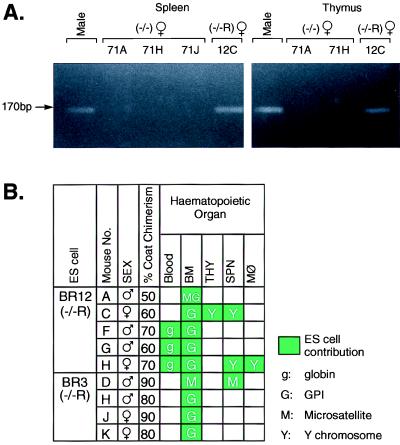
Because ES cells are male (18), the presence of the Y chromosome identifies the ES-derived cells in female chimeras. This permitted a Y chromosome-specific PCR-based assay for the ES cell derivatives in chimeras. When DNA was prepared from peritoneal macrophages isolated from two high-level female chimeras derived from the Lmo2 −/− clone 71, no Y chromosome-specific PCR product (17) was found because no hematopoiesis occurred from these Lmo2 null cells (Fig. 4A, mouse 71H about 90% chimeric and mouse 71J about 60% chimeric). However, PCR with DNA of a female chimera made from the Lmo2-expressing −/−R clone BR12, produced a Y chromosome-specific ES-cell-derived band, demonstrating the presence of ES cell derivatives in macrophages of the chimeras and thus that Lmo2 expression rescued the macrophage/monocyte developmental lineage in vivo.
Figure 4.
Hematopoietic rescue by reexpression of Lmo2 −/− ES cells. (A) PCR amplifications were performed with Y chromosome-specific primers (giving the Uty product) and DNA isolated from 24-h cultures of peritoneal macrophages from female chimeric mice produced with the Lmo2 −/− ES clone 71 (mice 71H and 71J) or the BR12 Lmo2-expressing −/− clone (−/−R; mouse 12H). The PCR products were separated on 4% Nusieve agarose. DNA quality for each PCR was tested simultaneously with the microsatellite primer pair D10Mit180 (data not shown). Electrophoresis of control PCRs using DNA samples from a male mouse and from ES cells are also shown. (B) Modified hemoglobin analysis of blood samples (13) of Lmo2 −/− clone 71 (71E and 71F) and clone BR12 (12F, 12G, and 12H) chimeric mice. Hemoglobins were separated on cellulose acetate thin-layer plates. Hbbs is specific for C57BL/6 (blastocysts) and Hbbd is specific for 129 (ES cells) as indicated by arrows. Note the relatively faint ES cell-specific bands seen in BR12 samples; this phenomenon was also observed in the Tal1/Scl rescue experiments (20) and is possibly due to the lack of proper regulation of gene expression (e.g., perhaps a promoter efficiency effect or a chromosome location effect on Lmo2 expression).
Similarly, globin synthesis was rescued in vivo by reexpressing Lmo2 in the Lmo2 −/− ES clones. Previous work (6) and in vitro differentiation data (Fig. 1) indicated that globinized blood cells could not form in the absence of Lmo2 protein. In high-level chimeras made with the Lmo2 −/− ES cells, no globin synthesis originating from these cells could be detected by cellulose acetate electrophoresis of blood samples (13) (Fig. 4B). In keeping with the in vitro data (Fig. 1), however, the ES cell contribution to globinized cells was found in three BR12 Lmo2 −/−R chimeras (Fig. 4B), demonstrating phenotypic rescue of adult erythropoiesis by reexpression of Lmo2 in vivo.
Rescue of in vivo lymphopoiesis from Lmo2 −/−R ES cells was also demonstrated with female chimeras and a Y chromosome PCR on DNA prepared from purified splenic and thymic cells from high-level chimeras (Fig. 5A). Although splenocytes and thymocytes derived from chimeras made with Lmo2 −/− clone 71 (mice 71A, 71H, and 71J) showed no Y chromosome contribution (Fig. 5A), those prepared from female chimeras generated with an Lmo2 −/−R clone BR12 consistently gave a Y chromosome-specific ES-cell-derived signal (e.g., Fig. 5A, chimera 12C), illustrating that lymphoid cells are formed in vivo from the phenotypically rescued ES cells. Fig. 5B summarizes the results of phenotype rescue experiments using the two independent Lmo2 reconstituted ES clone (BR3 and BR12) transfected the Lmo2 expression vector. In BM, recovery of the ES cell contribution was observed in all chimeric mice derived from BR12 and BR3. In BR12H mouse (75% coat chimerism), the ES cell contribution was detected in red blood cells (by hemoglobin analysis of peripheral blood), BM, splenic lymphocytes, and peritoneal macrophage, which confirmed the broad range function of Lmo2 protein in individual mice.
Figure 5.
Rescues in all components of hematopoiesis by Lmo2 reconstitution. (A) Similar PCR analyses were performed, as in Fig. 4, with DNA from purified splenic cells and purified thymic cells, and the products were fractionated on 4% Nusieve agarose gels. Three chimeras were analyzed from the Lmo2 −/− ES clone 71 (71A, 71H, and 71J) and one chimera made from ES clone BR12 (−/−R; 12C). (B) Summary of ES cell contribution of the Lmo2 −/− clones in which Lmo2 expression has been restored by using an Lmo2 expression vector (BR3 and BR12). Individual chimeric mice were analyzed, as shown, and the sex and estimated level of coat color chimerism are listed. Green boxes indicate ES contribution was detectable and the method applied is shown (M, microsatellite PCR; g, globin; G, GPI; Y, Y chromosome PCR). White boxes indicate no ES contribution was detectable. THY, thymic cells; SPN, splenic cells; MØ, peritoneal macrophage; KID, kidney; HRT, heart; LIV, liver; BRN, brain.
DISCUSSION
The Lmo2 Gene Is Required for Adult Hematopoiesis.
These experiments show that null mutation of the Lmo2 gene causes a cell autonomous defect leading to the failure of erythroid, myeloid, or lymphoid lineages to develop, demonstrating an essential role of the Lmo2 protein in the development of all components of hematopoiesis.
Our data show that Lmo2 is required at a point in hematopoiesis before bifurcation of myeloid and lymphoid precursors in adult mouse hematopoiesis. Possible sites of crucial Lmo2 function are in the BM pluripotential precursor cells (either in the proliferative phase or in self-renewal) or even earlier in the ventral mesoderm, which provides the hematopoietic stem cells. Lmo2 null mutation affects only hematopoiesis in mouse, because the vascular endothelial system develops normally in Lmo2 −/− knockout embryonic mice (6). The present study showed the Lmo2 requirement for macrophage development in adult mice. Previously it was reported that macrophages appeared to differentiate in yolk sac tissue culture and in vitro differentiation assay (6). Our in vivo studies demonstrate that development of macrophages is impaired in the absence of Lmo2 protein, suggesting that the cells identified by microscopic findings (not combined with molecular marker analysis) represented the proliferation of other components.
The Phenotype of Lmo2 Null Mutation and Protein Interactions of Lmo2.
Gene targeting experiments of the hematopoietic transcription factors Lmo2, Gata-1, and Tal1/Scl have demonstrated that null mutations of these genes result in lack of yolk sac erythropoiesis (6, 19) that, in the case of Lmo2 and Tal1/Scl, causes similar embryonic lethality (6, 20, 21). It is known that the LIM-only protein Lmo2 binds to both Tal1/Scl and to Gata-1 (7–9) in a complex that also involves E47 and Ldb1 (9, 10). Within this complex, it appears that the Lmo2 protein makes contacts with at least two of the components forming a bridge to maintain a dual DNA binding complex that recognizes a bipartite DNA sequence (10). It therefore seems plausible that this complex, via its binding to DNA, is important in controlling red cell developmental pathways, both in yolk sac and definitive erythropoiesis.
Our present data show an additional role for Lmo2 in adult hematopoiesis (which includes all lineages), indicating that there is a role for Lmo2 in early stages, before the erythroid precursors themselves. This role cannot easily be explained by Lmo2-associated complexes involving Gata-1. However, it has been found that null mutation of Gata-2 has an effect on development of all lineages of blood cells by chimeric mice analysis (22). Because Lmo2 can bind Gata-2 (9), it is possible that complexes involving Lmo2 and Gata-2, or indeed other proteins, might be effecting functional activity in the early stages of hematopoiesis.
In this study, we demonstrated the role of Lmo2 as master gene in hematopoiesis, coding for a protein forming a backbone of transcription factor complex. This complex can bind to DNA, presumably to regulate (positively or negatively) target gene transcription, and may involve different protein interactions at different stages of hematopoiesis to decide developmental fate of blood cells (a model whereby Lmo2 functions by modulating proteins by interactions cannot be ruled out). These observations present a potentially unique paradigm for regulating hematopoiesis and thus our observations on the role of Lmo2 in hematopoiesis further extends the understanding of the properties of protein factors in the hematopoietic network. Detailed studies of precisely what function Lmo2 contributes to hematopoietic stem cell function will, however, require new analytical approaches due to their low numbers in whole BM.
The role of Lmo2 as a key element in hematopoiesis potentially helps to explain the role of LMO2 in T cell acute leukemia after the gene activation by chromosomal translocations. If LMO2 protein can interact with different sets of proteins and regulate the hematopoietic pathway, it is likely that analogous interactions may ensue after the ectopic LMO2 expression in T cells. Thus the LMO2 protein may contact proteins for which it has an affinity but which it does not normally meet because it is not normally to be expressed in T cells. These interactions may thus be responsible for the disturbance of T cell differentiation observed in transgenic models of Lmo2 overexpression (4, 5, 23, 24). This paradigm may reflect a more generalized effect seen in T cell acute lymphoblastic leukemia that has somewhat uniform clinical features despite of the occurrence of different chromosomal translocations and activation of various transcription factors (for review, see ref. 1).
Acknowledgments
Y.Y. is supported by Graduate School of Medicine, Kyoto University, Kyoto, Japan. A.J.W. is the recipient of a Medical Research Council (MRC) Clinical Scientist Fellowship, and C.D. is the recipient of an MRC studentship. The work was supported by the Medical Research Council.
ABBREVIATIONS
- ES cell
embyronic stem cell
- BM
bone marrow
- LIM
Lin-11, Isl-1, Mec-3-like
References
- 1.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Boehm T, Foroni L, Kaneko Y, Perutz M P, Rabbitts T H. Proc Natl Acad Sci USA. 1991;88:4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Royer-Pokora B, Loos U, Ludwig W-D. Oncogene. 1991;6:1887–1893. [PubMed] [Google Scholar]
- 4.Larson R C, Osada H, Larson T A, Lavenir I, Rabbitts T H. Oncogene. 1995;11:853–862. [PubMed] [Google Scholar]
- 5.Larson R C, Lavenir I, Larson T A, Baer R, Warren A J, Wadman I, Nottage K, Rabbitts T H. EMBO J. 1996;15:1021–1027. [PMC free article] [PubMed] [Google Scholar]
- 6.Warren A J, Colledge W H, Carlton M B L, Evans M J, Smith A J H, Rabbitts T H. Cell. 1994;78:45–58. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 7.Valge-Archer V E, Osada H, Warren A J, Forster A, Li J, Baer R, Rabbitts T H. Proc Natl Acad Sci USA. 1994;91:8617–8621. doi: 10.1073/pnas.91.18.8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wadman I, Li J, Bash R O, Forster A, Osada H, Rabbitts T H, Baer R. EMBO J. 1994;13:4831–4839. doi: 10.1002/j.1460-2075.1994.tb06809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osada H, Grutz G, Axelson H, Forster A, Rabbitts T H. Proc Natl Acad Sci USA. 1995;92:9585–9589. doi: 10.1073/pnas.92.21.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wadman I A, Osada H, Grutz G G, Agulnick A D, Westphal H, Forster A, Rabbitts T H. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller G, Kennedy M, Papayannopoulou T, Wiles M V. Mol Cell Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy A, Rossant J. In: Gene Targeting A Practical Approach. Joyner A L, editor. Oxford: IRL; 1993. pp. 147–179. [Google Scholar]
- 13.Whitney J B. Biochem Genet. 1978;16:667–672. doi: 10.1007/BF00484723. [DOI] [PubMed] [Google Scholar]
- 14.Dietrich W, Katz H, Lincoln S E, Shin H S, Friedman J, Dracopoli N C, Lander E S. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich W F, Miller J C, Steen R, G, Merchant M, Damron D, Nahf R, Gross A, Joyce D C, Wessel M, Dredge R D, et al. Nat Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- 16.Yamada Y, Matsushiro H, Ogawa M S, Okamoto K, Nakakuki Y, Toyokuni S, Fukumoto M, Hiai H. Cancer Res. 1994;54:403–407. [PubMed] [Google Scholar]
- 17.Greenfield A, Scott D, Pennisi D, Ehrmann I, Ellis P, Cooper L, Simpson E, Koopman P. Nat Genet. 1996;14:474–478. doi: 10.1038/ng1296-474. [DOI] [PubMed] [Google Scholar]
- 18.Papaioannou V, Johnson R. In: Gene Targeting: A Practical Approach. Joyner A J, editor. Oxford: Oxford Univ. Press; 1993. pp. 107–146. [Google Scholar]
- 19.Robb L, Lyons I, Li R, Hartley L, Kontgen F, Harvey R P, Metcalf D, Begley C G. Proc Natl Acad Sci USA. 1995;92:7075–7079. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt F W, Orkin S H. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 21.Robb L, Elwood N J, Elefanty A G, Kontgen F, Li R, Barnet L D, Begley C G. EMBO J. 1996;15:4123–4129. [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai F-Y, Keller G, Kuo F C, Weiss M, Chen J, Rosenblatt M, Alt F W, Orkin S H. Nature (London) 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 23.Fisch P, Boehm T, Lavenir I, Larson T, Arno J, Forster A, Rabbitts T H. Oncogene. 1992;7:2389–2397. [PubMed] [Google Scholar]
- 24.Neale G A, Rehg J E, Goorha R M. Blood. 1995;86:3060–3071. [PubMed] [Google Scholar]
- 25.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]