Abstract
The sulfonation of 17β-estradiol (E2) by human liver and recombinant sulfotransferases is influenced by environmental contaminants such as hydroxylated metabolites of polychlorinated biphenyls (OH-PCBs), which are potent inhibitors, and the therapeutic drug, celecoxib, which affects positional sulfonation of E2. In some locations, the aquatic environment is contaminated by PCBs, OH-PCBs and widely used therapeutic drugs. The objectives of this study were to investigate the sulfonation kinetics of E2 in liver cytosol from channel catfish (Ictalurus punctatus); to examine the effect of OH-PCBs on E2 sulfonation; and to determine if celecoxib altered the position of E2 sulfonation, as it does with human liver cytosol. E2 was converted to both 3- and 17-sulfates by catfish liver cytosol. At E2 concentrations below 1 μM, formation of E2-3-sulfate (E2-3-S) predominated, but substrate inhibition was observed at higher concentrations. Rates of E2-3-S formation at different E2 concentrations were fit to a substrate inhibition model, with K′m and V′max values of 0.40 ± 0.10 μM and 91.0 ± 4.7 pmol/min/mg protein, respectively and Ki of 1.08 ± 0.09 μM. The formation of E2-17-S fit Michaelis-Menten kinetics over the concentration range 25 nM to 2.5 μM, with Km and Vmax values of 1.07 ± 0.23 μM and 25.7 ± 4.43 pmol/min/mg protein, respectively. The efficiency (Vmax/Km) of formation of E2-3-S was 9.8-fold higher than that of E2-17-S. Several OH-PCBs inhibited E2 3-sulfonation, measured at an E2 concentration of 1 nM. Of those tested, the most potent inhibitor was 4′-OH-CB79, with two chlorine atoms flanking the OH group (IC50: 94 nM). The inhibition of estrogen sulfonation by OH-PCBs may disrupt the endocrine system and thus contribute to the known toxic effects of these compounds. Celecoxib did not stimulate E2-17-S formation, as is the case with human liver cytosol, but did inhibit the formation of E2-3-S (IC50: 44 μM) and to a lesser extent, E2-17-S (IC50: >160 μM), suggesting the previously found effect of celecoxib on E2-17-S formation may be specific to human SULT2A1.
Keywords: Sulfotransferase, Sulfonation, Channel catfish, Inhibition, OH-PCBs, Celecoxib
1. Introduction
Sulfonation has evolved as a key step in xenobiotic metabolism, but also plays a critical role in modulating the biological activity and facilitating the inactivation and elimination of potent endogenous chemicals including steroids, thyroid hormones and catechols (Coughtrie, 1996; Falany, 1991; Strott, 2005). Sulfonation reactions are catalyzed by sulfotransferase (SULT) enzymes which transfer the sulfuryl group from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to an appropriate acceptor. In humans, at least 11 SULT isoforms have been identified and divided into several gene families based on their amino acid sequences. The SULT1 and SULT2 families are the largest and are responsible for sulfonation of a vast number of endogenous and xenobiotic compounds (Kauffman, 2004).
Sulfonation of 17β-estradiol (E2) has been reported in human liver (Hernandez et al., 1992) and platelet phenol sulfotransferase (Harris et al., 2000), human mammary epithelial cells and breast cancer cell lines (Falany and Falany, 1996), as well as in recombinant SULTs including SULT1E1, SULT2A1, Phenol-SULT (Falany et al., 1994; Falany et al., 1995; Wang and James, 2005; Zhang et al., 1998). However, knowledge of sulfonation in fish species is rather limited. Sulfonation of E2 has been reported in carp and zebrafish, but not whether the 3- or 17-sulfate was formed (Thibaut and Porte, 2004; Ohkimoto et al., 2003, 2004). Jurgella et al (2006) showed that incubation of E2 with lake trout liver and kidney tissues resulted in the formation of several metabolites, including E2-17-sulfate and E2 3- and 17-glucuronides. Several SULTs from the zebrafish have been characterized. Sugahara et al. (2003a) studied an expressed recombinant zebrafish with sequence similarity to the SULT1 family, which displayed activity towards a number of endogenous compounds including estrone, dopamine and thyroid hormones as well as xenobiotic compounds including some flavonoids, isoflavonoids, and other phenolic compounds. Further studies showed that this zebrafish SULT had activity with E2, however the Km was 13 μM, considerably higher than mammalian SULT1E1 (Ohkimoto et al., 2004). A putative hydroxysteroid SULT2 enzyme has been identified in zebrafish, and this isoform displayed 44%, 43% and 40% amino acid identity with mouse SULT2B1, human SULT2B1 and human SULT2A1, respectively (Sugahara et al. 2003b). This enzyme displayed activity towards DHEA and several neurosteroids but did not show activity with other phenolic compounds such as estrone, para-nitrophenol and quercetin. A phenol SULT of molecular mass 41 KDa was purified from channel catfish liver and intestine. The purified enzyme had an exceptionally high affinity for and activity with benzo[a]pyrene phenols, but also showed activity with estrone, E2 and DHEA (Tong and James, 2000).
There is evidence that xenobiotics found in polluted environments can inhibit the phase II conjugating enzymes in fish, including those that catalyze sulfation and glucuronidation of steroids, and potentially disrupt steroid hormone homeostasis (Wang and James, 2006). Aquatic species such as omnivorous fish living in polluted environments may be exposed to a variety of organic pollutant chemicals. Exposure may be water-borne, from sediments, or though the diet. Van den Hurk et al. (2002) found that several polychlorobiphenylols (OH-PCBs) inhibited both the sulfonation and glucuronidation of 3-hydroxybenzo[a]pyrene in channel catfish, indicating that both SULT and glucuronosyltransferase (UGT) are targets for inhibition by xenobiotics in fish. Ohkimoto et al.(2003) reported that bisphenol A, 4-n-nonylphenol (NP), and 4-n-octylphenol (OP) exerted concentration-dependent inhibition of the sulfonation of E2 in zebrafish, implying a potential role of these environmental estrogens in interfering with the sulfonation of endogenous estrogens. In addition, alkylphenols have a strong inhibitory effect on the sulfonation of estrone in a cyprinid fish, the chub (Leuciscus cephalus) (Kirk et al., 2003), and tributyltin (TBT), triphenyltin (TPT), and nonylphenol (NP) were inhibitors of the sulfonation of E2 by carp (Cyprinus carpio) cytosolic fractions (Thibaut and Porte, 2004), with IC50 values of 17–41 μM. Jurgella et al (2006) showed that bisphenol A and chlorinated and brominated bisphenol A as well as 4,4′-dihydroxy-3,3′,5,5′-tetrachlorobiphenyl and three halogenated phenols inhibited E2 metabolism in the lake trout liver and kidney. In these studies, however, the position of sulfonation of E2 was not examined.
The present study was designed to evaluate the kinetics of formation of E2-3-sulfate and E2-17-sulfate in channel catfish (Ictalurus punctatus) liver cytosol, and to determine if environmental compounds known to perturb E2 sulfonation by human SULT enzymes would have similar effects on the catfish enzymes. The compounds examined were OH-PCBs and celecoxib. OH-PCBs are found in the environment as metabolites of PCBs and celecoxib is a widely used non-steroidal anti-inflammatory drug with COX-2 selectivity. Although celecoxib has not yet been reported in environmental samples, it has been in the top 100 prescribed drugs for several years and is widely used to treat arthritis (Bensen, 2000), thus it is likely to be present in waste water. The OH-PCBs selected were 4-OH-3,3′,4′,5-tetrachlorobiphenyl (4′-OH-CB79), 4-OH-3,3′,4′-trichlorobiphenyl (4′-OH-CB35) and 4-OH-2′,3,3′,4′,5′-pentachlorobiphenyl (4′-OH-CB106), which were reported to have different inhibitory potencies against human recombinant SULT1E1, with IC50 of 0.21–0.61 nM, 4.3–7.8 nM and 100–120 nM, respectively (Kester et al., 2000). An unusual heterotropic modulation of human SULT2A1, a hydroxysteroid SULT, was reported with celecoxib (Cui et al., 2004; Wang and James, 2005). Celecoxib was able to switch the major position of sulfonation of ethynylestradiol (Cui et al., 2004) and E2 (Wang and James, 2005) from the 3- to the 17- with SULT2A1 as well as in human liver cytosol. The switching ability by celecoxib was specific for SULT2A1, with no effects on other phenol SULTs including SULT1A1, SULT1A3, SULT1B1 and SULT1E1 (Wang and James, 2005). Therefore, it was of interest to investigate the effects of celecoxib on sulfotransferases of other species such as fish, to determine if this is a general effect, or selective to human SULT2A1.
2. Materials and Methods
2.1. Chemicals
[3H]β-Estradiol (49.7 Ci/mmol) was obtained from PerkinElmer Life Sciences, Inc (Boston, MA). 3′-phosphoadenosine-5′-phosphosulfate (PAPS) was obtained from Dr. S.S. Singer, University of Dayton, OH. Celecoxib (Figure 1) was obtained via extraction from a 100-mg commercial capsule formulation (Express Pharmacy Services, Largo, FL) as described previously (Wang and James, 2005). E2-3-S standard was obtained from Sigma, St Louis, MO. E2-17-S and E2 disulfate standards were obtained from Steraloids (Newport, RI). The 4′-OH-CB35 and 4′-OH-CB79 were a gift from Dr. Larry Robertson, University of Iowa. The 4′-OH-CB106 was purchased from AccuStandard (New Haven, CT). Figure 1 shows the structures of the compounds used as modulators of SULT activity. Other reagents were the highest grade available from Fisher Scientific, Atlanta, GA, and Sigma, St. Louis, MO.
Fig. 1.
Structure of celecoxib and OH-PCBs used in this study
2.2. Fish
Channel catfish used in these studies were one male and two females with body weight ranging from 454 to 790 g. All fish were kept in flow-through well water with temperature 20 ± 0.6°C, total hardness 26.4 ± 5.3 mg of CaCO3/liter and pH 7.96 ± 0.31 over the maintenance and experimental period. The fish were grown and maintained on a commercial chow diet (Silvercup, Murray, UT) for at least 2 weeks. Care and treatment of the animals was conducted according to the guidelines of the University of Florida Institutional Animal Care and Use Committee.
2.3. Preparation of cytosolic fractions
Catfish were immobilized in ice water and killed by severing the spinal cord. Livers were removed, rinsed three times in ice-cold buffer 1 [composed of 1.15% KCl, 0.05 M potassium phosphate (pH 7.4), 0.2 mM PMSF], weighed, and homogenized in 4 volumes of buffer 1. The homogenates were centrifuged at 13,300×g for 20 min and the resulting supernatants were centrifuged again at 155,000×g for 45 min. The high speed cytosolic fraction was divided into aliquots and stored at −80°C until use. The protein concentrations were measured by the Lowry method (Lowry et al., 1951) using bovine serum albumin as a standard.
2.4. Sulfotransferase assay
Assay conditions were optimized such that the rate of reaction was linear with protein and time, and was saturating for PAPS. All studies were conducted with liver cytosol fractions from three different individuals, with duplicate tubes for each assay condition.
For examining the enzyme kinetics, incubation mixtures contained 100 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 28–33 μg cytosolic protein, 20 μM PAPS, and a concentration series of 0.05 to 6 μM [3H] E2 in a total volume of 0.25 ml. For studying the effect of celecoxib on positional sulfonation of E2, the E2 substrate concentration was 0.8 μM. At this concentration both the E2 sulfates were formed with no substrate inhibition. Stock solutions of celecoxib were prepared in DMSO, such that the final concentration of celecoxib was in the range of 1.25–160 μM and the DMSO concentration did not exceed 0.5% (v/v). Reaction was initiated with the addition of PAPS after a 3-min preincubation at 35°C. This was the optimum temperature for the assay. After 30 min incubation, the reaction was stopped with 0.3 ml methanol followed by vortex-mixing and centrifugation. The resultant supernatant was transferred into new tubes and analyzed by HPLC.
For studying inhibition by OH-PCBs, the sulfonation of E2 was assessed by a radiochemical extraction method. The assay mixture contained 56–67 μg cytosolic protein, 100 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 1 nM [3H] E2 and 20 μM PAPS (added last to start the reaction) in a total volume of 0.5 ml. Stock solutions of OH-PCBs were prepared in DMSO, such that the final concentration of OH-PCB was in the range of 0.01–5 μM and the DMSO concentration did not exceed 0.5% (v/v). Samples were incubated at 35°C for 30 min. The reaction was stopped by addition of 200 μL of cold 3% trichloroacetic acid (TCA; w/v). Dichloromethane(1 ml) was added and the mixture was extracted to remove unreacted E2 substrate. The phases were separated by centrifuging for 10 min at 600xg. The aqueous phase was extracted twice more to ensure removal of the substrate. An aliquot of the aqueous phase, 200 μl, containing the E2-3-sulfate, was taken for liquid scintillation counting (Beckman LS 6000; Beckman Coulter, Fullerton, CA).
2.5. HPLC analysis
HPLC analyses were conducted on a Beckman Gold Nouveau system, equipped with UV and fluorescence detectors and an IN/US (β-ram, IN/US systems, Inc., Tampa, FL) radiochemical detector. Separation of parent substrate and its sulfate conjugates was achieved on a C18 reverse-phase column (4.6 mm × 25 cm) with a C18 pre-column (Discovery system, Supelco, Bellefonte, PA) at a constant flow of 1 ml/min with 0.005 M tetrabutylammonium sulfate in 55% MeOH. The flow of scintillation cocktail (In-flow 2, IN/US systems, Inc., Tampa, FL) was maintained at 3 ml/min. E2-3-S, E2-17-S and E2 disulfate were identified by comparing the retention times of the authentic standards.
2.6. Data analysis
Results for catfish liver cytosol are presented as mean values with standard deviation from the results of three different individuals. The IC50 values were obtained by fitting log OH-PCB concentration and percent of control activity to a sigmoidal curve (with variable slope) with the software package Prism (GraphPad Software 4.0). The kinetic parameters (Km, Vmax) were calculated using the Michaelis-Menten non-linear regression equation with the software package Prism (GraphPad Software 4.0). Where we found evidence of E2 substrate inhibition, we fit the data into equation (1) derived from a model in which two molecules of substrate enter the active site and cause partial inhibition of activity (Zhang et al., 1998), using Prism (GraphPad Software 4.0):
| (1) |
This equation denoted the constant for binding of the first substrate (S) molecule as K′m and the second substrate molecule as Ki. V′max is the maximum rate for the noninhibitory substrate concentration range, and Vmin is the minimum rate in the inhibitory substrate concentration range. Data was also fit to a two-substrate inhibition model where binding of the second substrate causes loss of activity (Cornish-Bowden, 2004), following the equation:
| (2) |
Results from fitting data to these two equations were compared and the model that showed the best fit to the data was selected.
3. Results
3.1. Kinetics of sulfonation of E2 with catfish liver cytosol
Incubation of E2 with catfish liver cytosol at concentrations above 10 nM produced two sulfates E2-3-S and E2-17-S. Below 10 nM, only E2-3-S was detected. As shown in Fig. 2A, inhibition of activity was observed for the formation of E2-3-S, when the E2 concentration was more than 0.8 μM. For each fish, the data for the formation of E2-3-S was fit to two models for substrate inhibition. For both models, initial estimates of Km were provided by fitting data obtained at non-inhibitory concentrations of E2, less than 0.8 μM, using the Michaelis-Menten equation. To obtain results from each individual fish, the values obtained for the Michaelis constant, Km, were constrained as constants K′m for fitting data to the two inhibition models (equations 1 and 2). Results from equation 1 estimated that Vmin was zero, thus collapsing equation 1 to equation 2, substrate inhibition to an inactive complex. Kinetic data obtained from equation 2 are shown in Table 1.
Fig. 2.
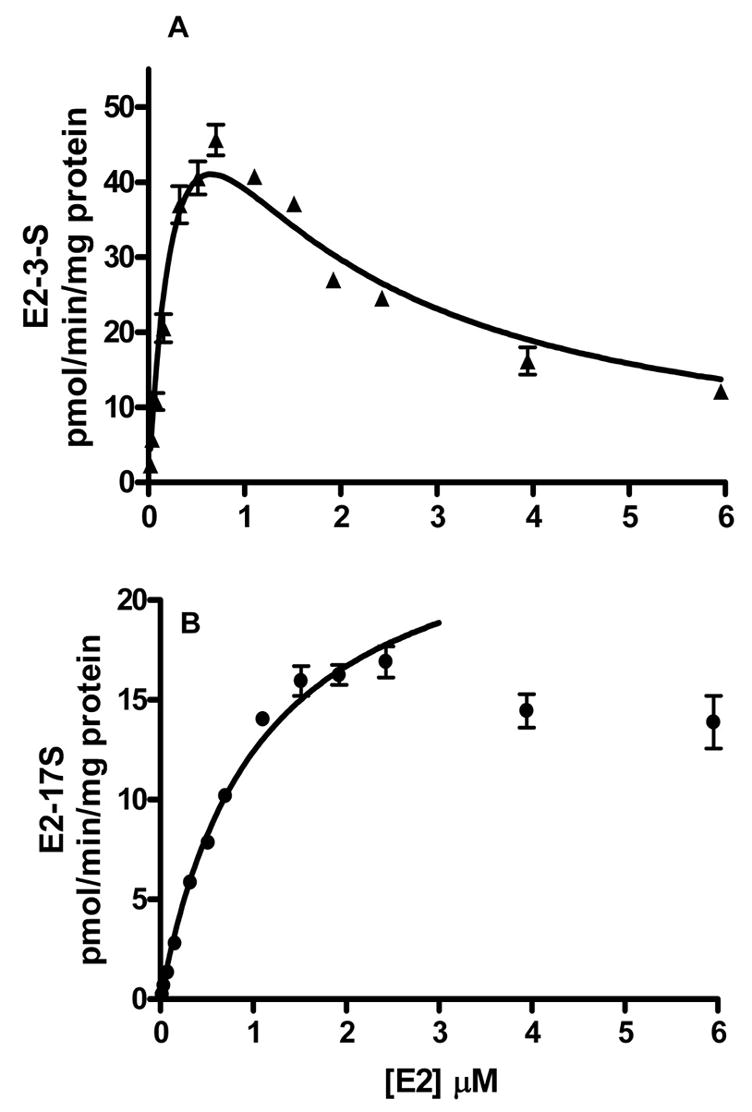
Results for rates of E2 sulfonation in catfish liver cytosol. Substrate inhibition of the formation of E2-3-S is shown in (A). The formation of E2-17-S is shown in (B). Data shown are the mean values from studies with three catfish, and error bars indicate standard deviation. The kinetic parameters are summarized in Table 1.
Table 1.
Apparent kinetic parameters for E2 sulfonation by catfish liver cytosol.
| E2 sulfate | Km(μM) | Ki(μM) | Vmaxpmol/min/mg protein) | Vmax/Km(μl/min/mg protein) | R2 |
|---|---|---|---|---|---|
| E2-3-S | 0.40 ± 0.10 | 1.08 ± 0.09 | 91 ± 4.6 | 238 ± 75 | 0.95 ± 0.02 |
| E2-17-S | 1.07 ± 0.23 | - | 25.7 ± 4.4 | 24.3 ± 2.0 | 0.99 ± 0.00 |
Values shown are mean ± S.D., n=3 individuals.
The formation of E2-17-S followed Michaelis-Menten kinetics in the substrate concentration range 0.05 to 2.5 μM E2 (Figure 2B). At concentrations above 2.5 μM, inhibition was observed, however not enough data points were studied to obtain the inhibitory constants. The apparent Km of formation of E2-17-S (1.07 ± 0.2 μM) was 2.6-fold higher than that of E2-3-S (0.40 μM), and the maximum rate of formation of E2-17-S (25.7 pmol/min/mg protein) was 3.5-fold less than that of E2-3-S (91 pmol/min/mg protein). The efficiency (Vmax/Km) of formation of E2-3-S was 9.8-fold higher than that of E2-17-S (Table 1).
3.2. Inhibition of E2 sulfonation by OH-PCBs
Studies showed that with 1 nM E2 as substrate, only E2-3-S was detected by HPLC. Therefore, the extraction assay was used to examine inhibition of the formation of E2-3-S. Addition of OH-PCBs to assay tubes resulted in a concentration-dependent inhibition of E2 sulfonation using catfish liver cytosol (Fig. 3). Of the OH-PCBs used in this study, 4′-OH-CB79 showed the most potent inhibition of E2-3-S formation, with an estimated IC50 value of 93.7 nM (Table 2). The 4′-OH-CB35, with one less chlorine atom flanking the OH group than 4′-OH-CB79 (Figure 1), was 2.7-fold less potent than 4′-OH-CB79 as an inhibitor, with an estimated IC50 value of 253 nM. Of the OH-PCBs tested in this study, 4′-OH-CB106 exhibited the lowest inhibitory potency (IC50: 552 nM).
Fig. 3.
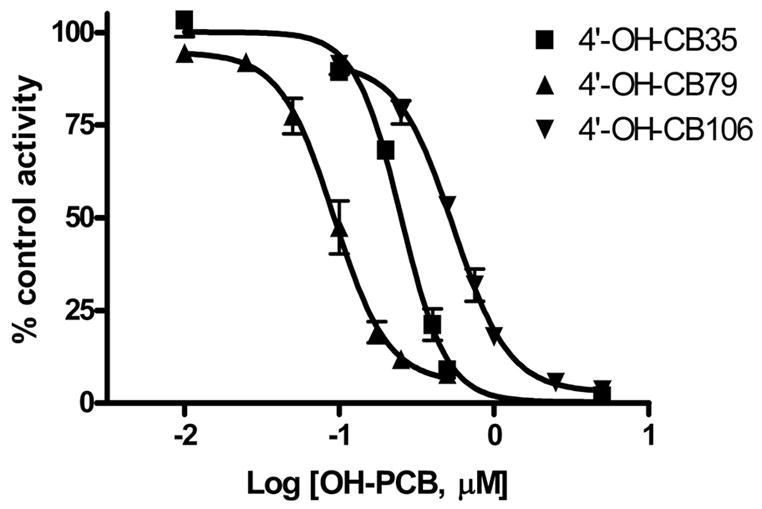
Inhibition of E2 sulfonation with catfish liver cytosol by OH-PCBs. The sulfotransferase activity is given as percentage of control activity. Data given are the mean ± S.D. of experiments with three fish.
Table 2.
In vitro inhibition of E2 (1 nM) sulfotransferase activity by the tested OH-PCBs using catfish liver cytosol
| IC50 (nM)
|
||
|---|---|---|
| Compound | Catfish liver cytosol | SULT1E1a |
| 4′-OH-CB35 | 252.9 ± 30.4 | 4.3–7.8 |
| 4′-OH-CB79 | 93.7 ± 20.2 | 0.21–0.61 |
| 4′-OH-CB106 | 552.4 ± 66.3 | 100–120 |
Data were taken from Kester et al. (2000). The enzyme used was human SULT1E1 with 1 nM E2.
3.3. Inhibition of E2 sulfonation by celecoxib
Celecoxib inhibited the formation of E2-3-S in a concentration-dependent manner over a range of celecoxib concentrations from 1.25 to 160 μM (Figure 4). The IC50 value for the formation of E2-3-S was 44.5 ± 3.7 μM. At low concentrations of celecoxib (1.25–10 μM), the formation of E2-17-S was slightly stimulated (102–103% of control). At celecoxib concentrations ranging from 20 μM to 160 μM, up to 3-30% inhibition of E2-17-S formation was observed.
Fig. 4.
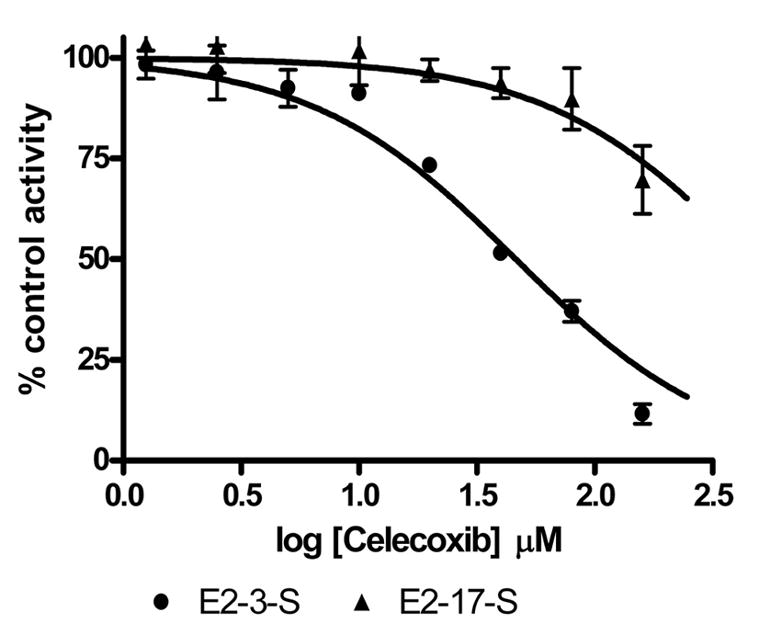
Inhibition of E2 sulfonation with catfish liver cytosol by celecoxib. The sulfotransferase activity is given as percentage of control activity with 0.8 μM E2. Data given are the mean ± S.D. of experiments with three fish.
4. Discussion
The sulfonation of estrogens by SULTs is an important route for the removal of the active hormones from the body. In mammals, there is evidence that a circulating pool of estrogen sulfate is re-converted to active estrogen by sulfatase in peripheral tissues (Kirk et al., 2001). The most significant SULTs with respect to estrogen metabolism are estrogen sulfotransferase (SULT1E1), which has a Km for E2 in the nanomolar range (Zhang et al., 1998), the thermostable phenol sulfotransferase (SULT1A1), which has a Km for E2 of approx. 2 μM (Harris et al., 2000) and hydroxysteroid sulfotransferase (SULT2A1), which has a Km of 1.5 μM for E2-3-sulfation and 3 μM for E2-17-sulfation (Wang and James, 2005). In the catfish liver, there is evidence for at least two forms of SULT (Tong and James, 2006; Merritt and James, 2006), however as yet the activities of the pure SULTs with E2 are not known. The possible existence of two SULT isoforms metabolizing E2, with Km values of 17 nM and 3.2 μM has been reported in carp (Thibaut and Porte, 2004). A putative estrogen-sulfating zebrafish SULT has been characterized and its Km values for estrone and E2 were 12.5 and 13 μM, respectively (Ohkimoto et al., 2004). SULTs that have Km values in the nanomolar range for estrogen sulfonation have been observed in freshwater (Kirk et al., 2003) and marine fish liver (Martin-Skilton et al., 2006). The above findings suggest that several SULTs, with differing abilities to sulfonate estrogens, are present in liver cytosol fractions from various fish species.
The sulfonation kinetics of E2 has been studied with human recombinant SULT1E1. Maximum sulfonation of E2 was observed at a concentration of approx. 20 nM and substrate inhibition was observed with higher E2 concentrations, which were explained by two-substrate partial inhibition model (Falany et al., 1995; Zhang et al., 1998). The kinetics for E2-3-S formation with catfish liver cytosol exhibited inhibition with increasing E2 concentrations in the present study (Fig. 2A), but the data fit a model in which the second molecule of substrate binding in the active site led to an inactive complex. The kinetic constant for E2 with catfish liver sulfotransferases (K′m: 0.40 μM) was significantly greater than that of E2 with human SULT1E1 (K′m: 5 nM), indicating catfish liver cytosol contains an enzyme with lower affinity for E2. The formation of E2-17-S with catfish liver cytosol followed Michaelis-Menten kinetics in the range of 0–2.5 μM (Fig 2B): this was similar to results for sulfonation of E2 catalyzed by human SULT2A1, which formed increasing amounts of E2-3-S, E2-17-S and E2-3,17-S with no inhibition over a range of E2 concentrations from 0–6 μM (Wang and James, 2005). These results suggest the possibility that the two observed products, E2-3-S and E2-17-S, are formed by different SULT isozymes in the catfish liver and that the substrate inhibition observed with E2 is isozyme dependent.
An interesting difference between catfish and human liver was that relatively more E2-17-S was formed in incubations with catfish hepatic cytosol. Although hepatic cytosol from both catfish and humans produced predominantly E2-3-S over the concentration range of E2 studied, the ratio of E2-3-S:E2-17S was higher in human liver cytosol than with catfish liver cytosol (Table 3). In human liver, SULT1E1 forms only E2-3-S, while SULT2A1 can form E2-3-S, E2-17-S and E2-3,17-S (Wang and James, 2005). The propensity of the catfish cytosolic SULT enzymes to sulfonate the aliphatic hydroxyl group of E2 suggests a different orientation of E2 binding to the catfish enzymes. Further studies with purified recombinant catfish enzymes would be needed to examine this possibility. Since a mixture of E2 sulfates was produced in catfish liver over a wide range of E2 concentrations, caution should be used in interpreting the effects of compounds on E2 sulfation if the extraction assay is used.
Table 3.
Ratio of E2-3-S to E2-17-S in catfish liver cytosol, SULT2A1 and human liver cytosol.
| Ratio (E2-3-S/E2-17-S)
|
|||
|---|---|---|---|
| E2 (μM) | Catfish liver cytosol | SULT2A1a | Human liver Cytosola |
| 0.05 | 8.3 ± 2.3 | 14.3 | 17 |
| 0.1 | 8.1 ± 2.5 | 14.8 | - |
| 0.2 | 7.6 ± 2.3 | 16.1 | - |
| 0.4 | 6.3 ± 1.4 | 15.1 | - |
| 0.6 | 5.3 ± 1.2 | - | - |
| 0.8 | 4.5 ± 0.9 | 14.2 | - |
Data were taken from Wang and James (2005).
It has been shown that OH-PCBs are potent inhibitors of E2-3-S formation catalyzed by human SULT1E1, and may therefore be indirect endocrine disruptors (Kester et al. 2000). Other xenobiotics, including alkyphenols and the organotin compounds tributyltin and triphenyltin showed inhibition of E2 sulfonation in fish species (Ohkimoto et al. 2003; Ohkimoto et al. 2005). The present study showed OH-PCBs were also inhibitors of E2-3S formation by catfish liver sulfotransferases. The concentration of E2 used in the inhibition assays was 1 nM, so that only E2-3-S would be formed. The most potent compounds of the three OH-PCBs tested was 4′-OH-CB79, with both sides of the hydroxyl group flanked by chlorine atoms. The 4′-OH-CB35 and 4′-OH-CB106, with one chlorine atom flanking the OH group, were less potent inhibitors than 4′-OH-CB79. With human SULT1E1, 4′-OH-CB79 showed very potent inhibitory activity (IC50: 0.21–0.61 nM), followed by 4′-OH-CB35 (IC50: 4.3–7.8 nM) and 4′-OH-CB106 (100–120 nM), which is in agreement with the order in potency with catfish liver cytosol (Table 2). Although the OH-PCBs studied exhibited IC50 values of less than 1 μM with catfish hepatic sulfotransferases, they were not as potent as with recombinant human SULT1E1. Further studies with expressed catfish sulfotransferases are needed to determine if OH-PCBs interact with human and catfish SULTs in a similar manner.
Recent studies showed that celecoxib was able to switch the dominant sulfate product of ethynylestradiol (EE) and E2 from 3-sulfate to 17-sulfate in human recombinant SULT2A1 as well as in human liver cytosol (Cui et al., 2004; Wang and James, 2005). The switch in dominant metabolite may lead to altered E2 levels in steroid target organs, since unlike E2-3-S, which is considered a transport form of E2 (Wood et al., 2003), E2-17-S is resistant to sulfatase hydrolysis (Chetrite et al., 2000; Pasqualini et al. 1989). As a widely used pharmaceutical agent, celecoxib may be present in waste water, thus it is of interest to examine its effect on E2 sulfation in the catfish. Although catfish liver SULTs produced relatively more E2-17-S sulfate than human liver SULTs, celecoxib did not alter the ratio of E2-3-S to E2-17-S formed, rather, it inhibited formation of both sulfate conjugates. The formation of E2-3-S was more readily inhibited than that of E2-17-S. The lack of effect of celecoxib on the relative amount of E2-17-S formed from E2 by catfish hepatic SULTs suggested that the effect of celecoxib on human SULT2A1 may be selective for the human enzyme.
5. Conclusion
The results presented here indicate that in catfish liver, both E2-3-S and E2-17-S are formed over a range of E2 concentrations. The formation of E2-3-S exhibited substrate inhibition at concentrations above 0.8 μM and the formation of E2-17-S followed Michaelis-Menten kinetics up to 2.5 μM. The three OH-PCBs used in this study were inhibitors of E2 sulfation by catfish liver, with IC50 values of less than 1 μM. Celecoxib did not stimulate E2-17-S formation with catfish liver cytosol, suggesting the effect of celecoxib on human SULT2A1 may be species dependent.
Acknowledgments
This study was supported by grants P42 ES007375 from the National Institute of Environmental Health Sciences, National Institutes of Health (NIH). This article reflects the authors’ views and not any official views of NIH. The authors would like to thank Drs. Larry W. Robertson and Hans-Joachim Lehmler for providing the OH-PCBs through grant P42 ES013661.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bensen WG. Anti-inflammatory and analgesic efficacy of COX-2 specific inhibition: from investigational trials to clinical experience. J Rheumatol Suppl. 2000;60:17–24. [PubMed] [Google Scholar]
- Chetrite GS, Cortes-Prieto J, Philippe JC, Wright F, Pasqualini JR. Comparison of estrogen concentrations, E1 sulfatase and aromatase activities in normal, and in cancerous, human breast tissues. J Steroid Biochem Mol Biol. 2000;72:23–27. doi: 10.1016/s0960-0760(00)00040-6. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Fundamentals of enzyme kinetics. 3. Portland Press Ltd; London: 2004. [Google Scholar]
- Coughtrie MWH. Sulphation catalysed by the human cytosolic sulphotransferases-chemical defence or molecular terrorism? Hum Exp Toxicol. 1996;15:547–555. doi: 10.1177/096032719601500701. [DOI] [PubMed] [Google Scholar]
- Cui D, Booth-Genthe CL, Carlini E, Carr B, Schrag ML. Heterotropic modulation of sulfotransferase 2A1 activity by celecoxib: product ratio switching of ethynylestradiol sulfation, Drug Metab. Dispos. 2004;32:1260–1264. doi: 10.1124/dmd.32.11.. [DOI] [PubMed] [Google Scholar]
- Falany CN. Molecular enzymology of human liver cytosolic SULTs. Trends Pharmacol Sci. 1991;12:255–259. doi: 10.1016/0165-6147(91)90566-b. [DOI] [PubMed] [Google Scholar]
- Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Mol Biol. 1995;52:529–539. doi: 10.1016/0960-0760(95)00015-r. [DOI] [PubMed] [Google Scholar]
- Falany CN, Wheeler J, Oh TS, Falany JL. Steroid sulfation by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 1994;48:369–375. doi: 10.1016/0960-0760(94)90077-9. [DOI] [PubMed] [Google Scholar]
- Falany JL, Falany CN. Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res. 1996;56:1551–1555. [PubMed] [Google Scholar]
- Harris BM, Waring RH, Kirk CJ, Hughes PJ. Sulfation of “estrogenic” alkylphenols and 17β-estradiol by human platelet phenol sulfotransferases. J Biol Chem. 2000;275:159–166. doi: 10.1074/jbc.275.1.159. [DOI] [PubMed] [Google Scholar]
- Hernandez JS, William R, Watson G, Wood TC, Weinshilboum RW. Sulfation of E1 and 17β-estradiol in human liver. Catalysis by thermastable phenol sulfotransferase and by dehydroepiandrosterone sulfotransferse. Drug Metab Dispos. 1992;20:413–422. [PubMed] [Google Scholar]
- Jurgella GF, Marwah A, Malison JA, Peterson R, Barry TP. Effects of xenobiotics and steroids on renal and hepatic estrogen metabolism in lake trout. Gen Comp Endocrinol. 2006;148:273–81. doi: 10.1016/j.ygcen.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Kauffman FC. Sulfonation in pharmacology and toxicology. Drug Metab Rev. 2004;36:823–843. doi: 10.1081/dmr-200033496. [DOI] [PubMed] [Google Scholar]
- Kester MHA, Bulduk S, Tibboel D, Meinl W, Glatt H, Falany CN, Coughtrie MWH, Bergman A, Safe SH, Kuiper GGJM, Schuur AG, Brouwer A, Visser TJ. Potent inhibition of estrogen sulfotransferase by hydroxylated PCB metabolites: A novel pathway explaining the estrogenic activity of PCBs. Endocrinology. 2000;141:1897–1900. doi: 10.1210/endo.141.5.7530. [DOI] [PubMed] [Google Scholar]
- Kirk CJ, Bottomley L, Minican N, Carpenter H, Shaw S, Kohli N, Winter M, Taylor EW, Waring RH, Michelangeli F, Harris RM. Environmental endrocrine disrupters dysregulate estrogen metabolism and Ca2+ homeostasis in fish and mammals via receptor-independent mechanisms. Comp Biochem Physiol A. 2003;135:1–8. doi: 10.1016/s1095-6433(02)00366-5. [DOI] [PubMed] [Google Scholar]
- Kirk CJ, Harris RM, Wood DM, Waring RH, Hughes PJ. Do dietary phytoestrogens influence susceptibility to hormone-dependent cancer by disrupting the metabolism of endogenous oestrogens? Biochem Soc Trans. 2001;29:209–216. doi: 10.1042/0300-5127:0290209. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Martin-Skilton R, Coughtrie MW, Porte C. Sulfotransferase activities towards xenobiotics and estradiol in two marine fish species (Mullus barbatus and Lepidorhombus boscii): characterization and inhibition by endocrine disrupters. Aquat Toxicol. 2006;79:24–30. doi: 10.1016/j.aquatox.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Merritt KK, James MO. Cloning and expression of sulfotransferases from channel catfish liver. Marine Env Res. 2006;62:S168. [Google Scholar]
- Ohkimoto K, Liu MY, Suiko M, Sakakibara Y, Liu MC. Characterization of a zebrafish estrogen-sulfating cytosolic sulfotranferase: inhibitory effects and mechanism of action of phytoestrogens. Chem Biol Interact. 2004;141:1–7. doi: 10.1016/j.cbi.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Ohkimoto K, Sakakibara Y, Suiko M, Yoshikawa H, Liu MC, Tamura H. Biocides, tributyltin and triphenyltin, as possible inhibitors of the human sulfotransferase involved in the estrogen homeostasis. Pesticide Biochem Physiol. 2005;81:32–38. [Google Scholar]
- Ohkimoto K, Sugahara T, Sakakibara Y, Suiko M, Liu MY, Carter G, Liu MC. Sulfonation of environmental estrogens by zebrafish cytosolic sulfotransferases. Biochem Biophs Res Commun. 2003;309:7–11. doi: 10.1016/s0006-291x(03)01524-9. [DOI] [PubMed] [Google Scholar]
- Pasqualini JR, Gelly C, Nguyen BL, Vella C. Importance of estrogen sulfates in breast cancer. J Steroid Biochem. 1989;34:155–163. doi: 10.1016/0022-4731(89)90077-0. [DOI] [PubMed] [Google Scholar]
- Strott CA. Sulfonation and molecular action. Endocrine Rev. 2005;24:703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- Sugahara T, Liu CC, Carter G, Pai GT, Liu MC. Molecular cloning, expression, and functional characterization of a novel zebrafish cytosolic sulfotransferase. Biochem Biophys Res Commun. 2003a;300:725–730. doi: 10.1016/s0006-291x(02)02915-7. [DOI] [PubMed] [Google Scholar]
- Sugahara T, Liu CC, Carter G, Pai GT, Liu MC. Sulphonation of dehydroepiandrosterone and neurosteroids: molecular cloning, expression, and functional characterization of a novel zebrafish SULT2 cytosolic sulphotransferase. Biochem J. 2003b;375:785–791. doi: 10.1042/BJ20031050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaut R, Porte C. Effects of endocrine disrupters on sex steroid synthesis and metabolism pathways in fish. J Steroid Biochem Mol Biol. 2004;92:485–494. doi: 10.1016/j.jsbmb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Tong Z, James MO. Purification and characterization of hepatic and intestinal phenol sulfotransferase with high affinity for benzo[a]pyrene phenols from Channel catfish, Ictalurus punctatus. Arch Biochem Biophys. 2000;376:409–419. doi: 10.1006/abbi.2000.1746. [DOI] [PubMed] [Google Scholar]
- van den Hurk P, Kubiczak GA, Lehmler HJ, James MO. Hydroxylated polychlorinated biphenyls as inhibitors of the sulfation and glucuronidation of 3-hydroxybenzo[a]pyrene. Environ Health Perspect. 2002;110:343–348. doi: 10.1289/ehp.02110343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LQ, James MO. Sulfotransferase 2A1 forms estradiol-17-sulfate and celecoxib switches the dominant product from estradiol-3-sulfate to estradiol-17-sulfate. J Steroid Biochem Mol Biol. 2005;96:367–374. doi: 10.1016/j.jsbmb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Wang LQ, James MO. Inhibition of sulfotransferases by xenobiotics. Current Drug Metabolism. 2006;7:83–104. doi: 10.2174/138920006774832596. [DOI] [PubMed] [Google Scholar]
- Wood CE, Gridley KE, Keller-Wood M. Biological activity of 17beta-estradiol-3-sulfate in ovine fetal plasma and uptake in fetal brain. Endocrinology. 2003;144:599–604. doi: 10.1210/en.2002-220764. [DOI] [PubMed] [Google Scholar]
- Zhang H, Varmalova O, Vargas FM, Falany CN, Leyh TS. Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. J Biol Chem. 1998;273:10888–10892. doi: 10.1074/jbc.273.18.10888. [DOI] [PubMed] [Google Scholar]


