Abstract
Mononuclear phagocytes (bone marrow monocyte-derived macrophages, alveolar macrophages, perivascular macrophages, and microglia) are reservoirs and vehicles of dissemination for the human immunodeficiency virus type-1 (HIV-1). How virus alters mononuclear phagocyte immunoregulatory activities to complete its life cycle and influence disease is incompletely understood. In attempts to better understanding the influence of virus on macrophage functions, we used one-dimensional electrophoresis, and liquid chromatography tandem mass spectrometry to analyze the secretome of HIV-1 infected human monocyte-derived macrophages. We identified 111 proteins in culture supernatants of control (uninfected) and virus-infected cells. Differentially expressed cytoskeletal, enzymes, redox, and immunoregulatory protein classes were discovered and validated by Western-blot tests. These included, but were not limited to, cystatin C, cystatin B, chitinase 3-like 1 protein, cofilin-1, L-plastin, superoxide dismutase, leukotriene A4 hydrolase, and α-enolase. This study, through the use of a unique proteomics platform, provides novel insights into virus-host cell interactions that affect the functional role of macrophages in HIV disease.
Keywords: proteomics, secretome, monocyte-derived macrophages, human immunodeficiency virus, HIV-1-associated dementia
Introduction
Mononuclear phagocytes [(MP): monocytes, dendritic cells, tissue macrophages, connective tissue histiocytes, Langerhans cells of skin, Kupffer cells of liver, and microglial cells of brain] comprise the principle elements in the clearance and inactivation for microbial pathogens. Paradoxically, these same cells represent the major target and infectious reservoir for HIV-1 (Bhoopat et al., 2006; Collman et al., 2003; Gartner et al., 1986; Gendelman and Morahan, 1992; Gendelman et al., 1989; Meltzer and Gendelman, 1992). In distinct ways, HIV alters MP innate and adaptive immune functions enabling the virus to persist for long time periods (Kramer-Hammerle et al., 2005; Maslin et al., 2005; Olivetta and Federico, 2006; Palmieri et al., 2004). As a result of circumventing cell-based anti-retroviral mechanisms, reservoirs for HIV are established in bone marrow progenitor cells, circulating monocytes, alveolar and perivascular macrophages, and microglia (Collman et al., 2003; Pomerantz, 2004). Defective phagocytic, intracellular killing, secretory activity, and antigen presentation have been reported in monocytes recovered from virus-infected individuals or following in vitro infections (Capsoni et al., 1992; Evans and Wansbrough-Jones, 1996; Gordon and Read, 2002; Kedzierska and Crowe, 2002; Zhou et al., 1998).
MP-virus interactions and immune activation ultimately leads to tissue destruction. This is commonly seen in the brain, gut, lung, and spinal cord where MP soluble factor secretion contributes to viral spread and host tissue injury and inflammation (Ansari, 2004; Giulian, Vaca, and Noonan, 1990; Gordon and Read, 2002; Gupta and Gollapudi, 1993; Ichikawa et al., 2003; Kaul et al., 2005; Lim, Condez, and Poulter, 1993; McArthur, Brew, and Nath, 2005; Satomi et al., 2005; Stevenson and Gendelman, 1994). The latter events can occur by inducing cell death or changes in immune and homeostatic functions as a result of cell secretions. The functional and biological outcomes of HIV-1 MP infection also hinge on cell differentiation, as the viral life cycle is dependent on it. Understanding how such events occur is pivotal in understanding how virus can affect disease while at the same time overcome potent innate anti-retroviral immune responses.
After entering tissue, monocytes differentiate into macrophages where they maintain homeostasis, eliminate microbial pathogens, and clear debris. At the same time the cells respond to a variety of environmental cues. Environmental factors, as well as infection itself, lead to the upregulation of secreted pro-inflammatory cytokines, reactive oxygen species, quinolinic acid, glutamate, arachidonic acid and its metabolites. How these and other cellular factors contribute to a wide range of “primary” HIV-associated diseases is a subject of intense research (Colton, 1994; Colton and Gilbert, 1987; Gelbard et al., 1994; Klegeris and McGeer, 1997). To gain a better understanding of the impact by which HIV-infected macrophage affects its environment we profiled its secreted proteins. A foundation for these studies was made through the establishment of an initial list of both the human macrophage proteome and secretome (Dupont et al., 2004; Mor-Vaknin et al., 2003). We posit that HIV-1 infected macrophages could affect the secretion of new proteins or change protein abundance expressed at otherwise very low levels and under normal physiological conditions. Therefore, our study was designed to profile secretome of infected monocyte-derived macrophages (MDM) and compare the profiles seen with uninfected cells. The intent of these works was to discover proteins that are linked to disease and could, in some manner, be applied as biomarkers for the infected human host.
To achieve these goals, we utilized one-dimensional separation (1D SDS-PAGE) and tandem MS to identify MDM proteins secreted as a consequence of HIV-1 infection. Differential expression of selected proteins was further validated using Western-blot analysis. Presented data provide novel insights into the delicate homeostatic changes within the macrophage evolving from persistent viral replication and cytopathicity.
Results
After 7 days in culture, human monocytes differentiate into macrophage-like cells and readily elicit productive infection following exposure to the macrophage tropic viral strain HIV-1ADA. Productive HIV-1 replication was demonstrated by RT activity and reflected progeny virion production released into culture fluids from the infected macrophages (Table 1) (Ciborowski et al., 2004; Gendelman et al., 1988). Another measure of progressive viral infection is formation of multinucleated giant cells (MNGC) expressed as a ratio of the number of nuclei per cell, also called Giant Cell Index (GCI). Fig. 1A illustrates photomicrographs of control and HIV-1-infected MDM. Fig. 1B depicts the GCI index. Formation of MNGC occurs as early as 3 days after infection. On day 10 most MDM form large syncytia. Kinetics, but not susceptibility, of progression of HIV-1 infection is donor dependent. In contrary to T-cells, which are susceptible to apoptosis shortly after being infected with HIV-1, macrophages are more resistant to cell-death and continuously support viral replication (Stevenson and Gendelman, 1994; Vazquez et al., 2005, Wahl, Feldman, and McCarthy, 1996). HIV has been reported to turn-off macrophage apoptotic pathways following infection (Greenway and McPhee, 1997).
Table 1.
Levels of reverse transcriptase (RT) activity in 6 culture supernatants
| RT ×104 | |||||||
|---|---|---|---|---|---|---|---|
| Donor | 1 | 2 | 3 | 4 | 5 | 6 | |
| Days | 1 | ND* | ND | ND | ND | ND | ND |
| 2 | - | - | - | 29.72 | |||
| 3 | - | - | - | 46.61 | 11.42 | 18.21 | |
| 5 | - | - | - | 6.50 | 13.27 | 47.71 | |
| 7 | 5.43 | 44.32 | 52.73 | 3.90 | 32.23 | 52.76 | |
| 9 | 14.53 | 85.12 | 44.35 | - | 34.95 | 57.16 | |
| 12 | 17.30 | 64.02 | 36.61 | - | - | - | |
| 14 | 15.81 | 49.24 | 20.46 | - | - | - | |
not detected
not tested
Figure 1.
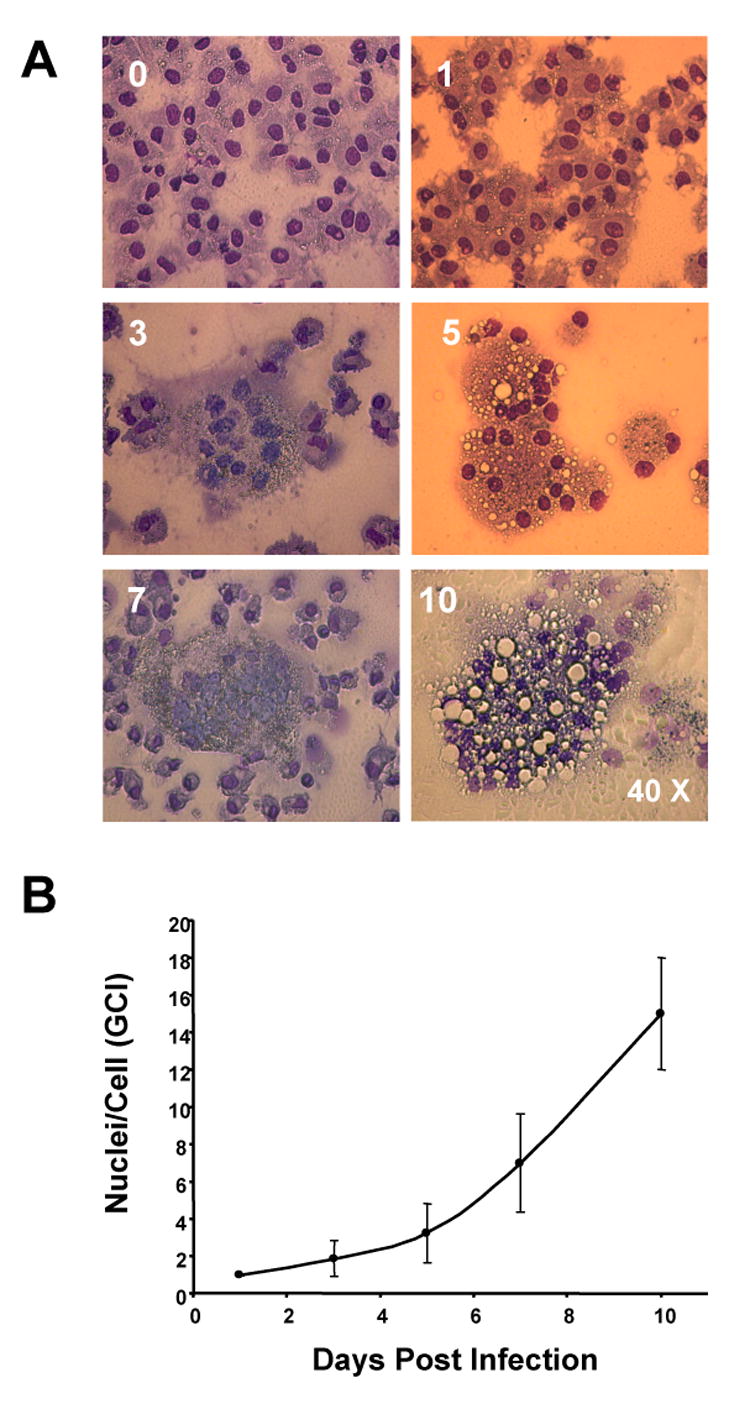
Cythopatic effects of HIV-1ADA- infected MDM. (A) Photographs of MDM culture infected with HIV-1 showing progressive MNGC formation from initiation of infection to day 10. (B) An index of the number of nuclei per cell increases along with culture progression and secondary infection of cells that were not infected during initial exposure to the virus.
Culture supernatants were collected at days 2, 3, 5, 7, 9, 12, and 14 after infection. Removal of the viral inoculum 24 hr after infection was performed by three consecutive rinses. These multiple rinses also removed any proteins that were introduced into the culture with viral inoculum during the initial period of infection. Typically, protein concentration in culture supernatants of HIV-1-infected MDM was ∼50% higher than what was observed in controls (data not shown). This difference could reflect changes in cell metabolism. Proteomic analyses were performed on selected culture fluids collected at maximal RT levels which coincided with substantial HIV-1p24 protein secreted into the culture fluids (Fig. 4). This varied amongst donors between days 3 and 9. Although in one instance the RT level was highest on day 12, this and later time points were excluded as cytosolic proteins from dying cells could be present in culture fluids (Kadiu et al., 2006).
Figure 4.
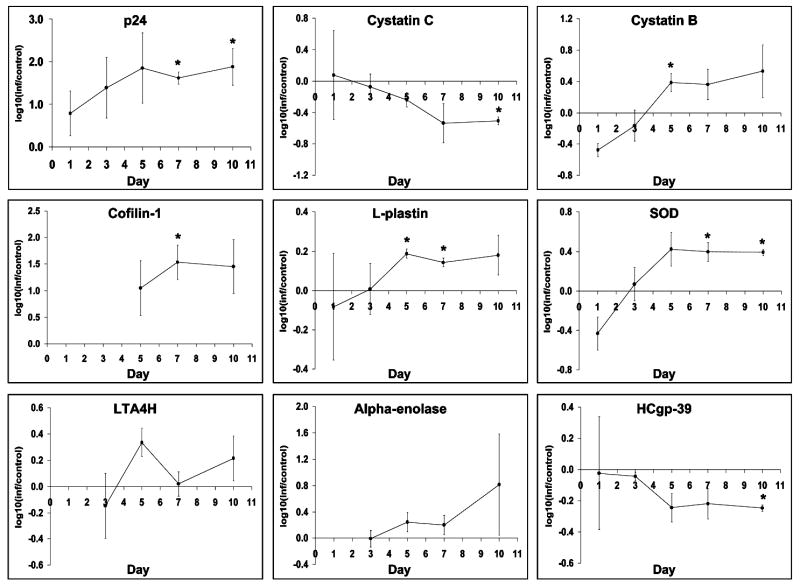
Quantitative changes in expression of 9 proteins (HIVp24, cystatin B, cystatin C, cofilin-1, L-plastin, SOD, LTA4H, α-enolase, and HCgp-39) in culture supernatants of HIV-1ADA infected MDM. Graph represent densitometric measurements of Western-blots and are expressed as log10 transformed value of change comparing to the corresponding control. Statistical significant differences (P< 0.05) in levels of expression are marked with *.
Despite exchanges of media with serum-free DMEM prior to sample collection residual serum albumin, immunoglobulins, and/or other most abundant serum proteins were present and likely retained on the cell surface. A representative 12% PAGE gel 24 cm long gel was arbitrarily divided to 20 bands indicated by black boxes (Fig. 2A). Proteins from 20 gel cubes were subjected to in-gel trypsin digestion and subsequently identified by liquid chromatography tandem mass spectroscopy (LC-MS/MS). Table 2 lists 83 proteins identified within high confidence limits. For database searches Seaquest algorithm (Bioworks 3.1SR software, ThermoElectron, Inc.) was employed. Listed proteins represent data combined from four independent experiments. Results of database searches were filtered and confidence hits accepted if sequenced peptides reached or exceeded the criteria of Unified Score of 3000 with Sp>500, Xcorr> 2.5 for doubly charged and 3.5 for triply charged precursor ions, and Delta Cn> 0.3. Identification of protein within high confidence limits was accepted as positive when a minimum of two peptides were sequenced meeting established criteria in at least two independent experiments. Peptides of 30 amino acids or longer with scattered y- or b- series ions that had assigned the Unified Score greater than 3000 but low Xcorr and DeltaCn values were excluded. HIV-1 peptides were poorly fragmented by LC-MS/MS and only few were sequenced (Table 4), which can result from either incomplete trypsin fragmentation or poor ionization. Serum albumin and immunoglobulins were excluded from the summary tables. Proteins which could originate as a residual “contamination” with human serum are marked in the table with “#.”
Figure 2.
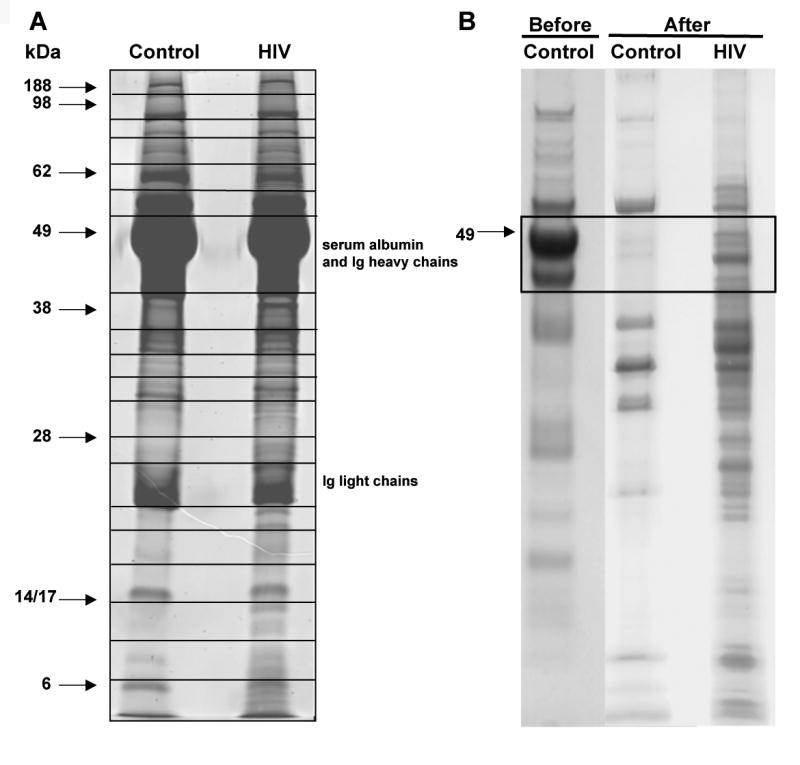
1DE SDS-PAGE analysis of secreted proteins from MDM culture supernatants. Gels were stained with SyproRuby® fluorescent dye. (A) Samples of 100 μg of protein were loaded per lane. 1DE SDS-PAGE analysis of secreted proteins before and after removal of 6 most abundant proteins: serum albumin, IgG, IgA, α-antitrypsin, haptoglobin, and transferring. (B) Samples of 60 μg of protein were loaded per lane. Black box shows region which was re-analyzed after serum albumin was removed.
Table 2.
Proteins identified in secretome of HIV-1-infected and control MDM (LC-MS/MS identification)
| HIV- infected | |||||
|---|---|---|---|---|---|
| N - uninfected | |||||
| COM - common | Protein | NCBI | Swiss Prot | M.W. | Postulated Function |
| HIV | 14-3-3 gamma protein | 5726310 | P61981 | 28,171 | Signal Transduction, Cytoskeletal organization and biogensis |
| HIV | 14-3-3 protein zeta/delta | 33872678 | P63104 | 27,745 | transcription factor binding |
| HIV | 14-3-3 protein/cytosolic phospholipase A2 | 4262000 | Q6LD62 | 7,957 | protein domain specific binding |
| 3-monooxgenase/tryptophan 5-monooxygenase | |||||
| HIV | activation protein | 31543976 | P61981 | 28,171 | regulation of mitosis, regulation of signal transduction |
| COM | Actin various forms | * | * | * | |
| COM | Aldolase A protein | 28595 | P04075 | 39,289 | fructose-bisphosphate aldolase activity, catalytic activity |
| #COM | Alpha 2 macroglobulin | 46812315 | P01023 | 163,278 | intracellular protein transport |
| COM | Alpha enolase | 2661039 | P06733 | 47,038 | phosphopyruvate hydratase activity, plasminogen activator activity |
| #COM | Alpha-1 antitrypsin | 28637 | Q13747 | 22,828 | serine-type endopeptidase inhibitor activity |
| chaperone-mediated protein complex assembly, acute-phase response, | |||||
| COM | Amyloid P component, serum | 30582339 | P02743 | 25,387 | protein folding |
| COM | Annexin A2 | 16306978 | P07355 | 38,473 | phospholipase inhibitor activity |
| COM | Apo-B100 precursor | 28780 | P04114 | 515,563 | lipid transporter activity, receptor binding |
| #COM | Apolipoprotein A-I | 37499465 | Q6Q785 | 30,778 | lipid transporter activity, protein binding |
| #COM | Apolipoprotein A-II preproprotein [Homo sapiens] | 4502149 | P02652 | 11,175 | lipid transporter activity, protein binding |
| COM | Apolipoprotein A-IV precursor [Homo sapiens] | 178757 | P06727 | 45,371 | lipid transporter activity, protein binding |
| N | Apolipoprotein C-II precursor [Homo sapiens] | 32130518 | P02655 | 11,284 | lipoprotein lipase activity |
| N | Apolipoprotein C-III precursor [Homo sapiens] | 4557323 | P02656 | 10,852 | lipoprotein lipase activity |
| COM | Apolipoprotein D precursor [Homo sapiens] | 4502163 | P05090 | 21,276 | lipid metabolism, transport |
| COM | Apolipoprotein E precursor [Homo sapiens] | 4557325 | P02649 | 36,154 | lipid transporter activity, lipid binding, tau protein binding |
| COM | Apolipoprotein H (beta-2-glycoprotein I) [Homo sapiens] | 32165624 | P02749 | 38,298 | lipid transporter activity, heparin binding |
| HIV | Cap-G [Homo sapiens] | 63252913 | P40121 | 38,518 | actin binding |
| COM | Cathepsin B preproprotein [Homo sapiens] | 22538437 | P07858 | 37,822 | intracellular degradation and turnover of proteins |
| HIV | cathepsin D preproprotein [Homo sapiens] | 4503143 | P07339 | 44,552 | aspartic-type endopeptidase activity |
| HIV | cathepsin S [Homo sapiens] | 179957 | P25774 | 37,510 | cysteine-type endopeptidase activity |
| COM | Ceruloplasmin (ferroxidase) [Homo sapiens] | 4557485 | P00450 | 122,205 | ferroxidase activity, copper ion transporter activity, oxidoreductase activity |
| extracellular matrix structural constituent, hydrolase activity | |||||
| COM | Chitinase 3-like 1 [Homo sapiens] | 25058041 | P36222 | 42,613 | hydrolyzing O-glycosyl compounds, chitinase activity |
| HIV | cofilin 1 (non-muscle) [Homo sapiens] | 30582531 | P23528 | 18,371 | actin binding, protein binding |
| HIV | cofilin 2 [Homo sapiens] | 33946278 | Q9Y281 | 18,737 | protein binding |
| COM | Cystatin B (stefin B) [Homo sapiens] | 68783 | Q76LA1 | 11,140 | endopeptidase inhibitor activity, cysteine protease inhibitor activity |
| N | Cystatin C precursor [Homo sapiens] | 4503107 | P01034 | 15,799 | cysteine protease inhibitor activity, protein homodimerization activity |
| HIV | cytosolic malate dehydrogenase [Homo sapiens] | 5174539 | P40925 | 36,295 | oxidoreductase activity; malate dehydrogenase activity |
| HIV | cytovillin 2 [Homo sapiens] | 6063151 | Q9UJZ2 | 18,419 | cytoskeletal protein binding |
| HIV | ezrin [Homo sapiens] | 6063147 | Q9UJZ6 | 19,237 | cytoskeletal protein binding |
| HIV | filamin [Homo sapiens] | 1203969 | P21333 | 280,761 | actin binding |
| HIV | filamin 2 [Homo sapiens] | 8885790 | Q14315 | 290,959 | actin binding |
| HIV | filamin A [Homo sapiens] | 53791221 | Q60FE5 | 278,226 | actin binding |
| N | FNHU fibronectin precursor [validated] - human | 279675 | P02751 | 262,607 | extracellular matrix structural constituent, protein binding |
| HIV | fructose-1,6-bisphosphatase [Homo sapiens] | 3293553 | P09467 | 36,683 | carbohydrate metabolism |
| HIV | gag protein [Human immunodeficiency virus 1] | 55416335 | % | % | |
| HIV | gag protein [Human immunodeficiency virus 1] | 33145913 | % | % | |
| HIV | gag protein [Human immunodeficiency virus 1] | 33145909 | % | % | |
| HIV | gag-pol polyprotein [Human immunodeficiency virus 1] | 21671035 | % | % | |
| COM | Gelsolin isoform a [Homo sapiens] | 4504165 | P06396 | 85,697 | actin filament polymerization, calcium ion binding |
| COM | Gelsolin isoform b [Homo sapiens] (cytoplasmic) | 38044288 | P06396 | 85,698 | actin filament polymerization, calcium ion binding |
| HIV | glutathione S-transferase omega 1-1 [Homo sapiens] | 55925946 | Q5TA03 | 27,566 | glutathione transferase activity |
| HIV | glyceraldehyde-3-phosphate dehydrogenase [Homo sapiens] | 31645 | P04406 | 35,922 | glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity |
| COM | GM2-activator protein [Homo sapiens] | 31857 | P17900 | 20,822 | sphingolipid activator protein activity |
| #COM | Haptoglobin [Homo sapiens] | 1620396 | P00739 | 39,008 | serine-type endopeptidase activity, hemoglobin binding |
| #COM | Haptoglobin [Homo sapiens] | 3337390 | P00738 | 45,205 | serine-type endopeptidase activity, hemoglobin binding |
| N | Histidine-rich glycoprotein precursor [Homo sapiens] | 4504489 | P04196 | 59,578 | cysteine protease inhibitor activity, heparin activity |
| HIV | hp2-alpha [Homo sapiens] | 296653 | Q6LBY9 | 41525 | serine-type endopeptidase activity, hemoglobin binding |
| HIV | Human Thioredoxin | 1827674 | P52566 | 11,705 | Rho GDP-dissociation inhibitor activity |
| HIV | ITH4_HUMAN Inter-alpha-trypsin inhibitor heavy chain H4 precursor | 13432192 | Q14624 | 103,358 | endopeptidase inhibitor activity |
| COM | Lactate dehydrogenase A [Homo sapiens] | 5031857 | Q6ZMR3 | 36,507 | L-lactate dehydrogenase activity, oxidoreductase activity |
| HIV | lactate dehydrogenase B [Homo sapiens] | 4557032 | Q9BYZ2 | 41,979 | L-lactate dehydrogenase activity |
| HIV | L-lactate dehydrogenase | 34527427 | P00338 | 36,558 | L-lactate dehydrogenase activity |
| HIV | Macrophage capping protein | 21730367 | P40121 | 38,524 | actin binding |
| COM | MMP9 [Homo sapiens] | 10443048 | P14780 | 78,427 | macrophage differentiation, metalloendopeptidase activity |
| HIV | moesin [Homo sapiens] | 4505257 | P26038 | 67,689 | cytoskeletal protein binding |
| HIV | NADP-dependent isocitrate dehydrogenase [Homo sapiens] | 11374664 | O75874 | 46,659 | isocitrate dehydrogenase (NADP+) activity |
| #COM | Orosomucoid 1, precursor [Homo sapiens] | 20070760 | P02763 | 23,512 | modulating the activity of the immune system during the acute-phase reaction |
| #COM | Orosomucoid 2 [Homo sapiens] | 4505529 | Q5T538 | 23,603 | signal transduction |
| COM | Peroxiredoxin 1 | 55959887 | Q06830 | 22,110 | oxidoreductase activity, peroxidase activity, antioxidant activity |
| HIV | phosphoglycerate kinase 1 [Homo sapiens] | 4505763 | P00558 | 44,483 | phosphoglycerate kinase activity |
| COM | PREDICTED: similar to FKSG30 | 51493213 | Q9BYX7 | 42,016 | structural constituent of cytoskeleton, protein binding, ATP binding |
| COM | Profilin 1 | 30582841 | Q53Y44 | 15,054 | cytoskeleton organization and biogenesis |
| HIV | proteasome activator subunit 1 | 5453990 | Q06323 | 28,723 | proteasome complex |
| HIV | proteasome activator subunit 2 | 30410792 | Q9UL46 | 27,230 | proteasome complex |
| HIV | prothymosin alpha [Homo sapiens] | 307352 | Q15203 | 8,161 | unknown |
| HIV | Rho GDP dissociation inhibitor beta [Homo sapiens] | 20379030 | P52566 | 22,988 | Rho GDP-dissociation inhibitor activity |
| endopeptidase inhibitor activity, serine-type endopeptidase inhibitor activity, | |||||
| COM | Serine (or cysteine) proteinase inhibitor | 30584253 | P01009 | 55,299 | protein binding |
| N | SH3 domain binding glutamic acid-rich protein like 3 [Homo sapiens] | 55960189 | Q5T123 | 9,381 | secreted protein |
| HIV | smooth muscle and non-muscle myosin alkali light chain isoform 1- 4 | 17986262 | P60660 | 16,799 | motor activity; calcium ion binding |
| N | Superoxide Dismutase [Homo sapiens] | 27065835 | P00441 | 15,805 | superoxide dismutase activity |
| HIV | transaldolase 1 [Homo sapiens] | 5803187 | P37837 | 37,540 | carbohydrate metabolism |
| #COM | Transferrin | 37747855 | P02787 | 77,050 | ion transport; iron ion binding |
| HIV | transferrin receptor | 4507457 | P02786 | 84,901 | transferrin receptor activity |
| HIV | Triosephosphate isomerase 1 | 17389815 | P60174 | 26,538 | triose-phosphate isomerase activity |
| HIV | tropomyosin 3 | 55665780 | Q5VU59 | 27,175 | cytoskeletal protein binding |
| HIV | Tropomyosin 4 | 12803959 | P67936 | 28,391 | cytoskeletal protein binding |
| COM | TTR | 48145933 | Q6IB96 | 15,873 | thyroid hormone-binding protein |
| tyrosine 3/tryptophan 5 -monooxygenase activation | |||||
| HIV | protein | 21735625 | P63104 | 27,745 | transcription factor binding |
| HIV | VIM | 47115317 | P08670 | 53520 | protein binding; structural constituent of cytoskeleton |
for details see Table 4
& and $ - NCBI and SwissProt accesion numbers respectively.
Multiple accesion numbers of various forms (including fragments) were obtained from dadabase searches.
Top 12 most abundant proteins found in human serum
Table 4.
LC-MS/MS identified HIV-1 proteins and found in culture supernatants of HIV-1 secreted proteins.
| Protein | NCBI accession number | Peptide sequence | MH+ | z | XCorr | DeltaCn |
|---|---|---|---|---|---|---|
| Reverse transcriptase | gi|55709508|gb|AAV58679.1| [55709508] | YQYNVLPMGWXGSPAIFQSSMTXVLEPFR | 3346.90 | 3 | 2.621 | 0.322 |
| Envelope glycoprotein | gi|62906511|gb|AAY20909.1 |[62906511] | QAYCNINATEWNNTLQEVGRELEK | 2825.06 | 3 | 2.937 | 0.353 |
| Gag protein | gi|5668939|gb|AAD46087.1| AF077336_1[5668939] | QLQPSLQTGSEELK | 1558.72 | 2 | 3.920 | 0.397 |
Two “intensely” stained protein bands ∼ 49 and 25 kDa (Fig. 2A) represented serum albumin and heavy and light chains of immunoglobulins (data not shown) in concentrated samples. Therefore, we removed albumin and the other five proteins (IgG, IgA, anti-a-trypsin, haptoglobin, and transferring) from the samples and this region was re-analyzed (Fig. 2B). After removal of high abundant proteins another 27 proteins were identified by LC-MS/MS. One of them, L-plastin, was significantly up-regulated in HIV-1 infected MDM fluids. Table 3 shows the summaries of identified proteins. The total number of identified proteins was 110.
Table 3.
Proteins identified in the range of 50 to 60 kDa in 1DE after removal of 6 most abundant proteins.
| HIV - infected; | |||||
|---|---|---|---|---|---|
| N - uninfected; | |||||
| COM - common | Protein | NCBI& | SwissProt$ | M.w. | Postulated functions |
| N | Alpha 2 macroglobulin | 46812315 | P01023 | 163,278 | Endopeptidase inhibitor activity, enzyme binding, II-1 binding II-8 binding, TNF binding, protein homooligomerization |
| HIV | ATPase, H+ transporting, lysosomal 70kDa | 15341906 | P38606 | 68,304 | Proton-transporting two-sector ATPase complex |
| HIV | BiP | 1143492 | P11021 | 72,333 | ATP binding, calcium ion binding, caspase inhibitor activity, negative regulation of caspase activity, anti-apoptosis |
| COM | Complement component 3 | 40786791 | P01024 | 187,164 | G-protein coupled receptor protein signaling pathway, immune response, signal transduction |
| COM | Gelsolin | 38044288 | P06396 | 85,698 | Actin filament polymerization, actin filament severing, barbed-end actin filament capping. |
| HIV | Heat shock 70kDa protein 8 isoform 2 | 24234686 | Q53HF2 | 53,500 | ATP binding, nucleotide binding, protein folding, response to unfolded protein |
| HIV | Heat shock 90kDa protein 1, alpha | 83699649 | Q2PP14 | 98,143 | ATP binding, nitric-oxide synthase regulator, protein homodimerization, TPR domain binding, mitochondrial trans regulation of nitric oxide biosynthesis, protein refolding, signal transduction |
| HIV | Heat shock 90kDa protein 1, beta | 56204416 | Q5T9W7 | 83,264 | Nitric-oxide synthase regulator, TPR domain binding, positive regulation of nitric oxide biosynthesis, protein refolding, unfolded protein |
| HIV | Heat shock protein | 4204880 | P54652 | 70,021 | Unfolded protein binding, male meiosis, response to unfolded protein, spermatid development |
| HIV | HSP70-2 | 4529892 | P08107 | 70,052 | Anti-apoptosis, mRNA catabolism, response to unfolded protein |
| HIV | Interferon-induced Mx protein | 4505291 | P20591 | 75,403 | GTP binding, defense response, induction of apoptosis, signal transduction |
| HIV | IQGAP1 protein | 40674640 | Q6P1N4 | 107,539 | Mediates VE-cadherin-based cell-cell contacts and VEGF signaling at adherence junctions |
| HIV | Karyopherin beta 1 | 19923142 | Q14974 | 97,170 | Zinc ion binding, NLS-bearing substrate import into nucleus, protein import into nucleus, translocation |
| COM | Leukotriene A4 hydrolase | 4505029 | P09960 | 69,154 | Epoxide hydrolase activity, peptidase activity, zinc ion binding, inflammatory response, leukotriene biosynthesis |
| COM | L-plastin | 14043359 | P13796 | 70,289 | Actin binding, calcium ion binding |
| HIV | MARCKS protein | 33877895 | Q6NVI1 | 14,859 | Interacts with the metabotropic glutamate receptor type 7 and modulates G protein-mediated constitutive inhibition |
| COM | Matrix metalloproteinase 9 | 22532481 | P14780 | 78,427 | Collagenase activity, gelatinase B activity, zinc ion binding, macrophage differentiation |
| HIV | Moesin | 5419633 | P26038 | 67,689 | Structural constituent of cytoskeleton, cell motility, receptor binding |
| HIV | Myristoylated alanine-rich protein kinase C substrate | 56203645 | P29966 | 31,423 | Actin filament binding, calmodulin binding |
| HIV | Radixin | 28436809 | P35241 | 68,564 | Binding barbed end of actin filaments, binds SLC9A3R1 |
| HIV | SEC23-related protein A | 38202214 | Q15436 | 86,147 | Vesicle-mediated transport |
| N | Transferrin receptor | 4507457 | P02786 | 84,901 | Transferrin receptor, activity iron ion homeostasis |
| COM | Transketolase | 31417921 | Q53EM5 | 67,906 | Oxygen-independent glucose metabolism |
| HIV | UNC-112 related protein 2 long form | 41281905 | Q86UX7 | 75,953 | Cell adhesion |
| HIV | Vacuolar sorting protein 35 | 9622850 | Q96QK1 | 91,707 | Retrograde transport, endosome to Golgi |
| HIV | Villin 2 (ezrin) | 66347778 | Q4VX75 | 69,413 | Cytoskeletal protein binding |
| HIV | Zyxin | 58530845 | Q15942 | 61,277 | Cell adhesion, cell-cell signaling, signal transduction |
& and $ - NCBI and SwissProt accesion numbers respectively
Although our proteomic approach was not quantitative per se, we observed differences in the relative abundance of peptides sequenced by LC-MS/MS. This indicated that these proteins could be differentially expressed as a result of viral infection. Based on this interpretation we performed quantitative Western-blot analysis in order to confirm our LC-MS/MS results. This strategy was applied successfully in our previous works (Ciborowski et al., 2004). A few examples of differentially expressed proteins are worth noting. Mass spectrometry indicated cystatin C as differentially expressed and found in culture supernatants of uninfected MDM, but not in infected cells (Table 2). Although Western-blot analysis detected the protein in both control and infected MDM, cystatin C was downregulated in infected cells making it concordant with mass spectrometry data. Cofilin-1 was found upregulated in infected MDM by mass spectrometry and confirmed by Western blot tests (Fig. 3). Using the same paradigm six differentially expressed proteins were found. This included: cystatin B, cystatin C, L-plastin, leukotrine A4 hydrolase (LTA4H), α-enolase, and chitinase 3-like 1 protein (HC-gp39). Presence of HIV-1 p24 included in Fig. 3 served as an internal control and substantiating virus production. Among selected proteins, levels of HC-gp39 were decreased, and L-plastin, LTA4H and α-enolase were increased following HIV-1 infection. The exception was superoxide dismutase (SOD), a protein shown to be up regulated in HIV-infected MDM and confirmed by Western-blot tests although LC MS/MS showed conflicting results. We posit that such results were seen as SOD co-migrates with other identified proteins such as cofilin-1 and cystatin B when analyzed on 1D SDS-PAGE. Thus, protein co-migration may lead to such discrepancies between LC-MS/MS and Western-blot results.
Figure 3.
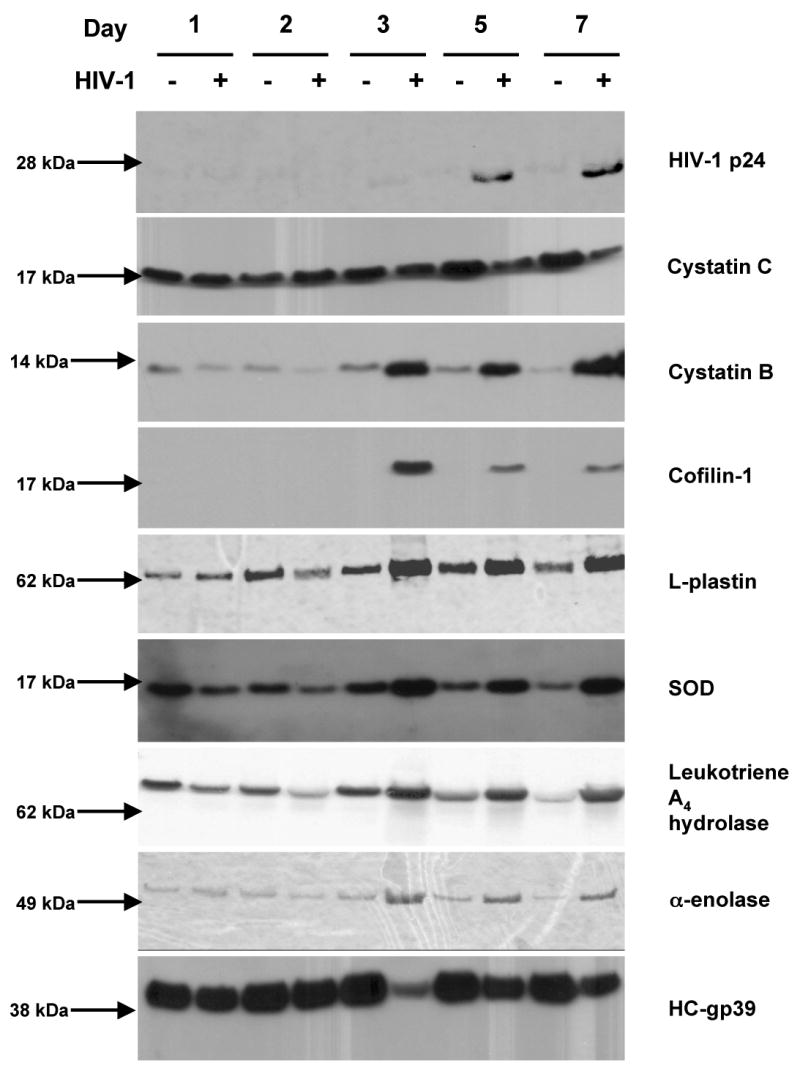
Western blot analysis of HIVp24, cystatin B, cystatin C, cofilin-1, L-plastin, SOD, LTA4H, α-enolase, and HCgp-39 in culture supernatants of a representative experiment. Samples of culture supernatants from HIVADA-1-infected and control MDM (5 to 25μg of protein per lane) were loaded on 4-12% NuPAGE gel. Separated proteins were electroblotted onto PVDF membrane and immunodetected as described in the Material and methods.
As mentioned above, the dynamic of viral infection will vary from donor to donor. In consequence, levels of differentially expressed proteins will vary and give rise to substantive standard deviations amongst data sets. Nevertheless, our densitometry analyses (Fig. 4) demonstrated that with the exception of LTA4H, all proteins showed consistent trends in expression with many time points being different with statistical significance (p<0.05) and are marked with “*”. We also expect that differential expression of more proteins will be identified using this strategy enhanced by additional steps of sample preparation and fractionation.
Discussion
HIV-1 infected MP affect the human host including, most prominently, neuronal dysfunction. This occurs by inflammatory factors and viral protein secretions (Giulian et al., 1993). Nonetheless, little is known about how viral infection triggers immunomodulatory mechanisms affecting its function in disease. The mechanisms of MNGC formation are also not well understood. To gain a better understanding of macrophage function as a consequence of HIV-1 infection, we used profiled secreted proteins by proteomics tests. Our results show that the majority of differentially expressed proteins reported in this study represent a wide range of functional groups and include structural (cytoskeleton), redox, regulatory, and enzymes. In some instances one protein was included in two groups as SOD is an enzyme and is an important component of redox regulatory mechanisms. It is important to note that following HIV-1 infection, the macrophage is stimulated and the number of secreted proteins increased. This observation supports the notion that the virus accelerates production and secretion of macrophage proteins to its advantage, and perhaps represents a coping mechanism to best contain ongoing viral replication.
Using Western-blot analysis, we confirmed differential expression of a number of proteins, which could be linked to viral replication. The roles of these proteins in HIV-1 macrophage infection are not yet fully understood and their contribution to disease can now only be speculated. With regards to HIV-1-associated cognitive impairment, it is well known that virus-infected macrophages and microglia directly contribute to disease as a consequence of inflammatory toxic secretions (Kadiu et al., 2005). The nature of these factors and their relative contribution to disease is complex and involves interactions between primary neuronal cells, glia and blood-borne macrophages (Rostasy, 2005).
Many proteins identified in this study are directly involved or associated with MP inflammatory responses. SOD follows macrophage activation as it is regulated by pro-inflammatory cytokines and independent of substrate concentrations (Ferlat and Favier, 1993; Marikovsky et al., 2003). If mechanism of SOD and other redox enzymes goes out of delicate balance, macrophage are unable to control the levels of toxic radical species which leads to tissue injury. Decreased levels of cystatin C in culture supernatants of in vitro HIV-1-infected MDM correlates with previous reports showing decreased levels of this enzyme in cerebrospinal fluid (CSF) during inflammatory processes (Nagai et al., 2000). In CSF of multiple sclerosis patients, the levels of cystatin C were decreased compared to healthy controls (Irani et al., 2006); while in CSF of patients with Alzheimer's or Creutzfeld-Jacobs diseases, cystatin C were increased (Carrette et al., 2003, Sanchez et al., 2004). Taking this together we postulate that the mechanism regulating cystatin C expression is modulated in a disease specific manner rather than being a general characteristic of inflammatory activation of macrophage in the brain. This might be indicative of a compensatory response of macrophage to ongoing productive HIV-1 infection and subsequent inflammation. HC-gp39 is a 42 kDa glycoprotein secreted by articular chondrocytes, synoviocytes and macrophages found to play a role in modulation of cytokine responses following TNF-α and IL-1 stimulation. Therefore, decreased expression of HC-gp39 may lead to enhancement of pro-inflammatory effects and profound toxicities. Leukotrienes are made predominantly by polymorphonuclear leukocytes, macrophages, and mast cells. LTA4H catalyzes the hydrolysis of leukotriene A4 (LTA4) into leukotrine B4 (LTB4). LTB4 is a lipid mediator with roles in immune defense, inflammation, and disease. It is a potent chemo-attractant and mediator of inflammation (Funk, 2001), stimulator of adhesion to endothelium (Tonnesen et al., 1989), and degranulation (Dahlen et al., 1981). It also stimulates generation of superoxide anion (Claesson and Feinmark, 1984) and phagocytosis. Overproduced LTB4 contributes to the pathogenesis of a variety of inflammatory disorders (Goetzl, An, and Smith, 1995).
It is intriguing that cellular proteins, such as cofilin-1 and L-plastin, are found secreted by HIV-1-infected macrophages. Cofilin-1 was not detected or detected at very low levels in culture fluids of control MDM and is present in significant levels in culture supernatants of infected counterparts. Ott and colleagues (Ott, 2002) postulated that structural proteins such as actin, cofilin, ezrin, and moesin are involved in virus assembly and budding, are carried specifically with viral particles, and were not found in vesicles. An H9 T-cell lymphoma cell lines was used as a model for these studies in which virus is released by budding from cell membrane. In our studies, MDM were used where virus budding occurs through the exosomal pathway (Nguyen et al., 2003). Both studies indicate that cofilin-1 is involved in viral assembly and budding suggest that although virus release has different pathways, there are common but yet unknown elements regulating this process. Inhibition of viral assembly utilizing such elements can be used for targeting both types of cells in which HIV-1 replicates.
Presented work provides a systematic proteomic approach leading to a better understanding of the impact of HIV-1 on regulation of MP, which are critical components of innate immunity. In a previously published study of macrophage secretome using 2-dimensional electrophoresis and MALDI-TOF protein fingerprinting, the authors reported 66 identifications based on accession numbers; but many of these identifications were duplicates such as actin, serum albumin, apolipoproteins, etc. (Dupont et al., 2004). In our current study, we did not find all previously reported proteins; however we found proteins that were not reported by Dupont and colleagues (Dupont et al., 2004). It is important to note that our validation Western-blot analyses showed significant and important differences in expression of several proteins which were identified in samples from infected and control MDM. Identification of functional roles of proteins, which seems to be typical cytoplasmic such as α-enolase and cofilin-1 and which we found to be at relatively higher levels in the extracellular milieu due to HIV-1 infection of MDM, may lead to better understanding mechanisms of how macrophage respond to viral infection. These proteins may have yet unknown functions, specific to extracellular localization, or can be specifically incorporated in HIV-1 particles. Moreover, the fact that proteins belonging to various classes can be regulated in macrophages following viral infection highlights the broad range of cellular processes controlled by the virus.
Proteomic analysis of MDM secretions induced by HIV-1 infection identifies a spectrum of proteins that can be explored for potential use as a broader panel of biomarkers. Multidimensional fractionation protocols of samples lead to identification of constantly increasing number of proteins. Nevertheless, huge dynamic range of proteins concentrations (Anderson and Anderson, 2002) and sensitivity of mass spectrometers remain problems in proteomic studies. Although, in this study we did not detect low abundant proteins such as cytokines and/or chemokines, we did identify differential regulation of many proteins that have not been reported in the literature. Determining whether an individual differentially expressed protein will serve as a biomarker of an inflammatory process in the brain or will be a part of a group of biomarkers with value in predicting clinical outcome will require a larger study using CSF samples from individuals whose clinical status of neuropathology is well defined. Such study is currently undergoing in our laboratory. The application of proteomic approaches are likely to be of continued importance in studies of macrophage function and gene regulation leading to a better understanding of disease processes and the means to combat them.
Material and Methods
Isolation, cultivation, and HIV-1 infection of human MDM
Peripheral blood mononuclear cells were isolated from HIV-1, 2 and hepatitis seronegative donors by leukophoresis and purified by countercurrent centrifugal elutriation (Gendelman et al., 1988). Purified monocytes were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, Carlsbad, CA) supplemented with 10% heat-inactivated pooled human serum, 1% glutamine, 50 μg/ml gentamicin (Sigma Chemical Co., St. Louis, MO), 10μg/ml ciprofloxacin (Sigma) and 1000 U/ml highly purified recombinant human macrophage colony stimulating factor (MCSF; a generous gift from Wyeth, Inc., Cambridge, MA). Cells were maintained at 37°C with 5% CO2 and half medium exchange was performed every two days. After seven days in culture, MDM were infected with HIV-1ADA at a multiplicity of infection of 0.1 or left uninfected as controls. Cells were exposed to viral stock for 4 hr at 1/5 of the total medium volume followed by an additional incubation time of 20 hr in full volume of medium. After 24 hr of exposure to viral stock, inoculum was removed, cells were washed twice, and fresh full medium was added to the cultures. Twenty-four hours prior to media collection for proteomic analyses, full medium was removed, cells were rinsed twice with PBS, and freshly prepared serum-free DMEM was added to the cultures (Ciborowski et al., 2004; Ghorpade et al., 2001).
Cytopathicity
To determine levels of cytopathicity by determining the GCI in HIV-1 infected MDM, monocytes were cultured in 4-well Lab-Tech chamber slides (Invitrogen, Carlsbad, CA) and treated as adherent cultures described above. Giemsa staining was performed on HIV-1ADA infected and control MDM according to the manufacturer's protocol (Fisher Scientific, Pittsburgh, PA) to assess number of nuclei per cells.
Sample processing
Collected culture fluids were centrifuged at 6000 × g for 10 min to remove any detached cells which might be collected with the fluids. Such prepared culture supernatants were supplemented with a cocktail of protease inhibitors (Sigma, St. Louis, MO) and 0.1% Triton X-100 (Fisher Scientific, Pittsburg, PA). For reverse transcriptase activity measurements, aliquots of culture supernatants were set aside without adding detergent and inhibitors. Protein concentrations were determined using Bio-Rad DC Protein Assay (Bio-Rad, Hercules, CA) and then samples were concentrated to ∼2 mg/ml with an Ultrafree-CL centrifugal filter device with 5000 Da cutoff membrane (Millipore Corporation, Bedford, MA) unless stated otherwise. Concentrated supernatants were dialyzed 18 hr against water at 4°C using QuixSep® micro dialyzers (Membrane Filtration Products Inc., Seguin, TX) and protein concentrations were measured again using a Bio-Rad DC Protein Assay.
Multiple Affinity Column, 4.6 × 50 mm Hu-6 (Agilent Technologies, Inc., Santa Clara, CA) was used to remove the 6 most abundant proteins (serum albumin, IgG, antitrypsin, IgA, transferrin, haptoglobin) from concentrated culture supernatants following manufacturer recommendations. Briefly, 700 μl of concentrated culture supernatant was mixed with Buffer A and loaded onto the column. A flow through fraction was collected and column was regenerated using Buffer B. To collect sufficient amounts of protein for analysis, a total amount of 2.1 ml (3 injections, 700 μl each) was processed for each sample.
Measures of RT activity
RT measured levels measured the numbers of progeny virus and is reflective of productive viral replication. RT activity was determined in culture fluids in triplicates. Reaction mixture consisting of 0.05% Nonidet P-40 (Sigma), 10 μg/ml poly(A) oligonucleotide, 0.25 μg/ml oligo(dT) (Amersham/Pharmacia Biotech, Piscataway, NJ), 5 mM dithiothreitol (Amersham Pharmacia), 150 mM KCl, 15 mM MgCl2, and 10μCi/ml [3H] dTTP (Perkin Elmer, Wellesley, MA) in 50 mM Tris-HCl buffer (pH 7.9) was incubated with samples for 18 hr at 37°C. Radiolabeled nucleotides were precipitated with cold 10% trichloroacetic acid and 95% ethanol using an automatic cell harvester (Skatron Inc., Sterling, VA) and were quantitated by liquid scintillation spectroscopy (Kalter et al., 1991).
One Dimensional Electrophoresis (1-DE) and Western-blot assays
1-DE SDS-PAGE separations were performed using BioRad Proteome IIxi system and 4-20% gradient gel. For sequencing, a sample of 200 μg of denaturated and reduced proteins were loaded per lane. Gels were stained with Brilliant Blue G-Colloidal Coomassie (Sigma), de-stained following the protocol provided by the manufacturer, and digitized using a Typhoon 9410 Phosphoimager (Amersham Biosciences). For Western-blot analyses, 5 to 25μg of protein was loaded per lane of 4-12% NuPAGE gel (Invitrogen, Carlsbad, CA). Electrophoresis followed by transfer and immunodetection was performed as previously described (Ciborowski et al., 2004).
Antibodies
The following primary antibodies were those directed against cystatin B (Biovision, Mountain View, CA), HIV-1 p24 (DakoCytomation, Carpinteria, CA), cystatin C (U.S. Biological, Swampscott, MA), HC-gp39 (R&D Systems, Minneapolis, MN), cofilin-1 (BD Transduction Laboratories, Lexington, KY), L-plastin (Biovision, Mountain View, CA), SOD (Calbiochem, La Jolla, CA), Leukotriene A4 hydrolase (Cayman Chemical, Ann Arbor, MI), and α-enolase (Santa Cruz, Santa Cruz, CA). Secondary antibodies included HRP-conjugated F(ab')2 fragment of Goat anti-Mouse Fcγ and Cy5-conjugated F(ab')2 fragment of Donkey anti-Mouse IgG (Jackson Immunoresearch, West Grove, PA), HRP-conjugated anti-Rabbit IgG (Cell Signaling Technology, Beverly, MA), and HRP-conjugated anti-goat IgG and HRP-conjugated anti-Sheep IgG (U.S. Biological). All antibodies were used at concentrations recommended by manufacturers.
In-gel trypsin digestion
Gel pieces excised from 1-DE were cut into 20 fragments and destained at room temperature using 200 μl 50% ACN/50 mM NH4HCO3 for 1 hr. Gel pieces were dried, and 1μg trypsin in 50 mM NH4HCO3 buffer (Promega, Madison, WI) was added to each piece. Tryptic digestion was carried out overnight at 37°C. Peptides were extracted by washing gel pieces twice with 60% acetonitrile and 0.1% trifluoroacetic acid, transferred to clean vials, dried, and re-suspended in 12 μl of water with 0.1% formic acid for mass spectrometric analysis. All samples were purified using ZipTip® (Millipore Corporation) prior to mass spectrometric analysis.
ESI-LC-Tandem Mass Spectrometry and search parameters
Samples of in-gel trypsin digested proteins were fractionated on microcapillary RP-C18 column (New Objectives, Woburn, MA) and resulting peptides were sequenced using ESI-LC-MS/MS system (ProteomeX System with LCQDecaPlus mass spectrometer, ThermoElectron, Inc., San Jose, CA) with a nano-spray configuration. The spectra obtained from LC-MS/MS analysis were searched against the NCBI.fasta protein database narrowed to a subset of human proteins using Sequest search engine (BioWorks 3.1SR software from ThermoElectron, Inc.). In the TurboSEQUEST Search Parameters, Threshold for Dta generation was 10000, Precursor Mass Tolerance for .Dta Generation was 1.4. For .Dta Search, Peptide Tolerance was 1.5 and Fragment Ions Tolerance was 0.00. Charge state was set on “Auto.” Database nr.fasta was retrieved from ftp.ncbi.nih.gov and used to create “in-house” an indexed human.fasta.idx with the following 5 key words: Homo, sapiens, human, man, primate and excluding keratins and cytokeratins. We have excluded keratins from our database search based on observations that these proteins can be just contaminants acquired during gels and samples processing. Database was updated every other week throughout of this study. Oxidated methionine was allowed in searches.
Densitometry and statistical analyses
Differential expression of 9 proteins was analyzed by Western-blot (Western Pico, Pierce, Inc. Rockford, IL) and results were transformed into numerical values using densitometry measurement of X-ray films. Densitometry was performed using ImageQuant 5.2 (GE Healthcare, Inc.). Samples were from 3 donors, in infected and control samples on days 1, 3, 5 7, and 10. Sample from one donor was measured twice and the results were averaged and treated as one biologic replicate for analysis. Data was normalized from each analysis by taking the fold change as infected/control on each day for each experiment. The data was then transformed on the log10 scale so that a ratio of 1 on the original scale becomes 0 on the log10 scale, ratios greater than 1 are then positive numbers on the log10 scale and ratios less than 1 are negative on the log10 scale. The experiments from Donor 5 were then averaged giving one set of values to be analyzed. One sample t-tests were then used to compare the average log10(ratio) to 0 for each day. P-values less than 0.05 are considered to be statistically significant.
Acknowledgments
The work was supported by NIH Grants 5 R37 NS36126-07, 5 R01 NS034239-10, 1 P01 NS43985-01A1, 5 P01NS31492-11, 5 R01 MH64570-03, P01 NS11766-28 (to H.E.G.); 20 RR15635 from the COBRE Program of the National Center for Research Resources (to P.C. and H.E.G.); and 1R21 MH075662-01 (to P.C.). The authors are grateful to Ms. Robin Taylor for outstanding administrative and computer support.
Abbreviations
- MP
mononuclear phagocytes
- HIV-1
human immunodeficiency virus type-1
- MDM
monocyte derived macrophages
- SOD
superoxide dismutase
- HCgp-39
chitinase 3-like 1 protein
- LTA4H
leukotriene A4 hydrolase
- DMEM
Dulbecco's Modified Eagle Medium
- MCSF
macrophage colony stimulating factor
- RT
reverse transcriptase
- 1-DE
one-dimensional electrophoresis
- MS/MS
tandem mass spectrometry
- ROS
reactive oxygen species
- LTA4
leukotriene A4
- LTB4
leukotrine B4
- MNGC
multinucleated giant cells
- GCI
Giant Cell Index
- mAb
monoclonal antibody
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- Ansari AA. Autoimmunity, anergy, lentiviral immunity and disease. Autoimmun Rev. 2004;3(78):530–40. doi: 10.1016/j.autrev.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Bhoopat L, Rithaporn TS, Khunamornpong S, Bhoopat T, Taylor CR, Thorner PS. Cell reservoirs in lymph nodes infected with HIV-1 subtype E differ from subtype B: identification by combined in situ polymerase chain reaction and immunohistochemistry. Mod Pathol. 2006;19(2):255–63. doi: 10.1038/modpathol.3800527. [DOI] [PubMed] [Google Scholar]
- Capsoni F, Minonzio F, Ongari AM, Rizzardi GP, Lazzarin A, Zanussi C. Monocyte-derived macrophage function in HIV-infected subjects: in vitro modulation by rIFN-gamma and rGM-CSF. Clin Immunol Immunopathol. 1992;62(2):176–82. doi: 10.1016/0090-1229(92)90070-5. [DOI] [PubMed] [Google Scholar]
- Carrette O, Demalte I, Scherl A, Yalkinoglu O, Corthals G, Burkhard P, Hochstrasser DF, Sanchez JC. A panel of cerebrospinal fluid potential biomarkers for the diagnosis of Alzheimer's disease. Proteomics. 2003;3(8):1486–94. doi: 10.1002/pmic.200300470. [DOI] [PubMed] [Google Scholar]
- Ciborowski P, Enose Y, Mack A, Fladseth M, Gendelman HE. Diminished matrix metalloproteinase 9 secretion in human immunodeficiency virus-infected mononuclear phagocytes: modulation of innate immunity and implications for neurological disease. J Neuroimmunol. 2004;157(12):11–16. doi: 10.1016/j.jneuroim.2004.08.033. [DOI] [PubMed] [Google Scholar]
- Claesson HE, Feinmark SJ. Relationship of cyclic-AMP levels in leukotriene B4-stimulated leukocytes to lysosomal enzyme release and the generation of superoxide anions. Biochim Biophys Acta. 1984;804(1):52–7. doi: 10.1016/0167-4889(84)90098-3. [DOI] [PubMed] [Google Scholar]
- Collman RG, Perno CF, Crowe SM, Stevenson M, Montaner LJ. HIV and cells of macrophage/dendritic lineage and other non-T cell reservoirs: new answers yield new questions. J Leukoc Biol. 2003;74(5):631–4. doi: 10.1189/jlb.0703357. [DOI] [PubMed] [Google Scholar]
- Colton CA. Microglial oxyradical production: causes and consequences. Neuropathol Appl Neurobiol. 1994;20(2):208–9. [PubMed] [Google Scholar]
- Colton CA, Gilbert DL. Production of superoxide anions by a CNS macrophage, the microglia. FEBS Lett. 1987;223(2):284–8. doi: 10.1016/0014-5793(87)80305-8. [DOI] [PubMed] [Google Scholar]
- Dahlen SE, Bjork J, Hedqvist P, Arfors KE, Hammarstrom S, Lindgren JA, Samuelsson B. Leukotrienes promote plasma leakage and leukocyte adhesion in postcapillary venules: in vivo effects with relevance to the acute inflammatory response. Proc Natl Acad Sci U S A. 1981;78(6):3887–91. doi: 10.1073/pnas.78.6.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont A, Tokarski C, Dekeyzer O, Guihot AL, Amouyel P, Rolando C, Pinet F. Two-dimensional maps and databases of the human macrophage proteome and secretome. Proteomics. 2004;4(6):1761–78. doi: 10.1002/pmic.200300691. [DOI] [PubMed] [Google Scholar]
- Evans MR, Wansbrough-Jones MH. Alveolar macrophage activation in HIV infection. J Infect. 1996;33(2):91–4. doi: 10.1016/s0163-4453(96)92967-9. [DOI] [PubMed] [Google Scholar]
- Ferlat S, Favier A. Tumor necrosis factor (TNF) and oxygen free radicals: potential effects for immunity. C R Seances Soc Biol Fil. 1993;187(3):296–307. [PubMed] [Google Scholar]
- Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294(5548):1871–5. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233(4760):215–9. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- Gelbard HA, Nottet HS, Swindells S, Jett M, Dzenko KA, Genis P, White R, Wang L, Choi YB, Zhang D, et al. Platelet-activating factor: a candidate human immunodeficiency virus type 1-induced neurotoxin. J Virol. 1994;68(7):4628–35. doi: 10.1128/jvi.68.7.4628-4635.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendelman HE, Morahan PM. The macrophage in viral infections. In: Lewis CE, McGee J, editors. The Natural Immune System Series: The Macrophage. Oxford University Press; London: 1992. pp. 156–232. [Google Scholar]
- Gendelman HE, Orenstein JM, Baca LM, Weiser B, Burger H, Kalter DC, Meltzer MS. The macrophage in the persistence and pathogenesis of HIV infection. Aids. 1989;3(8):475–95. doi: 10.1097/00002030-198908000-00001. [DOI] [PubMed] [Google Scholar]
- Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, Wahl LA, Lane HC, Fauci AS, Burke DS, et al. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med. 1988;167(4):1428–41. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorpade A, Persidskaia R, Suryadevara R, Che M, Liu XJ, Persidsky Y, Gendelman HE. Mononuclear phagocyte differentiation, activation, and viral infection regulate matrix metalloproteinase expression: implications for human immunodeficiency virus type 1-associated dementia. J Virol. 2001;75(14):6572–83. doi: 10.1128/JVI.75.14.6572-6583.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulian D, Corpuz M, Chapman S, Mansouri M, Robertson C. Reactive mononuclear phagocytes release neurotoxins after ischemic and traumatic injury to the central nervous system. J Neurosci Res. 1993;36(6):681–93. doi: 10.1002/jnr.490360609. [DOI] [PubMed] [Google Scholar]
- Giulian D, Vaca K, Noonan CA. Secretion of neurotoxins by mononuclear phagocytes infected with HIV-1. Science. 1990;250(4987):1593–6. doi: 10.1126/science.2148832. [DOI] [PubMed] [Google Scholar]
- Goetzl EJ, An S, Smith WL. Specificity of expression and effects of eicosanoid mediators in normal physiology and human diseases. Faseb J. 1995;9(11):1051–8. doi: 10.1096/fasebj.9.11.7649404. [DOI] [PubMed] [Google Scholar]
- Gordon SB, Read RC. Macrophage defences against respiratory tract infections. Br Med Bull. 2002;61:45–61. doi: 10.1093/bmb/61.1.45. [DOI] [PubMed] [Google Scholar]
- Greenway A, McPhee D. HIV1 Nef: the Machiavelli of cellular activation. Res Virol. 1997;148(1):58–64. doi: 10.1016/s0923-2516(97)81915-2. [DOI] [PubMed] [Google Scholar]
- Gupta S, Gollapudi S. P-glycoprotein (MDR 1 gene product) in cells of the immune system: its possible physiologic role and alteration in aging and human immunodeficiency virus-1 (HIV-1) infection. J Clin Immunol. 1993;13(5):289–301. doi: 10.1007/BF00920237. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Sugita M, Takahashi M, Satomi M, Takeshita T, Araki T, Takahashi H. Breast milk macrophages spontaneously produce granulocyte-macrophage colony-stimulating factor and differentiate into dendritic cells in the presence of exogenous interleukin-4 alone. Immunology. 2003;108(2):189–95. doi: 10.1046/j.1365-2567.2003.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani DN, Anderson C, Gundry R, Cotter R, Moore S, Kerr DA, McArthur JC, Sacktor N, Pardo CA, Jones M, Calabresi PA, Nath A. Cleavage of cystatin C in the cerebrospinal fluid of patients with multiple sclerosis. Ann Neurol. 2006;59(2):237–47. doi: 10.1002/ana.20786. [DOI] [PubMed] [Google Scholar]
- Kadiu I, Ciborowski P, Ricardo-Dukelow M, Gendelman HE. 13th CROI; Denver, CO. 2006. [Google Scholar]
- Kadiu I, Glanzer JG, Kipnis J, Gendelman HE, Thomas MP. Mononuclear phagocytes in the pathogenesis of neurodegenerative diseases. Neurotox Res. 2005;8(12):25–50. doi: 10.1007/BF03033818. [DOI] [PubMed] [Google Scholar]
- Kalter DC, Nakamura M, Turpin JA, Baca LM, Hoover DL, Dieffenbach C, Ralph P, Gendelman HE, Meltzer MS. Enhanced HIV replication in macrophage colony-stimulating factor-treated monocytes. J Immunol. 1991;146(1):298–306. [PubMed] [Google Scholar]
- Kaul M, Zheng J, Okamoto S, Gendelman HE, Lipton SA. HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death Differ. 2005;12 1:878–92. doi: 10.1038/sj.cdd.4401623. [DOI] [PubMed] [Google Scholar]
- Kedzierska K, Crowe SM. The role of monocytes and macrophages in the pathogenesis of HIV-1 infection. Curr Med Chem. 2002;9(21):1893–903. doi: 10.2174/0929867023368935. [DOI] [PubMed] [Google Scholar]
- Klegeris A, McGeer PL. beta-amyloid protein enhances macrophage production of oxygen free radicals and glutamate. J Neurosci Res. 1997;49(2):229–35. doi: 10.1002/(sici)1097-4547(19970715)49:2<229::aid-jnr11>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Kramer-Hammerle S, Rothenaigner I, Wolff H, Bell JE, Brack-Werner R. Cells of the central nervous system as targets and reservoirs of the human immunodeficiency virus. Virus Res. 2005;111(2):194–213. doi: 10.1016/j.virusres.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Lim SG, Condez A, Poulter LW. Mucosal macrophage subsets of the gut in HIV: decrease in antigen-presenting cell phenotype. Clin Exp Immunol. 1993;92(3):442–7. doi: 10.1111/j.1365-2249.1993.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikovsky M, Ziv V, Nevo N, Harris-Cerruti C, Mahler O. Cu/Zn superoxide dismutase plays important role in immune response. J Immunol. 2003;170(6):2993–3001. doi: 10.4049/jimmunol.170.6.2993. [DOI] [PubMed] [Google Scholar]
- Maslin CL, Kedzierska K, Webster NL, Muller WA, Crowe SM. Transendothelial migration of monocytes: the underlying molecular mechanisms and consequences of HIV-1 infection. Curr HIV Res. 2005;3(4):303–17. doi: 10.2174/157016205774370401. [DOI] [PubMed] [Google Scholar]
- McArthur JC, Brew BJ, Nath A. Neurological complications of HIV infection. Lancet Neurol. 2005;4(9):543–55. doi: 10.1016/S1474-4422(05)70165-4. [DOI] [PubMed] [Google Scholar]
- Meltzer MS, Gendelman HE. Mononuclear phagocytes as targets, tissue reservoirs, and immunoregulatory cells in human immunodeficiency virus disease. Curr Top Microbiol Immunol. 1992;181:239–63. doi: 10.1007/978-3-642-77377-8_9. [DOI] [PubMed] [Google Scholar]
- Mor-Vaknin N, Punturieri A, Sitwala K, Markovitz DM. Vimentin is secreted by activated macrophages. Nat Cell Biol. 2003;5(1):59–63. doi: 10.1038/ncb898. [DOI] [PubMed] [Google Scholar]
- Nagai A, Murakawa Y, Terashima M, Shimode K, Umegae N, Takeuchi H, Kobayashi S. Cystatin C and cathepsin B in CSF from patients with inflammatory neurologic diseases. Neurology. 2000;55(12):1828–32. doi: 10.1212/wnl.55.12.1828. [DOI] [PubMed] [Google Scholar]
- Nguyen DG, Booth A, Gould SJ, Hildreth JE. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J Biol Chem. 2003;278(52):52347–54. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- Olivetta E, Federico M. HIV-1 Nef protects human-monocyte-derived macrophages from HIV-1-induced apoptosis. Exp Cell Res. 2006;312(6):890–900. doi: 10.1016/j.yexcr.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Ott DE. Potential roles of cellular proteins in HIV-1. Rev Med Virol. 2002;12(6):359–74. doi: 10.1002/rmv.367. [DOI] [PubMed] [Google Scholar]
- Palmieri C, Trimboli F, Puca A, Fiume G, Scala G, Quinto I. Inhibition of HIV-1 replication in primary human monocytes by the IkappaB-alphaS32/36A repressor of NF-kappaB. Retrovirology. 2004;1(1):45. doi: 10.1186/1742-4690-1-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RJ. Ludwik Hirszfeld Memorial Lecture: HIV-1 reservoirs: major molecular obstacles to viral eradication. Arch Immunol Ther Exp (Warsz) 2004;52(5):297–306. [PubMed] [Google Scholar]
- Rostasy KM. Inflammation and neuroaxonal injury in multiple sclerosis and AIDS dementia complex: implications for neuroprotective treatment. Neuropediatrics. 2005;36(4):230–9. doi: 10.1055/s-2005-865864. [DOI] [PubMed] [Google Scholar]
- Sanchez JC, Guillaume E, Lescuyer P, Allard L, Carrette O, Scherl A, Burgess J, Corthals GL, Burkhard PR, Hochstrasser DF. Cystatin C as a potential cerebrospinal fluid marker for the diagnosis of Creutzfeldt-Jakob disease. Proteomics. 2004;4(8):2229–33. doi: 10.1002/pmic.200300799. [DOI] [PubMed] [Google Scholar]
- Satomi M, Shimizu M, Shinya E, Watari E, Owaki A, Hidaka C, Ichikawa M, Takeshita T, Takahashi H. Transmission of macrophage-tropic HIV-1 by breast-milk macrophages via DC-SIGN. J Infect Dis. 2005;191(2):174–81. doi: 10.1086/426829. [DOI] [PubMed] [Google Scholar]
- Stevenson M, Gendelman HE. Cellular and viral determinants that regulate HIV-1 infection in macrophages. J Leukoc Biol. 1994;56(3):278–88. doi: 10.1002/jlb.56.3.278. [DOI] [PubMed] [Google Scholar]
- Tonnesen MG, Anderson DC, Springer TA, Knedler A, Avdi N, Henson PM. Adherence of neutrophils to cultured human microvascular endothelial cells. Stimulation by chemotactic peptides and lipid mediators and dependence upon the Mac-1, LFA-1, p150,95 glycoprotein family. J Clin Invest. 1989;83(2):637–46. doi: 10.1172/JCI113928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez N, Greenwell-Wild T, Marinos NJ, Swaim WD, Nares S, Ott DE, Schubert U, Henklein P, Orenstein JM, Sporn MB, Wahl SM. Human immunodeficiency virus type 1-induced macrophage gene expression includes the p21 gene, a target for viral regulation. J Virol. 2005;79(7):4479–91. doi: 10.1128/JVI.79.7.4479-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl SM, Feldman GM, McCarthy JB. Regulation of leukocyte adhesion and signaling in inflammation and disease. J Leukoc Biol. 1996;59(6):789–96. doi: 10.1002/jlb.59.6.789. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Kurihara T, Ryseck RP, Yang Y, Ryan C, Loy J, Warr G, Bravo R. Impaired macrophage function and enhanced T cell-dependent immune response in mice lacking CCR5, the mouse homologue of the major HIV-1 coreceptor. J Immunol. 1998;160(8):4018–25. [PubMed] [Google Scholar]