Abstract
Bacteriophage Φ6 has a genome of three segments of double-stranded RNA. The segments are designated S, M, and L. Each segment has a unique packaging site, pac, near the 5′ end of the plus strand. The plus strands of the segments are normally packaged in the order S, M, L. Chimeras of segment M and S in which segment M is at the 5′ end of the plus strand can be stably incorporated into the virion; however, an independent segment S must be included along with normal segment L, even if it contains no active genes. A chimera of segment M and S in which segment S is at the 5′ end of the plus strand can be stably incorporated into the virion along with normal segment L to form a two-segment genome. A chimera of segments S, M, and L in which the packaging sequence is that of S can also form a stable nonsegmented genome. These findings are consistent with a model that we have proposed for the packaging of the Φ6 genome.
Keywords: genomic packaging, Reoviridae
Most plus strand RNA viruses contain one or two molecules of single-stranded (ss) RNA, and it is believed that the capsid assembles around the RNA (1). Several classes of virus carry a larger number of chromosomes within the same particle. The Reoviridae contain 10, 11, or 12 unique double-stranded (ds) RNA chromosomes in each virion (2). There is a consensus that the genomic content of each particle is uniform and stoichiometric (3). Influenza virus contains about 12 RNA segments per particle, but the complete complement is 8. The packaging of flu RNA is believed to be random (4) (but see ref. 5). Φ6 is the only bacteriophage that contains dsRNA (6). It has three unique chromosomes per particle, and the packaging seems to be very precise (7). Plus strand copies of the genomic segments are packaged into preformed procapsids (8). Each of the three plus strands has a unique sequence of about 200 nucleotides near the 5′ end that, along with the 5′ end, is necessary and sufficient for packaging. These are called pac sites, and they have no sequence similarity with each other. Although the packaging of the normal sized genomic segments is precise, we have found that a segment that contains a deletion that reduces its size to half that of normal is packaged as two segments so as to result in a constant mass for each segment class (9, 10). A segment such as L can be divided into two segments, each with different genes but both with the pac sequence of L. In that case virions contain four segments, one each of S and M and two of L.
We have proposed a model for the genomic packaging of Φ6 (11). Packaging of the genomic segments is serially dependent in that segment S is packaged first, then segment M is packaged, and finally segment L is packaged. The model states that the empty procapsid has binding sites for segment S on the outside. The plus strand of segment S is the first to be incorporated into the procapsid. When it is completely packaged, the binding sites for S on the outside of the particle are lost and binding sites for M appear. The trigger for the change in binding sites is the packaging of RNA of a mass equal to that of normal S. If S is smaller than normal then multiple copies are packaged until the correct mass is attained. When segment M is packaged, the binding sites for M disappear and the binding sites for L are exposed. Once segment L is packaged, minus strand synthesis commences. If segment S is constructed to be equal in size to the sum of S and M, then packaging of this segment will lead to the exclusion of additional segment M and the subsequent packaging of segment L. If segment S is constructed to be equal in size to the entire genome of Φ6 it should be packaged, and it should turn on minus strand synthesis. These predictions were shown to hold for in vitro packaging of segments (11). In the present paper, we show that the predictions hold for viable phage.
MATERIALS AND METHODS
Bacterial Strains and Plasmids.
Escherichia coli strain JM109 (12) was used for the propagation of all plasmids. Pseudomonas syringae p. phaseolicola HB10Y (HB) is the normal host of Φ6 (6). Plasmid pLM450 contains a cDNA copy of genomic segment L of the virus Φ6, encoding the four procapsid proteins P1, P2, P4, and P7 (13). Plasmids pLM659, pLM656, and pLM687 contain cDNA copies of the genomic segments S, M, and L, respectively, in the pT7T3 19U vector (14). Plasmids that produce transcripts with deletions or transcripts that are chimeras of segments S, M, and L were constructed by using standard techniques (15) and are shown in Fig. 1. Plasmid pKT230 is a wide host range plasmid that replicates in HB (16). The derivatives of pT7T3 19U were ligated to pKT230 at the EcoRI site for propagation in HB.
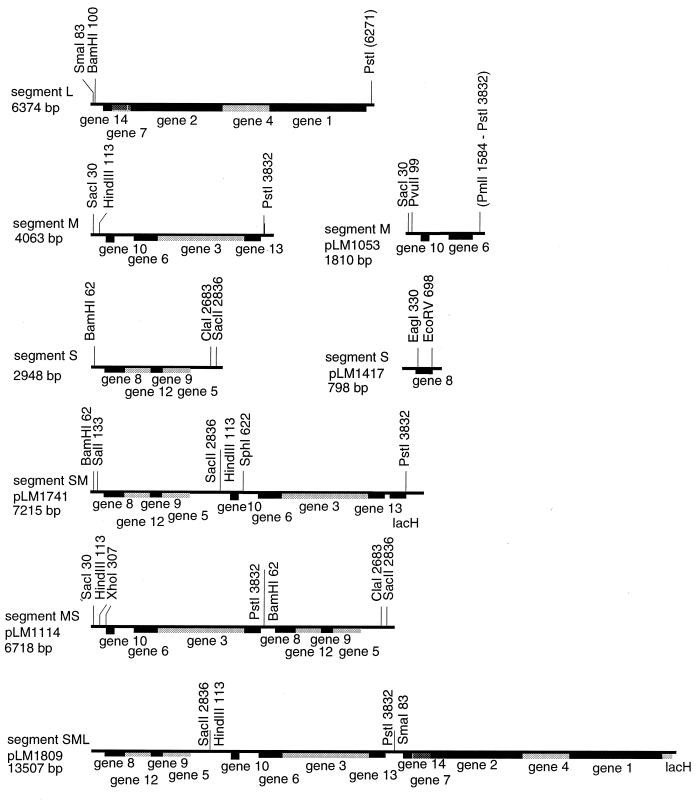
Figure 1.
Restriction maps of the cDNA copies of the genomic segments of Φ6, L, M, and S as well as diagrams of the chimeric constructions utilized in this study. The sequences are all embedded in plasmid pT7T319U, and the transcripts produced by T7 RNA polymerase have the correct base sequence at the 5′ ends. pLM1114 was prepared by ligating segment M cut at the PstI site to segment S cut at the BamHI site to form an MS chimera. Plasmid pLM1741 was constructed by cutting pLM659 with SacII and ligating to the HindIII site of segment M after blunting with T4 DNA polymerase. The segment M in this case had a type of Lacα inserted into the PstI site to adjust the size of the chimera to closely approximate the sum of segments S and M. Plasmid pLM1417 is a derivative of segment S in which the EcoRV site is ligated to a copy of the terminus of segment M to yield a segment S that is functional for packaging and replication but that contains no complete genes. Plasmid pLM1809 was constructed by ligating a cDNA copy of segment L cut at the SmaI site to pLM1741 cut and blunted at the PstI site. Plasmid pLM2064 was prepared from pLM1809 by exchanging the PstI/XbaI fragment at the 3′ terminus for the EcoRV/XbaI fragment from segment M.
In Vitro Transcription of Plasmids with T7 Polymerase.
Plasmids were cut with the restriction endonuclease XbaI. The XbaI site is located at the 3′ end of the cDNA inserts. The 5′ overhang resulting from the XbaI digestion was blunted with mung bean nuclease so that the runoff transcript would have an identical 3′ end to that of the viral single-stranded RNA middle transcript. The plasmids were transcribed with T7 RNA polymerase as described previously (14).
In Vitro Genomic Packaging of T7 Transcripts and Nucleocapsid Infection.
An in vitro genomic packaging system for bacteriophage Φ6 utilizing RNA transcripts of plasmids carrying cDNA copies of the viral genomic segments has been described (14). Briefly, Φ6 procapsids formed in E. coli strain JM109 were prepared according to Gottlieb et al. (8, 13). The in vitro packaging reaction conditions were performed as described by Olkkonen et al. (14). Each 25-μl reaction mixture contained 0.5–1.5 μg of a T7 transcript. Control reactions contained either no RNA or Φ6 single-stranded RNA. The assembly of the coat protein P8 onto packaged procapsids was performed as described by Olkkonen et al. (14). Purified P8 was a generous gift of D. H. Bamford (University of Helsinki). Spheroplasts were made from strains HB10Y, and infection was done as described previously by Ojala et al. (17). The transfected cells were collected and resuspended in a mixture of fresh cells and soft agar. The total mixture was poured onto a fresh LC plate and allowed to sit overnight at room temperature. Single plaques were picked and purified. Procapsids that had packaged Φ6 RNA generally yielded several hundred plaques.
Acquisition of Genomic Segments by Pickup of Plasmid Transcripts in Vivo.
Φ6 is capable, in vivo, of picking up transcripts of plasmids that contain normal 5′ and 3′ sequences even if originally buried inside the transcript (10). This involves selection of modified transcripts because packaging requires precise placement of the pac sequences relative to the 5′ ends. The chimeric constructions shown in Table 1 were prepared in plasmid pT7T3 19U. This plasmid does not replicate in pseudomonads. We prepared cointegrates of these plasmids and pKT230 (16), a plasmid with a wide host range, by cutting each plasmid at a unique EcoRI site and ligating. The resulting plasmids could be propagated in P. phaseolicola, phages containing deletions could be complemented by the genes carried by the plasmids, and transcripts could be acquired in place of the deletion segments. The frequency of phage that has acquired a transcript as a genomic segment ranged from 10% for transcripts that were exact copies of segment M with proper 5′ and 3′ sequences embedded in the transcripts to frequencies as low as 10−9 or 10−10 for transcripts that were chimeras of segments S and M.
Table 1.
cDNA plasmids
| Plasmid | Segment | Remarks | Ref. or source |
|---|---|---|---|
| pKT230 | Vector for pseudomonads | 16 | |
| pT7T3 19U | amp, PT7, PT3, lacZ; cloning and transcription vector | Pharmacia | |
| pLM656, 4,063-bp transcript | M | amp, PT7; exact copy of segment M in pT7T3 19U | 14 |
| pLM659, 2,948-bp transcript | S | amp, PT7; exact copy of segment S in pT7T3 19U | 28 |
| pLM687, 6,374-bp transcript | L | amp, PT7; copy of segment, L in pT7T3 19U | 29 |
| pLM1053, 1,810-bp transcript | M | pLM656 with a deletion from PmII to PstI | 10 |
| pLM1114, 6,718-bp transcript | MS 5′ M | Segment M cut at PstI and ligated to BamHI site of S | This study |
| pLM1241, 7,215-bp transcript | SM 5′ S | Segment S cut at SacII and ligated to HindIII of M; lacH is in PstI site | This study |
| pLM1417, 798-bp transcript | S 5′S | Segment S to EcoRV ligated to 100-base | This study |
| pLM1809, 13,507-bp transcript | SML 5′S | Segment S to SacII ligated to HindIII of M; PstI of M ligated to SmaI of L, with lacH in PstI site of L | 11 |
| pLM2064, 14,059-bp transcript | SML 5′S | pLM1809 with EcoRV/XbaI piece of M replacing PstI/XbaI | This study |
Production of Virus from a Plasmid Transcript Containing the Entire Genome of Φ6 in One Molecule.
Plasmid pLM2064 (Fig. 1) produces a transcript containing all the genes of Φ6. It was ligated to pKT230 and used to transform JM109 with selection for kanamycin and ampicillin resistances. DNA was prepared from JM109 and used to transform HB with kanamycin and ampicillin selection. Transformant colonies were pooled and replated as a lawn on Luria–Bertani plates. Hundreds of small plaques were observed. Several plaques were picked and replated on normal HB. dsRNA was prepared from a plate lysate, and this exhibited a migration pattern consistent with a size of about 14 kbp. A phage from this group was designated as Φ2515.
Isolation of dsRNA.
High titer stocks were prepared by a variation of the “plate lysate” method (15). Six ml of each phage stock was centrifuged in a Ti50 rotor at 30,000 rpm for 2 h at 4°C. The phage pellet was resuspended in 200 μl of Buffer A (6), and the dsRNA was isolated by extraction with phenol/chloroform/isoamyl alcohol and concentrated 4-fold by ethanol precipitation. The dsRNA preparations were analyzed on 0.8% SK-agarose gels in Tris/borate/EDTA buffer (15).
Labeling of Viral Proteins.
Cultures of HB in M8 medium were infected with virus at a multiplicity of infection of about 10 at 0°C for 30 min. The cultures were then transferred to 26°C, and rifampicin was added at 250 μg/ml at various times (18). Ten μCi of tritiated leucine was added to 1 ml of culture 15 min later, and the cells were harvested after 5 min. Cells were lysed in sample buffer and applied to 15.5% acrylamide gels as described previously (18).
RESULTS AND DISCUSSION
We prepared plasmids with two kinds of chimeras of segments S and M. In the first case (MS) we placed the genes of segment M at the 5′ end of the plus strand so that its pac site was functional; the pac site of S was missing (Fig. 1, pLM1114). In the second case (SM) we placed segment S at the 5′ end of the plus strand so that its pac site was functional; the pac site of M was mostly missing (Fig. 1, pLM1741). It should be emphasized that even deletions as small as 4 nucleotides at the BamHI site in segment S or at the HindIII site in M will preclude the use of that pac site (19). In the first case, we isolated phage with the chimeric MS segment by plating phage Φ2007, which has a deletion of gene 3 (Fig. 1, pLM1053, and Fig. 2, lane a) (10) on HB carrying plasmid pLM1114 ligated to pKT230. This is the transcript acquisition method. Phage was collected and plated on normal HB and on the strain that complements gene 3. Plaques appeared on normal HB at a frequency of 10−4 relative to plating on a strain that produced protein P3. When RNA from the phages was analyzed, it was found that the chimeric MS segment was present, but the phages also contained a normal segment S (Fig. 2, lane b). The phages with the MS chimeric segment could be propagated, but they were somewhat unstable and tended to shorten the chimeric segment by heterologous recombination (20). Phages were also isolated by a different procedure, the transfection method (14). In this case, procapsids isolated from E. coli carrying plasmid pLM450 were isolated and used to package plus strand RNA transcripts of plasmids pLM687, which is identical to segment L, pLM1114, which is the MS chimera and plasmid pLM1417, which has the 5′ and 3′ sequences of segment S but no complete genes (Fig. 1, pLM1417). After RNA packaging and minus strand synthesis, the procapsids were used to infect spheroplasts of HB, and plaques were obtained on HB. These plaques contained phage that had normal segment L, the MS chimera, and a segment of 800 bp that corresponded to the transcript of pLM1417 (Fig. 2, lane c). These phages were very stable.
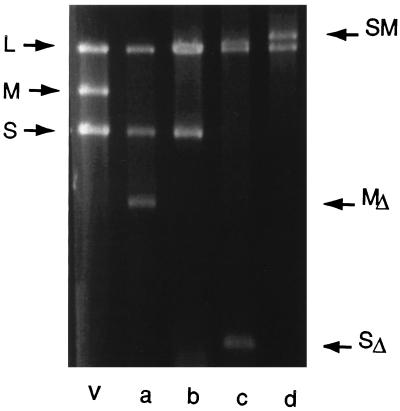
Figure 2.
Agarose gel electrophoresis of dsRNA isolated from virions. Lane v shows the distribution of normal segments L, M, and S. Lane a shows dsRNA from bacteriophage Φ2007, which has a deletion in segment M. Lane b shows RNA from Φ2064, which has normal L, an MS chimera picked up from pLM1114, and a normal segment S. Lane c shows RNA from Φ2323, which contains normal L, the MS chimera shown in b, and a deleted segment S that contains no genes and is only 798 bp. Lane d shows RNA from Φ2361, which contains normal L and a chimera of S and M but no normal segment M or S.
Phage containing the SM chimera was isolated by plating Φ2007 on a lawn of HB carrying the ligation product of pKT230 and pLM1741 (Fig. 1). The phage was collected and plated on normal HB and a strain that complemented gene 3. The frequency of plating on HB was between 10−9 and 10−10. However, 8 of 9 plaques analyzed showed the presence of only two genomic segments, one corresponding to normal L and the other to the SM chimera (Fig. 2, lane d).
On the basis of our packaging model we predicted that the SM chimera should replace segments S and M, and the MS chimera should still require a functional segment S to form viable phage. The chimera of S and M (SM) exists stably in the virion along with normal L (Fig. 2, lane d). In the case where the chimera has the packaging site of M at the 5′ end (MS), it behaves like an analog of segment M. It, therefore, cannot be packaged alone. It requires the prior packaging of segment S. We show that this chimera requires a segment S to form a stable virion. The extra segment S does not need to contain any active genes. It can be as small as several hundred nucleotides. It is interesting to see in Fig. 2, lane c, that the amount of the small S segment in the virion is clearly greater than a single copy. Our packaging model and previous results (21) would predict that the virion would incorporate about 3–4 copies of the segment S to approximate the weight of a normal S segment.
We considered the possibility that the chimeric segments cannot be packaged efficiently by normal procapsids. Perhaps the virus that we isolated contained mutations in the packaging apparatus that accommodated the abnormal genomic segments. To test this possibility we prepared purified phage and isolated nucleocapsids. The nucleocapsids were used for in vitro transcription to produce plus strand messages of the genomic segments. This RNA was then packaged into procapsids of normal Φ6, and spheroplasts were transfected with this material. We found that these preparations yielded hundreds of plaques, similar to the numbers found with normal Φ6 RNA. This indicated that the initial low frequencies of isolation of the phage with chimeric segments were not because of an intrinsic problem in packaging these abnormal constructions. It may have been because of inadvertent deleterious mutations in the RNA produced by the T7 RNA polymerase.
We then prepared plasmid pLM1809, which has the pac site of S at the 5′ end and the genes of segments S, M, and L in that order. This plasmid was ligated to plasmid pKT230 and introduced into JM109. DNA from JM109 was used to transform HB; however, the frequency of transformation was very low, and the transformants did not complement phages defective in gene 1. It has been clear for many years that gene 1 is toxic in HB (unpublished observations). We now found that pLM1809 toxicity could be reduced by either removing the sequence between the PstI site and the terminal XbaI site or by replacing this sequence with the 3′-terminal EcoRV/XbaI fragment of segment M (Fig. 1). The resulting plasmid with the 3′ end of segment M was designated pLM2064. The chimera of this plasmid with pKT230 was able to be introduced into HB at high frequency, and the resulting cells were able to complement phage that were defective in genes 1, 7, 2, 4, and 3, indicating that the genes of segments L and M were being expressed. In addition, we found that the lawn of these cells contained hundreds of plaques and that these plaques were produced by phage that contained only one genomic segment (Fig. 3). This phage is designated Φ2515. The cells were unstable genetically and lost the ability to produce plaques after a few passages, although it was still possible to isolate some phage from cultures. This is not very surprising as the production of virus would be expected to be lethal to the cells. The size of the dsRNA in the virions, 14.1 kbp, was consistent with that predicted from the structure of the Φ6-specific transcript of pLM2064. This genomic segment is the largest packaged replicating dsRNA molecule found so far. There are several larger dsRNA molecules of viruses that do not package their genomes (22).
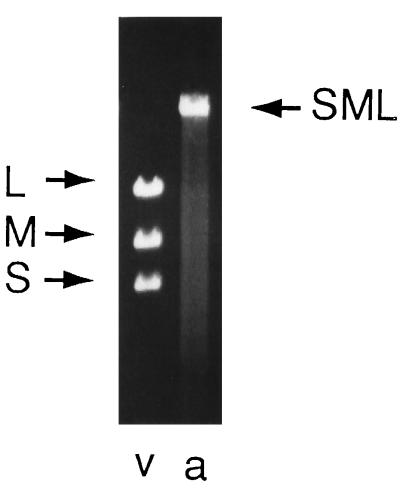
Figure 3.
Agarose gel electrophoresis of dsRNA isolated from virions. Lane v shows the distribution of normal segments L, M, and S. Lane a shows dsRNA from Φ2515, which contains the entire genome of Φ6 in one segment. The migration of the RNA indicates a size of about 14 kbp.
The ability of the trichimeric phage to propagate indicates that several of the features of normal Φ6 are not absolutely necessary. During normal infection, the genes for proteins 7, 2, 4, and 1 are expressed early (23), and the amount of transcription of segment L is closer to that of S and M (24). Later in infection, the amount of transcription of S and M is about 10-fold greater than that of L (24). In the case of the trichimeric genome, there is only one transcript. Normal segment L has GU as the first two nucleotides whereas S and M start with GG. It has been shown that this sequence difference leads to changes in the in vitro transcription levels of the segments (25). In the present case, the genomic segment has only the GG sequence.
The value of a segmented genome to Φ6 is probably a combination of the benefits of easier control of gene expression, easier packaging of shorter RNA molecules, and the ability to lose deleterious mutations through segment reassortment. Because Φ2515 has the entire genome in one segment, it should not be able to reassort or recombine, because recombination takes place inside the procapsid and only one molecule would fit. It would be interesting to see whether this phage has a more challenging prospect in dealing with deleterious mutations than does normal Φ6 (26). Normal Φ6 shows temporal control of gene expression in that genes 1, 2, 4, and 7 are expressed early in infection (23). The one-segment phage shows only a little less control (Fig. 4). Some of the late genes such as 8 and 9 are expressed rather early, and some of the late genes such as 5, 6, and 12 are expressed late. It is clear from Fig. 4 that the genes that are normally on segment L (1, 2, 4, and 7) are still being expressed early in the one-segment phage. The plus strand transcript of segment L appears to have a longer half-life in infected cells than the transcripts of segments S and M (unpublished observations). It is possible that the part of the transcript in the one-segment phage that contains the “early” genes may be more stable as well. The plaques of the one-segment phage are small, and we have not seen mutant forms that produce larger plaques. The evolutionary pathway from the earliest viruses to those found at the present time is not known. Arguments can be offered in support of the earliest viruses being segmented or polycistronic. It has been possible to construct viruses with more segments than found naturally. This has been done for Sindbis virus, which ordinarily has one segment and in which a two-segment virus was selected (27). We have prepared stable derivatives of Φ6 that contain four segments instead of three (10). In the present report we reduce the number of segments in Φ6 from three to two or one.
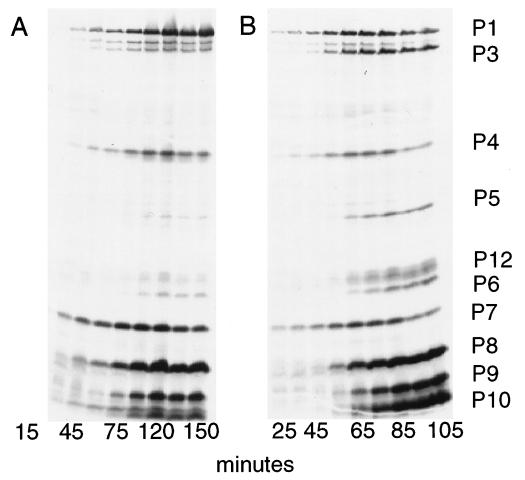
Figure 4.
Autoradiogram of labeled proteins synthesized in cells infected by Φ2515, which has only one genomic segment (A), or by normal Φ6 (B). Numbers below the lanes are the times at which labeled leucine was added. Unlabeled lanes are at interpolated times.
Our packaging model for Φ6 is unique in that it proposes that the steps in the program change for packaging are determined by the amount of RNA in the procapsid and not its specific sequence. Sequence recognition takes place on the outside of the procapsid before packaging. The packaging of the genomic segments must occur in the specific order S, M, L; however, molecules that have sizes equal to more than one segment can move the program past the next stage. We found that a molecule with the pac site of S, but of the size of S plus M, could facilitate the packaging of segment L in vitro (11). A molecule with the pac site of M, but of the size of M plus L, could facilitate the turning on of minus strand synthesis in vitro. We also showed that a molecule with the pac site of S, but of the size of the entire genome, could be packaged and would turn on minus strand and then plus strand synthesis in vitro. In the present paper we show that a viable virus can be produced that shows similar behavior.
Acknowledgments
This work was supported by Grant GM34352 from the National Institutes of Health (to L.M.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: ds, double-stranded; HB, Pseudomonas syringae pathovar phaseolicola HB10Y.
References
- 1.Casjens S. In: Virus Structure and Assembly. Casjens S, editor. Boston: Jones and Bartlett; 1985. pp. 75–147. [Google Scholar]
- 2.Tyler K L, Fields B N. In: Virology. Fields B N, Knipe D M, editors. New York: Raven; 1990. pp. 1271–1273. [Google Scholar]
- 3.Spendlove R S, McClain M E, Lennette E H. J Gen Virol. 1970;8:83–93. doi: 10.1099/0022-1317-8-2-83. [DOI] [PubMed] [Google Scholar]
- 4.Enami M, Sharma G, Benham C, Palese P. Virology. 1991;185:291–298. doi: 10.1016/0042-6822(91)90776-8. [DOI] [PubMed] [Google Scholar]
- 5.Duhaut S D, McCauley J W. Virology. 1996;216:326–337. doi: 10.1006/viro.1996.0068. [DOI] [PubMed] [Google Scholar]
- 6.Vidaver A K, Koski R K, Van Etten J L. J Virol. 1973;11:799–805. doi: 10.1128/jvi.11.5.799-805.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day L A, Mindich L. Virology. 1980;103:376–385. doi: 10.1016/0042-6822(80)90196-8. [DOI] [PubMed] [Google Scholar]
- 8.Gottlieb P, Strassman J, Qiao X, Frucht A, Mindich L. J Bacteriol. 1990;172:5774–5782. doi: 10.1128/jb.172.10.5774-5782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mindich L, Qiao X, Qiao J. Virology. 1995;212:213–217. doi: 10.1006/viro.1995.1470. [DOI] [PubMed] [Google Scholar]
- 10.Onodera S, Qiao X, Qiao J, Mindich L. Virology. 1995;212:204–212. doi: 10.1006/viro.1995.1469. [DOI] [PubMed] [Google Scholar]
- 11.Qiao X, Qiao J, Mindich L. Proc Natl Acad Sci USA. 1997;94:4074–4079. doi: 10.1073/pnas.94.8.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 13.Gottlieb P, Strassman J, Bamford D H, Mindich L. J Virol. 1988;62:181–187. doi: 10.1128/jvi.62.1.181-187.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olkkonen V M, Gottlieb P, Strassman J, Qiao X, Bamford D H, Mindich L. Proc Natl Acad Sci USA. 1990;87:9173–9177. doi: 10.1073/pnas.87.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 16.Bagdasarian M, Lurz R, Ruckert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 17.Ojala P M, Romantschuk M, Bamford D H. Virology. 1990;178:364–372. doi: 10.1016/0042-6822(90)90333-m. [DOI] [PubMed] [Google Scholar]
- 18.Sinclair J F, Cohen J, Mindich L. Virology. 1976;75:198–208. [PubMed] [Google Scholar]
- 19.Gottlieb P, Qiao X, Strassman J, Frilander M, Mindich L. Virology. 1994;200:42–47. doi: 10.1006/viro.1994.1160. [DOI] [PubMed] [Google Scholar]
- 20.Mindich L. Semin Virol. 1996;7:389–397. [Google Scholar]
- 21.Mindich L, Qiao X, Qiao J. Virology. 1995;212:213–217. doi: 10.1006/viro.1995.1470. [DOI] [PubMed] [Google Scholar]
- 22.Pfeiffer P, Jung J L, Heitzler J, Keith G. J Gen Virol. 1993;74:1167–1173. doi: 10.1099/0022-1317-74-6-1167. [DOI] [PubMed] [Google Scholar]
- 23.Sinclair J F, Tzagoloff A, Levine D, Mindich L. J Virol. 1975;16:685–695. doi: 10.1128/jvi.16.3.685-695.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coplin D L, Van Etten J L, Koski R K, Vidaver A K. Proc Natl Acad Sci USA. 1975;72:849–853. doi: 10.1073/pnas.72.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frilander M, Poranen M, Bamford D H. RNA. 1995;1:510–518. [PMC free article] [PubMed] [Google Scholar]
- 26.Chao L, Tran T T, Tran T T. Genetics. 1997;147:953–959. doi: 10.1093/genetics/147.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geigenmuller-Gnirke U, Weiss B, Wright R, Schlesinger S. Proc Natl Acad Sci USA. 1991;88:3253–3257. doi: 10.1073/pnas.88.8.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frilander M, Gottlieb P, Strassman J, Bamford D H, Mindich L. J Virol. 1992;66:5013–5017. doi: 10.1128/jvi.66.8.5013-5017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mindich L, Qiao X, Onodera S, Gottlieb P, Frilander M. Virology. 1994;202:258–263. doi: 10.1006/viro.1994.1341. [DOI] [PubMed] [Google Scholar]