Abstract
Molecular techniques based on the detection of genomic sequences by reverse transcription (RT)-PCR, nested PCR, or real-time PCR have made possible the rapid diagnosis of dengue virus (DENV) infections, and these approaches have been accepted by clinical laboratories as the new standard method for the detection of dengue virus in acute-phase serum samples. One of these PCR-based assays, the two-step RT nested PCR (RT-NPCR) technique is used routinely in laboratories worldwide. In the present study, the two-step RT-NPCR as described by Lanciotti and collaborators (1992) was adapted to a novel single-tube nested PCR (STNPCR) format, which is less prone to cross-contamination and reduces reaction cost and time. When standards for each dengue serotype were tested, the detection limit of the STNPCR was at least 10 copies for DENV-1 and 100 copies for DENV-2 and DENV-3, whereas the detection limit for the two-step RT-NPCR was 100 copies for each serotype. Sera from 22 patients with confirmed DENV-3 infections and from 14 healthy individuals were then tested in the STNPCR format using the system described by Lanciotti as the reference standard. The results indicated a sensitivity of 75.9% (CI 95%, 60.3–91.4) and a specificity of 100% for the RT-STNPCR. Athough RT-STNPCR was less sensitive than the conventional two-step RT-NPCR for the detection of virus in serum samples, it was still adequately sensitive, and the advantages associated with a single-tube format may outweigh the somewhat lower assay sensitivity, making it useful for diagnosis in the field.
Keywords: nested PCR, diagnosis, dengue, single-tube
Dengue virus is a mosquito-borne flavivirus and the most prevalent arbovirus in tropical and subtropical regions of Asia, Africa and Central and South America (Harris et al., 2000). There are four distinct serotypes, DENV-1, DENV-2, DENV-3 and DENV-4, all of which may cause an asymptomatic infection or a spectrum of diseases ranging from dengue fever (DF) to more severe clinical forms such as dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (Guzman and Kouri, 2004). In recent years, dengue virus infection has emerged as a major public health problem, with expanding geographical distribution and increasing epidemic activity (Guha-Sapir and Schimmer, 2005).
Although dengue virus infection confers protective immunity to the homologous serotype, there is only partial protection against subsequent secondary infections by other serotypes. Epidemiological studies have shown that secondary infection is a major risk for DHF and DSS (Rothman, 2004). Thus, discrimination between primary and secondary dengue virus infection and current infection is important for both patient management and epidemiological investigations.
Because of the extensive cross-reactivity among flaviviruses, and the fact that IgM antibody responses are often low during the first 5 days of symptoms, identification of the infecting virus by serology during the acute phase of the infection is not possible in many cases in which flaviviruses are circulating (Guzman and Kouri, 2004). Molecular techniques based on the detection of genomic sequences by reverse transcription (RT)-PCR, nested PCR, and real-time PCR allow for rapid diagnosis of dengue virus serotypes and have replaced virus isolation as the new standard method for the detection of dengue virus in acute-phase serum samples (Guzman and Kouri, 2004). One of these, the two-step nested RT-PCR (RT-NPCR) approach, is routinely employed in laboratories worldwide. Identification and typing of dengue virus serotypes is achieved by RT-PCR followed by nested PCR, using complex- and serotype-specific primers, respectively (Guzman and Kouri, 2004). Although real-time PCR offers the additional advantage of providing quantitation of the viral burden (Anwar et al., 2006; Chien et al., 2006; Kong et al., 2006), its use is still limited to well-structured reference laboratories.
In the present study, we have adapted the two-step nested RT-PCR described by Lanciotti et al. (1992), which requires the transfer of the product of the first-round amplification to a second tube to perform the second PCR reaction, to a novel single-tube nested PCR (STNPCR) format that is less prone to cross-contamination and more cost-effective. Recently, portable thermocylers have been developed for PCR reactions in the field, and the STNPCR strategy we introduce here is clearly more suitable for field use because it requires less manipulation.
The oligonucleotide primers used (D1, D2, TS1, TS2, TS3 and TS4) have been described previously (Lanciotti et al., 1992).
The STNPCR was carried out in 0.5-ml PCR tubes using an Eppendorf Master Cycler Gradient thermocycler TM, essentially as described previously (Abath et al., 2002). This STNPCR technology is based on partial immobilization of the inner oligonucleotide primers inside the tube cap by adsorption, followed by solubilization after the first reaction. In brief, the reaction was optimized in terms of the concentrations of deoxynucleoside triphosphates, Mg2+, outer and inner primers. Based on these experimental results, in all subsequent experiments, we used a concentration ratio of 1:50 (0.3 pmoles: 15 pmoles) for the outer primer set/internal primer set (OP/IP). Seven microliters containing 15 pmoles of inner primers (DI, TS1, TS2, TS3 and TS4) with traces of bromophenol blue were immobilized onto the microtube cap by incubating the tube at 37°C until the cap was dry. The conditions of the amplification were the same for the first (15 cycles) and second (45 cycles) rounds of amplifications (92°C for 30 s, 55°C for 1 min, and 72°C for 1 min). The 50-μl PCR mixture contained 10 mM Tris-HCl, 50 mM KCl, 0.1 mg/ml gelatin, 1.5 mM MgCl2, 0.2 mM each dNTP, 0.3 pmoles of each outer primer (D1 and D2), and 2.5 U of Taq DNA polymerase (GE Healthcare, Uppsala, Sweden). One μl of the serotype standard or 2 μl of processed serum sample was added to the reaction mixture. After the first-round PCR, the thermal cycler was paused at 92°C, and the closed tubes were inverted several times to dissolve the inner primers adsorbed on the caps, then returned to the machine for the second round of amplification. Products (10 μl) were separated by electrophoresis on 1% agarose gels, and ethidium bromide-stained gels were visualized and photographed over a UV light using the Kodak Molecular Imaging Software version 4.0. For generation of virus DNA quantitation standards, amplicons corresponding to different serotypes were produced by RT-PCR and purified using a Sephaglas BandPrep Kit (GE Healthcare, Uppsala, Sweden) according to the instructions of the supplier. The DNA products were quantified spectrophotometrically.
Twenty-two human serum samples were obtained from patients with clinically characterized and virologically confirmed DENV-3 infection, and 14 sera were obtained from healthy individuals. All serum samples studied were analyzed by RT-NPCR as described previously (Lanciotti et al., 1992); this assay was considered the gold standard for this evaluation. Viral RNA was purified from 200 μl of serum samples with a QI amp MiniElute Virus Spin Kit (Qiagen GmbH, Germany, Germany), following the manufacturer’s instructions. The purified RNA was resuspended in 60 μl of water, and 5 μl were used for full-length total cDNA synthesis using the first-strand synthesis system for RT-PCR (Superscript III, Invitrogen, CA, USA) containing random hexamers (final volume, 20 μl).
NPCR is a two-step procedure in which the products of a first PCR using outer primers are subjected to re-amplification with a second set of inner primers located within the previously amplified sequence. Although the two-step nested PCR is more sensitive than the conventional PCR, an inherent drawback is the need to open tubes after the first round of amplification to transfer amplicons to a second tube for another PCR amplification reaction that uses a different primer pair or to introduce new reagents and/or primers (Picken et al., 1996). This process significantly increases the risk of cross-contamination of negative samples with amplicons derived from positive specimens during the first round of amplification. A single-tube multiplex RT-PCR to detect and serotype dengue virus has been published (Harris et al., 1998; Kumaria and Chakravarti, 2005), but additional studies are necessary to determine whether this approach is as specific and sensitive as the conventional nested PCR format. We reasoned that it might be possible to minimize the risk of cross-contamination associated with the classical RT-NPCR of Lanciotti et al.. 1992 by designing a test in a single-tube nested PCR format in which the internal primers are separated from the components of the first round of amplification by immobilization onto the inside of the microtube. This approach has previously been used for the detection of Schistosoma mansoni and Plasmodium DNA (Abath et al., 2002; Melo et al., 2006; Montenegro et al., 2004), indicating that the method is appropriate for nested PCR targeting other sequences.
The test was initially carried out using one of the serotype standards in each reaction tube. The detection limit of the STNPCR was at least 10 copies for DENV-1 and 100 copies for DENV-2 and DENV-3 (Figure 1A); the sensitivity reported previously by Lanciotti et al.(1992) was 100 copies for each serotype. Although in epidemiological terms, co-infection with various serotypes is rare, we simulated this situation in our assay evaluation by testing the serotype standards simultaneously. This experiment resulted in a detection limit of 100 copies for each serotype (Figure 1B), perhaps because of competition between the targets for reagents. As our laboratory routinely performs PCR assays for the detection of dengue virus, and the presence of DENV-4 has not yet been detected in the Brazilian Northeast, standards for serotype 4 were not tested. The specificity of the four serotype-specific primers used was evaluated by Lanciotti et al. (1992) for detection of 4 dengue serotypes and against five flaviviruses (West Nile, Japanese encephalitis, St. Louis encephalitis, yellow fever, and Edge Hill). In order to evaluate the performance of our novel RT-STNPCR assay using clinical samples, we tested sera from 22 patients who were DENV-3 infection-positive by the RT-PCR described by Lanciotti and his group (Lanciotti et al., 1992), as well as sera from 14 healthy individuals negative for dengue. The data in Figure 2 are representative of the results obtained using serum samples. The sensitivity of the RT-STNPCR was 75.9% (CI 95%: 60.3–91.4), and the specificity was 100%. In our evaluation, 80.5% of the serum samples gave the same results in RT-NPCR and RT-STNPCR. One possibility for further advancing this technique would be to co-immobilize additional reagents such as dNTPs with the primers for the second PCR round; another possibility would be to optimize the RNA extraction. The results obtained using standards showed an equal sensitivity for both methods, whereas the STNPCR showed a lower level of sensitivity with clinical samples. It is possible that this lower sensitivity reflects the activity of inhibitors carried over from the RNA extraction, since they are not diluted further by the transfer of an aliquot to a fresh reaction mixture in the case of the STNPCR. New single-step viral RNA extraction protocols are under evaluation. Although the RT-STNPCR was somewhat less sensitive than the conventional two-step RT-NPCR for the detection of serum samples, the use of a single-tube format is an advantage, decreasing the possibility of carryover contamination and false-positives. Also, the one-step approach reduces the need to repeat assays in the case of suspected cross-contamination or false-positives.
Fig 1.
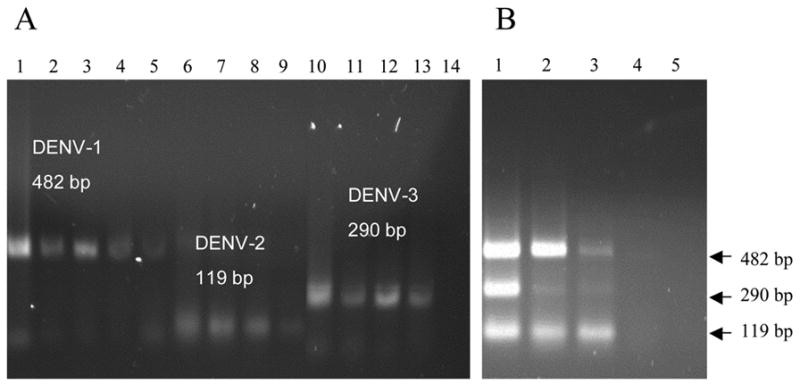
Agarose gel eletrophoresis showing the detection limit of STNPCR for three serotypes of dengue virus. Panel A, standards for each serotype were tested individually. Lanes 1–5, 105 to 10 copies of DENV-1 virus (482 bp); lanes 6–9, 105 to 102 copies of DENV-2 virus (119 bp); lanes 10–13, 105 to 102 copies of DENV-3 virus (290 bp), lane 14, no-template control. Panel B, standards for each serotype were tested simultaneously. Lanes 1–4, 104 to 10 copies of DEN-1, DEN-2 and DEN-3 viruses; lane 5, no-template control.
Fig 2.

Representative agarose gel electrophoresis showing the amplification of the diagnostic band for DENV-3 virus in sera of patients (lanes 1 to 7) and healthy individuals (lanes 8 to 12). The reactions were performed with 2 μl of the RNA extract solution obtained from 200 μl of sera.
Acknowledgments
This work was supported by FIOCRUZ internal funds (PDTIS) and National Institute of Allergy and Infectious Diseases (NIAID/NIH), under Grant U19 AI56541. FGCA is the recipient of a CNPq Research Fellowship.
Footnotes
This manuscript is dedicated to the family and former students of Dr. Abath.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abath FG, Melo FL, Werkhauser RP, Montenegro L, Montenegro R, Schindler HC. Single-tube nested PCR using immobilized internal primers. BioTechniques. 2002;33:1210–1214. doi: 10.2144/02336bm05. [DOI] [PubMed] [Google Scholar]
- Anwar A, August JT, Too HP. A stem-loop-mediated reverse transcription real-time PCR for the selective detection and quantification of the replicative strand of an RNA virus. Anal Biochem. 2006;352:120–8. doi: 10.1016/j.ab.2006.01.046. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Kumaria R. Molecular detection and serotypic characterization of dengue viruses by single-tube multiplex reverse transcriptase–polymerase chain reaction. Diagn Microbiol Infect Dis. 2005;52:311–316. doi: 10.1016/j.diagmicrobio.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Guha-Sapir D, Schimmer B. Dengue fever: new paradigms for a changing epidemiology. Emerg Themes Epidemiol. 2005;2:1. doi: 10.1186/1742-7622-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman MG, Kouri G. Dengue diagnosis, advances and challenges. Int J Infect Dis. 2004;8:69–80. doi: 10.1016/j.ijid.2003.03.003. [DOI] [PubMed] [Google Scholar]
- Harris E, Roberts TG, Smith L, Selle J, Kramer LD, Valle S, Sandoval E, Balmaseda A. Typing of dengue viruses in clinical speciments and mosquitoes by single-tube multiplex reverse transcriptase PCR. J Clin Microbiol. 1998;36:2634–2639. doi: 10.1128/jcm.36.9.2634-2639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E, Videa E, Perez L, Sandoval E, Tellez Y, Perez MA, Cuadra R, Rocha J, Idiaquez W, Alonso RE, Delgado MA, Campo LA, Acevedo F, Gonzalez A, Amador JJ, Balmaseda A. Clinical, epidemiologic, and virologic features of dengue in the 1998 epidemic in Nicaragua. Am J Trop Med Hyg. 2000;63:5–11. doi: 10.4269/ajtmh.2000.63.5. [DOI] [PubMed] [Google Scholar]
- Kong YY, Thay CH, Tin TC, Devi S. Rapid detection, serotyping and quantitation of dengue viruses by TaqMan real-time one-step RT-PCR. J Virol Methods. 2006;138:123–130. doi: 10.1016/j.jviromet.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Kumaria R, Chakravarti A. Molecular detection and serotypic characterization of dengue viruses by single-tube multiplex reverse transcriptase-polymerase chain reaction. Diagnostic Microbiology and Infectious Disease. 2005;52:311–316. doi: 10.1016/j.diagmicrobio.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid Detection and Typing of Dengue Viruses from Clinical Samples by Using Reverse Transcriptase-Polymerase Chain Reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien LJ, Liao TL, Shu PY, Huang JH, Gubler DJ, Chang GJJ. Development of Real-Time Reverse Transcriptase PCR Assays To Detect and Serotype Dengue Viruses. J Clin Microbiol. 2006;44:1295–1304. doi: 10.1128/JCM.44.4.1295-1304.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo FL, Gomes ALdV, Barbosa CS, Werkhauser RP, Abath FGC. Development of molecular approaches for the identification of transmission sites of schistosomiasis. Trans R Soc Trop Med Hyg. 2006;100:1049–1055. doi: 10.1016/j.trstmh.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Montenegro LML, Montenegro RA, Lima AS, Carvalho AB, Schindler HC, Abath FGC. Development of a single-tube hemi-nested PCR for genus specific detection of Plasmodium in oligoparasitemic patients. Trans R Soc Trop Med Hyg. 2004;98:616–625. doi: 10.1016/j.trstmh.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Picken MM, Picken RN, Han D, Cheng Y, Strle F. Single-tube nested polymerase chain reaction assay based on flagellin gene sequences for detection of Borrelia burgdorferi sensu lato. Eur J Clin Microbiol Infec Dis. 1996;15:489–498. doi: 10.1007/BF01691317. [DOI] [PubMed] [Google Scholar]
- Rothman AL. Dengue: defining protective versus pathologic immunity. J Clin Invest. 2004;113:946–951. doi: 10.1172/JCI21512. [DOI] [PMC free article] [PubMed] [Google Scholar]


