Abstract
The transcription factors Oct4 and Nanog are essential for the maintenance of an undifferentiated and pluripotent state in early embryonic cells, embryonic stem cells and embryonal carcinoma cells in humans and mice. These factors are co-localized to promoters of more than 300 genes, and synergistically regulate their activities. Currently, the molecular interaction between these two factors has not been well-characterized. During attempts to co-immunoprecipitate Oct4 and Nanog we found that cross-linking with dithiobis[succinimidylpropionate] was necessary to maintain their interaction. This result was supported by gel filtration analysis. Surprisingly, formaldehyde, a cross-linker commonly used during chromatin immunoprecipitation of Oct4 and Nanog, did not preserve the complex. Our findings demonstrate the effectiveness of using DSP to mitigate the instability of the interaction between these two particular proteins. Additionally, this solution may potentially allow us to identify novel members of the Oct4-Nanog complex, leading to better understanding of the regulatory mechanisms behind pluripotency.
Keywords: Cross-linking, dithiobis[succinimidylpropionate] (DSP), embryonal carcinoma cells, embryonic stem cells (ES cells), formaldehyde, gel filtration, immunoprecipitation, Nanog, Oct4, pluripotency
Introduction
Stem cell biology is currently attracting wide attention due to the potential application of stem cells for the generation of cells and tissues that could be used for cell-based therapies. A detailed understanding of the molecular mechanisms for differentiation and the maintenance of an undifferentiated state of stem cells is critically important, if we are to move forward in developing the use of stem cells for medical practice. The transcription factors Oct4 and Nanog, specifically expressed in early embryonic cells, embryonic stem cells (ES cells) and embryonal carcinoma cells in humans and mice, are essential for the maintenance of an undifferentiated and pluripotent state in these cells [1]. Blastocysts of the Oct4-null mouse contain only extraembryonic tissues, lack the pluripotent inner cell mass, and die around the time of implantation [2]. This phenomenon is due to the differentiation of the inner cell mass cells into extraembryonic cells in the absence of Oct4 [3]. Consistent with this finding, ES cells, which are derived from inner cell mass, differentiate into extraembryonic tissues (trophectodermal lineages) after depletion of Oct4 [4]. Without Nanog, inner cell mass cells and ES cells also differentiate into extraembryonic tissues (parietal endoderm lineage) and Nanog-null mice die around the time of implantation [5].
Oct4 and Nanog co-occupy more than 300 promoter regions in human and mouse ES cells and synergistically regulate the expression of the corresponding genes [6, 7]. Although a recent report demonstrated co-immunoprecipitation (co-IP) of transfected Nanog and endogenous Oct4 in mouse ES cells [8], the interaction between endogenous Nanog and endogenous Oct4 has not been well-studied. Without characterizing the interactions among endogenous Oct4, Nanog, and other members of the transcriptional machinery, it is difficult to fully understand how these key transcription factors are involved in the maintenance of pluripotency and how to manipulate them for the purpose of cell-based therapy. While trying to isolate an endogenous protein complex containing Oct4 and Nanog, we found that these proteins form a weak or transient complex, making isolation of the complex from cell extract difficult. In this study, through systematic comparison of different conditions for the preparation of cell extract and performance of immunoprecipitation (IP), we found that cross-linking was necessary to reproducibly co-immunoprecipitate these two proteins from embryonal carcinoma cell extract. The cross-linker dithiobis[succinimidylpropionate] (DSP) [9] was particularly useful for this purpose because it is cell-permeable and has a disulfide bond which is readily cleaved by a reducing reagent, such as β-mercaptoethanol in SDS sample buffer.
Materials and methods
Cell culture
Mouse embryonal carcinoma cells F9 (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (Invitrogen) in a 5% CO2 atmosphere at 37°C.
Immunoprecipitation
The following five conditions were compared in this study. The buffer components are summarized in Table 1.
Table 1.
Buffer compositions used for preparation of cell extract and performing IP. Each buffer contained 10 mM Hepes pH 7.8, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 2.0 μM leupeptin and 3.0 μM pepstatin A in addition to the chemicals listed in the table.
| Buffer | NP-40(%) | NaCl (mM) | Triton X-100(%) | Sodium deoxycholate (%) | SDS(%) |
|---|---|---|---|---|---|
| A | 0.5 | 150 | 1.0 | 0 | 0 |
| B | 0.025 | 150 | 0 | 0 | 0 |
| C | 1.0 | 150 | 0 | 0.5 | 0.1 |
| D | 0 | 150 | 0.003 | 0 | 0 |
Condition 1
For our first attempt at co-IP of Oct4’s interacting proteins, a mild buffer containing 0.5% NP-40 and 1.0% Triton X-100 was used (Buffer A in Table 1). Protein from 1×107 F9 cells was extracted in 1 ml Buffer A and collected by centrifugation at 14,000 × g for 15 min at 4 °C. The extract was precleared by incubation with 5 μg of normal goat IgG (sc-2028, Santa Cruz Biotechnology, Santa Cruz, CA) and 120 μl of GammaBind™ G Sepharose™ beads (Amersham Biosciences, Piscataway, NJ) for 2 hr at 4 °C on a rotator, and collected by centrifugation as described above. In a separate reaction, 20 μl of GammaBind G beads was incubated with 5 μg of goat anti-Oct4 antibody (sc-8628, Santa Cruz Biotechnology) or normal goat IgG in 1 ml of Buffer A for 2 hr at 4 °C. The antibody-bound beads were collected via centrifugation and incubated with the precleared extract for 16 hr at 4 °C on a rotator. The beads were recovered by centrifugation and washed six times with Buffer A. Finally, the immunoprecipitated proteins were eluted with SDS sample buffer [10] by incubating for 5 min at 98 °C.
Condition 2
The procedure was the same as that of Condition 1, except that a milder buffer (Buffer B in Table 1) was used in place of Buffer A.
Condition 3
The procedure was also the same as that of Condition 1, except for the usage of a strong denaturing buffer (Buffer C in Table 1) instead of Buffer A.
Condition 4
Before making extract F9 cell suspension was incubated with 2 mM DSP (Pierce Biotechnology, Rockford, IL) in phosphate buffered saline for 2 hr at 4°C and cross-linking was terminated with 20 mM Tris HCl pH 8.0 at 25°C. The rest of the procedure was the same as in Condition 3.
Condition 5
F9 cell suspension was incubated with 4% formaldehyde for 20 minutes at 25° C, after which the cell suspension was washed twice with phosphate buffered saline. The rest of the procedure was the same as in Condition 3.
Western blotting
The proteins eluted from the GammaBind G beads were resolved in a 12% SDS-PAGE gel and detected by silver staining (Owl Seperation Systems, Partsmouth, NH) and Western blotting. Rabbit anti-Oct4 antibody at 1:1000 dilution (sc-9081, Santa Cruz Biotechnology) and rabbit anti-Nanog antibody at 1:2000 dilution (AB5731, Chemicon, Temecula, CA) were used as the primary antibodies, and SuperSignalR West Dura (Pierce Biotechnology) was used as a substrate for Western blotting.
Gel filtration of F9 cell extract
Cell extract prepared under Conditions 1 and 4 were separately applied to a Superdex 200 gel filtration column (Amersham Biosciences) to compare the elution patterns of Oct4 and Nanog. The Superdex 200 column was attached to an AKTA system (Amersham Biosciences) and equilibrated with Buffer D (Table 1). Four hundred μl of the cell extract was applied to the column and fractionated proteins were precipitated with 20% trichloroacetic acid for 30 min on ice before being loaded onto SDS-PAGE gels. The resolved proteins were analyzed by Western blotting with antibodies against Oct4 and Nanog.
Results and discussion
We sought to find a co-IP condition that minimized non-specific binding, but retained the interaction between Oct4 and Nanog, using F9 embryonal carcinoma cells as our source material. Under a mild buffer condition (Condition 1), Oct4 was successfully immunoprecipitated but Nanog was not (Fig. 1B), indicating disruption of the Oct4-Nanog interaction during the preparation of cell extract or performance of IP. When the F9 cell extract prepared under Condition 1 was applied to a gel filtration column, Oct4 (38 kDa) was eluted in fractions No. 12, 13 and 14 (corresponding to a molecular mass in the range of 60-400 kDa) while the majority of Nanog (34 kDa) was eluted in fractions No. 8 through 11 (larger than 400 kDa) (Fig. 2A). The separation of Oct4 and Nanog was consistent with our failure to co-IP these proteins under Condition 1. The results from gel filtration also suggest that Oct4 and Nanog likely had already dissociated before IP, probably during the making of cell extract.
Figure 1. Comparison of different buffer conditions for co-IP of Oct4 and Nanog.
(A) Silver stained SDS-PAGE gels demonstrating co-immunoprecipitated proteins from F9 cell extract with normal IgG and Oct4 antibody under three different buffer conditions combined with two cross-linkers. Each lane contains proteins immunoprecipitated from 3×106 F9 cells. Arrows label bands that represent candidate Oct4-interacting proteins. These bands became prominent after cross-linking with DSP.
(B) Detection of immunoprecipitated Oct4 and co-immunoprecipitated Nanog by Western blotting. The lanes correspond to the lanes in Figure 1A, and each lane contains proteins immunoprecipitated from 3×106 cells. Arrows indicate bands of immunoprecipitated Oct4 for comparison of intensity. The input lane contains extract from 2.5×105 cells.
Figure 2. Gel filtration analysis of the Oct4-Nanog interaction.
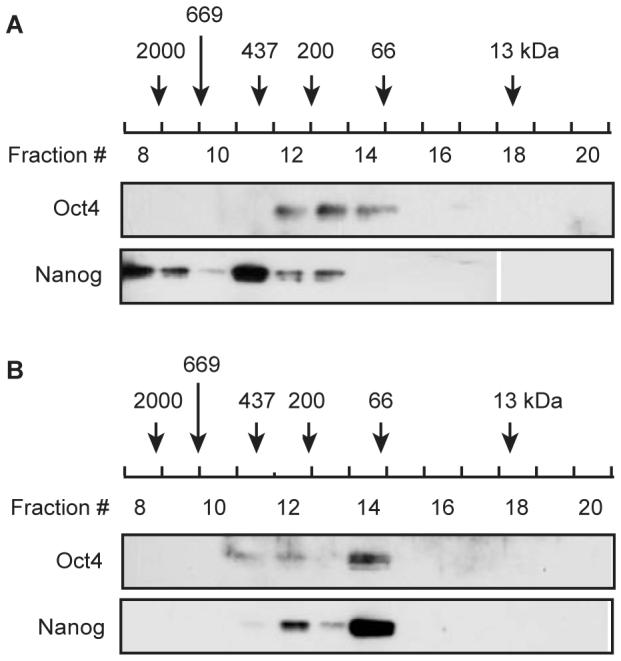
Immunoblotting of Oct4 and Nanog in fractions eluted during Superdex gel filtration column chromatography. The column was loaded with F9 cell extract prepared under Condition 1 (A), and Condition 4 (B). The elution peaks of standard marker proteins are indicated to allow estimation of the molecular mass of any eluted proteins.
To preserve the Oct4-Nanog interaction, the same procedure was carried out under a milder buffer condition, using Buffer B for making cell extract and performing IP (Condition 2). Buffer B has a lower concentration of NP-40 and lacks Triton X-100. This condition still failed to co-IP Nanog, suggesting that the interaction between Oct4 and Nanog is unstable (Fig. 1B). In addition, numerous non-specifically co-precipitated proteins were observed in a silver-stained gel due to the mildness of Condition 2 (Fig. 1A). At this point we considered cross-linking the cellular proteins before extraction to preserve the Oct4-Nanog complex, but were faced with the problem that cross-linking likely would also increase co-IP of non-specifically bound proteins. To address this dilemma, we tried to decrease non-specific binding by using a denaturing buffer (Buffer C, also known as RIPA buffer) containing two anionic detergents (sodium deoxycholate and SDS) in addition to a higher concentration of NP-40 (Table 1) for making cell extract and performing IP. Before performing the IP using cross-linking, we decided to test Buffer C alone to see how effective it was at removing non-specific protein-protein interactions (Condition 3). Given the strong denaturing effect of this buffer, we expected to see a substantial reduction in the amount of non-specific binding; however, we were surprised to find that there was only a slight difference between Buffers A and C (Fig. 1A). As expected, Nanog was not co-precipitated with Oct4 using Buffer C (Fig. 1B). Although co-IP was not successful, the Oct4 signal was stronger for IP performed using Buffer C compared with those done using Buffers A or B when immunoprecipitated proteins from these three conditions were run in the same Western blot, developed onto a single X-ray film (Fig. 1B, arrows). This result could be due to the increased accessibility of epitopes under the denaturing condition provided by Buffer C.
In our next experiment, Condition 4, we sought to maintain the Oct4-Nanog interaction by cross-linking proteins with DSP. Silver staining of a protein gel displayed several bands of proteins that seemed to have been specifically co-precipitated with the Oct4 antibody (Fig. 1A, arrows). Consistent with this result, Nanog was also co-precipitated (Fig. 1B). Accordant with the co-IP result, Oct4 and Nanog were eluted in the same fractions (mainly in Nos. 12 and 14) in gel filtration column chromatography of the cell extract prepared under this condition (Fig. 2B). Under denaturing conditions such as this, the unfolding of proteins makes size estimation by gel filtration chromatography inaccurate. Finally, we tested whether the use of a different cross-linking reagent would alter the results. This time, formaldehyde was used to cross-link proteins instead of DSP, utilizing Buffer C (Condition 5). Unexpectedly, under this condition Nanog was not observed to be co-immunoprecipitated, suggesting that the Oct4-Nanog interaction was not preserved by cross-linking with formaldehyde. This result could be explained by the fact that formaldehyde cross-links molecules at a shorter distance (approximately 2Å) than DSP does (12 Å) [11, 12].
Our results highlight the instability of the interaction between Oct4 and Nanog in F9 cell extract and demonstrate the effectiveness of using DSP to maintain this interaction. Furthermore, the detection of several proteins that were co-precipitated with the Oct4 antibody under this condition indicates that DSP potentially preserved the binding of other partners in this complex. More detailed studies of these proteins are required to confirm the interactions of these proteins with Oct4 and Nanog, as well as the functional meaning of the interactions. Our results assume increased importance when one considers them in light of recent findings by Yamanaka’s group [13]. They have shown that transduced Oct4, in combination with three other transcription factors, can convert mouse fibroblast cells into ES-like cells. The possibility that Oct4 could be used exogenously for stem cell therapy makes a deeper understanding of Oct4’s interactions within the cell all the more pressing for the advancement of stem cell biology.
Acknowledgments
This research was supported in part by the National Institutes of Health grants R01 GM068027 to N.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- [2].Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- [3].Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- [4].Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- [5].Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- [6].Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- [8].Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- [9].Lomant AJ, Fairbanks G. Chemical probes of extended biological structures: synthesis and properties of the cleavable protein cross-linking reagent [35S]dithiobis(succinimidylpropionate) J Mol Biol. 1976;104:243–261. doi: 10.1016/0022-2836(76)90011-5. [DOI] [PubMed] [Google Scholar]
- [10].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- [11].Fujita N, Wade PA. Use of bifunctional cross-linking reagents in mapping genomic distribution of chromatin remodeling complexes. Methods. 2004;33:81–85. doi: 10.1016/j.ymeth.2003.10.022. [DOI] [PubMed] [Google Scholar]
- [12].Pierce Biotechnology Inc. Instruction to DSP and DTSSP 2004
- [13].Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]



