Abstract
Centromeres are defining features of eukaryotic chromosomes, providing sites of attachment for segregation during mitosis and meiosis. The fundamental unit of centromere structure is the centromeric nucleosome, which differs from the conventional nucleosome by the presence of a centromere-specific histone variant (CenH3) in place of canonical H3. We have shown that the CenH3 nucleosome core found in interphase Drosophila cells is a heterotypic tetramer, a “hemisome” consisting of one molecule each of CenH3, H4, H2A, and H2B, rather than the octamer of canonical histones that is found in bulk nucleosomes. The surprising discovery of hemisomes at centromeres calls for a reevaluation of evidence that has long been interpreted in terms of a more conventional nucleosome. We describe how the hemisome structure of centromeric nucleosomes can account for enigmatic properties of centromeres, including kinetochore accessibility, epigenetic inheritance, rapid turnover of misincorporated CenH3, and transcriptional quiescence of pericentric heterochromatin. Structural differences mediated by loop 1 are proposed to account for the formation of stable tetramers containing CenH3 rather than stable octamers containing H3. Asymmetric CenH3 hemisomes might interrupt the global condensation of octameric H3 arrays and present an asymmetric surface for kinetochore formation. We suggest that this simple mechanism for differentiation between centromeric and packaging nucleosomes evolved from an archaea-like ancestor at the dawn of eukaryotic evolution.
Keywords: hemisome, histones, centromere CENP-A
DNA replication is much the same in all cellular life forms; however, segregation of duplicated chromosomes differs radically between prokaryotes and eukaryotes (1). Whereas prokaryotes generally pull chromosomes apart during replication, eukaryotic cells complete replication and then undergo mitosis, a process in which chromosomes condense, congregate, orient, attach to microtubules, and are pulled apart during a brief phase of the cell cycle. A key innovation that made mitosis possible is the kinetochore, a proteinaceous structure on the chromosome that captures spindle microtubules, which it uses to move rapidly to the pole after sister chromatids separate. The kinetochore forms at mitosis at a single dedicated site on the chromosome, referred to as the centromere. Although the vast majority of DNA in a chromosome is passively segregated during mitosis, centromeres are active participants, faithfully specifying the location of the kinetochore on every chromosome for every cell division in the lifetime of an organism. Furthermore, the centromere is inherited from one organismal generation to the next and has been conserved in position for tens of millions of years in lineages such as ours. Yet despite the central role of centromeres in this most fundamental process of eukaryotic biology, their underlying structure and specification are poorly understood.
One and only one centromere must function on every chromosome at anaphase to avoid chromosome loss, a dominant lethal event. It is therefore remarkable that the DNA sequence does not appear to play the primary role in the maintenance of centromeres of most organisms. Rather, centromeres are specified by the presence of a special nucleosome that contains a centromere-specific histone H3 variant (CenH3) in place of H3 (2, 3). Mammalian CenH3 (CENP-A) is present at all active centromeres, including neocentromeres that occur spontaneously on chromosome arms and can be inherited through mitosis and meiosis (4). Although many studies of CenH3s have been performed in a variety of organisms, the epigenetic mechanism responsible for the extraordinarily faithful inheritance of centromeres has remained mysterious. Here we review how the surprising structure and dynamics of the CenH3 nucleosome provide insights into the epigenetic inheritance of centromeres and on the possible origin of eukaryotic nucleosomes.
CenH3/H4/H2A/H2B Tetramers Form the Core of Centromeric Nucleosomes in Drosophila
Nucleosomes package the entire genomes of nearly all eukaryotes. The structure of the nucleosome is an octamer consisting of two each of the four core histones wrapping 1.7 turns of DNA (5). The solution of the high-resolution structure of the nucleosome core particle (6) has provided the structural basis for much of subsequent chromatin research. Because the core of all known CenH3s can be aligned reasonably well with canonical H3, the structure of the CenH3 nucleosome has been assumed to be a similar octamer, although in vivo evidence has long suggested an unusual structure (7, 8). The presumed octameric structure of the nucleosome core has provided the basis for models aimed at explaining the peculiar properties of centromeric nucleosomes (9–11). However, we recently made the unexpected observation that Drosophila CenH3 nucleosomes at interphase do not have stable octameric cores but rather heterotetrameric ones (12), and this finding requires a reevaluation of past studies of centromere properties that have drawn conclusions based on an octameric model. As described below, we suggest that the simplest interpretation of existing data is that interphase tetramers similar to what we have described in Drosophila cells are universal for CenH3 nucleosomes, and we discuss implications that this insight may have on centromere biology and nucleosome evolution.
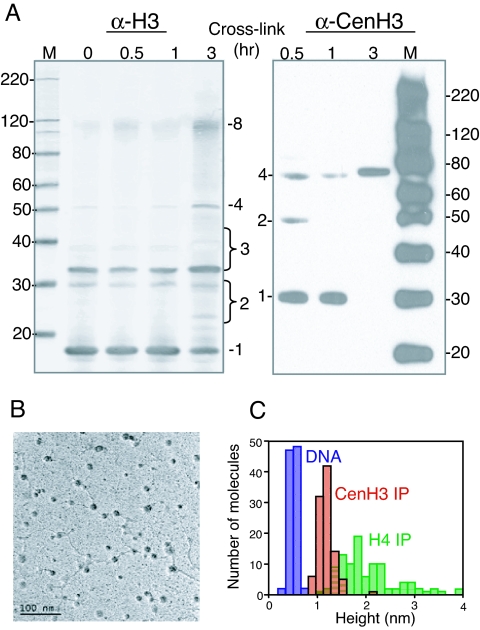
Direct evidence for a heterotetrameric CenH3 nucleosome derives from our recent biochemical characterization of nucleosomes containing Drosophila CenH3 (named Cid for centromere identifier) (12). We used a multifaceted approach, consisting of cross-linking to deduce core structure, purification to identify and quantify core protein components, nuclease digestion to assess the length of the DNA wrap, electron microscopy (EM) to estimate size distributions of core particles and DNA linkers, and atomic force microscopy to estimate relative core particle heights. Our results were unequivocal. Cross-linking and purification identified a CenH3 tetramer consisting of one molecule each of CenH3, H4, H2A, and H2B (12), referred to as a “hemisome” (13). Nuclease protection showed that this particle wraps <120 bp of DNA, and EM revealed that the particle is small, separated by long linkers, and resists ionic condensation. Finally, atomic force microscopy demonstrated that the CenH3 particle is approximately half as high as bulk nucleosomes (Fig. 1). It remains to be seen whether hemisomes are the only CenH3 species found at centromeres because we also detected a larger cross-linked species in mitotic cells that might represent a larger particle. Nevertheless, the existence of a stable nucleosome particle that is similar to a half-nucleosome of the type rarely detected in bulk eukaryotic chromatin is notable in being an exception to the octameric structure of the eukaryotic nucleosome that has been the dominant paradigm for more than 30 years (13). An intriguing possibility is that other histone variants assemble into noncanonical nucleosomes, such as the H2AL1/L2-TH2B-containing particle found in mouse heterochromatin during spermiogenesis (14) and unusual H3 and H2B variants of trypanosomes (15).
Fig. 1.
CenH3 nucleosomes are hemisomes in vivo (12). (A) Western blots of D. melanogaster S2 cell chromatin after cross-linking with dimethylsuberimidate. Anti-H3 cross-linked products include multiple dimeric (2) and trimeric (3) species, tetramers (4) and octamers (8) (Left), whereas an anti-CenH3 antibody detects only CenH3/H4 dimers (2) and CenH3/H4/H2A/H2B tetramers (4). M, markers. (B) EM shows the “beads-on-a-string” conformation of CenH3- immunoprecipitated chromatin, with extended linkers. (C) CenH3-immunoprecipitated nucleosomes display a tight distribution of heights determined by atomic force microscopy, averaging half that of bulk chromatin (H4 IP). DNA provides an internal marker. (Adapted from ref. 12 with permission.)
CenH3 Nucleosomes Found in Other Organisms
How broadly do our findings in Drosophila generalize to other eukaryotes? Most studies of CenH3s have been performed on human cells or budding yeast. Some of these studies have addressed the question of the structure of the CenH3 nucleosome, and so reexamination of the data in light of evidence from Drosophila should help us determine whether the heterotypic centromeric nucleosome could be a general feature of eukaryotic biology.
Previous in vivo studies have shown that nucleosomes containing CenH3 lack H3, but they have not directly addressed the question of whether the particle has one or two CenH3s. Production of nearly equal amounts of tagged and untagged CENP-A in HeLa cells and immunoprecipitation of tagged chromatin showed that both forms could be pulled down. However, the tagged form was in much greater excess over the untagged form than would be expected for equal inputs (refs. 16 and 17 and Y.D., unpublished data). We suggest that the low levels of untagged CENP-A resulted from using solubilized chromatin that consists of nucleosomal arrays in addition to mononucleosomes: some untagged CENP-A from neighboring nucleosomes would be pulled down together with the tagged nucleosome. In support of this interpretation, our studies of Drosophila Cid nucleosomes have indicated that no conditions of micrococcal nuclease digestion used for solubilization yielded mononucleosomes in the absence of nucleosomal arrays (12). Micrococcal nuclease digestion of fission yeast centromeric DNA also resulted in an indistinct digestion pattern (7, 18) rather than the classical 165- to 200-bp ladder with core protection of 150 bp seen for bulk chromatin (19).
In another study, yeast Cse4 alleles were found to display partial interallelic complementation, a phenomenon that is usually interpreted in terms of interactions within a homodimer (20). However, this interpretation seems unlikely in light of a subsequent study by the same group showing that one of the mutants used, with a deletion of the N-terminal tail, is by itself able to provide nearly wild-type Cse4 function if produced at high levels (21). Such suppression was interpreted by the authors as indicating a role for the N-terminal tail in stabilizing Cse4 before deposition. The partial interallelic complementation observed in the earlier experiment would suggest stabilization of preassembly forms rather than interaction within the assembled particle. In another study, the ability to enhance a presumed Cse4 homodimerization mutant by overexpressing H3 but not by overexpressing H4 was interpreted in terms of Cse4 homodimerization (22), but this negative result is also predicted for a hemisome. Thus, data from previous in vivo studies are consistent with the existence of unusual CenH3 nucleosomes in humans and yeast.
Budding yeast centromeres are exceptional in that specific DNA-binding complexes recruit Cse4 to form a single centromeric nucleosome that organizes the kinetochore (23). Budding yeast centromeres are defined by an ≈125-bp sequence, of which the outer DNA-binding motifs have been implicated in binding of the Cbf1 protein and the Ndc10 complex. Only the central ≈80-bp A/T-rich sequence appears to be available for interaction with a Cse4 nucleosome, which would seem to be too small to wrap an octameric nucleosome. This enigma has led to the proposal that CenH3 nucleosomes flank the functional centromere (24). However, single-nucleosome mapping has definitively localized CenH3 precisely to the functional centromere (23). A tetrameric nucleosome can accommodate this CenH3 mapping with the apparent availability of only 80 bp for wrapping. Although a budding yeast (Cse4/H4)2 form that lacks H2A/H2B has been proposed (25), this proposal was not based on direct characterization of the CenH3 nucleosome in vivo but rather on comparing relative levels of DNA pulled down by antibodies from formaldehyde-cross-linked and sonicated chromatin. The recovery of immunoprecipitated chromatin likely depends on the number and distribution of cross-linkable lysines on histones, which differ between Cse4 and H3. Indeed, Drosophila CenH3 does not detectably cross-link directly to H2A or H2B, but only to H4, yielding a single dimeric 45-kDa intermediate, whereas multiple dimeric intermediates are seen for cross-linked H3 (Fig. 1A and ref. 12). Another complication is the use of different epitopes for immunoprecipitation of H4 and of H2A and H2B in that study (25): the H4 epitope lacks lysines (N-DNIQ-C), whereas the H2A and H2B FLAG tag epitope (N-DYKDDDDK-C) is potentially subject to enhanced cross-linking of lysines within the kinetochore region, resulting in epitope masking. A (Cse4/H4)2 tetramer is also difficult to reconcile with the isolation of the Mif2 complex associated with Cse4, H4, H2A, and H2B but not H3 (26) and with genetic evidence for a role of H2A in centromere function (27). Mif2 is the budding yeast CENP-C ortholog (28); in Drosophila, its conserved C-terminal domain is cytologically near CenH3, whereas its N-terminal domain is near the Ndc80 complex, which interacts with spindle microtubules (29). Taken together, these observations suggest that CenH3/H4/H2A/H2B hemisomes are present at yeast centromeres.
The existence of a CenH3 hemisome in vivo is consistent with previously published studies of reconstituted CenH3 nucleosomes. Reconstitution of CENP-A nucleosomes in vitro showed that the particles are smaller than bulk nucleosomes; contain CENP-A, H4, H2A, and H2B; require twice as many nucleosomes to obtain equivalent supercoiling; and protect significantly less DNA relative to bulk nucleosomes, facts noted by the authors but not interpreted in terms of an inherently smaller particle (30). More recently, a detailed study of reconstituted CENP-A octameric nucleosomes showed that they are inherently less stable than H3 nucleosomes and specifically noted the inability to reconstitute DNA onto (CENP-A/H4)2 homotypic tetramers (31). Notably, reconstitution requires the use of 2 M salt to form core particles in the absence of DNA, whereas in vivo, histone subunits are assembled by chaperones directly onto DNA in an ≈0.15 M ionic strength environment. In our study of Drosophila nucleosomes, CenH3-containing protein species larger than tetramers could be obtained in 2 M salt and cross-linked in vitro in the absence of DNA (12). However, no comparable species could be detected in chromatin in vivo, which implies that forcing octamers to form under nonphysiological conditions, in the absence of DNA, might not be relevant to the process of chaperone-mediated assembly in vivo.
Reconstituted nucleosomes [or “tetrasomes” when only H3 and H4 are provided (32)] display features that are consistent with those obtained by purification of particles from native chromatin (33), which amply justifies their widely accepted use in chromatin studies. However, properties deduced from in vitro studies of CenH3 nucleosomes must be interpreted cautiously until direct observations of stable CenH3-containing octamers in vivo are presented. Thus far, we know of no compelling evidence for more than one type of centromeric nucleosome in vivo, and therefore it seems most likely that the CenH3/H4/H2A/H2B hemisome is the universal core that wraps centromeric DNA.
CenH3 Deposition and Rapid Removal from Chromosome Arms
One of the implications of a CenH3 hemisome is that it should be more easily evicted from DNA than an H3 octamer in vivo because much less DNA wraps around a tetramer. Nucleosome eviction and histone replacement are now well documented features of active chromatin, as measured by the accumulation of the universal replacement variant, histone H3.3 (34). Histone turnover rates have been measured directly over the yeast genome by using tagged versions of yeast H3, which is the yeast counterpart of H3.3 (35). As a result of these and other in vivo studies of histone dynamics, a consistent picture of histone turnover is emerging, whereby active genes and regulatory sites, including promoters, boundary elements, and cis-regulatory memory elements, are sites of conspicuous histone replacement (34). Furthermore, a precedent exists for nucleosome instability that depends on the histone 3 variant that is incorporated, because H3.3 nucleosomes are less stable in vitro than H3 nucleosomes, and this difference in stability is greatly amplified by the incorporation of another histone variant, H2A.Z (36). The relative instability of active chromatin, probably driven by nucleosome remodeling, leads to the expectation that CenH3 hemisomes misincorporated into regions of active chromatin will be rapidly evicted, which, indeed, appears to be the case.
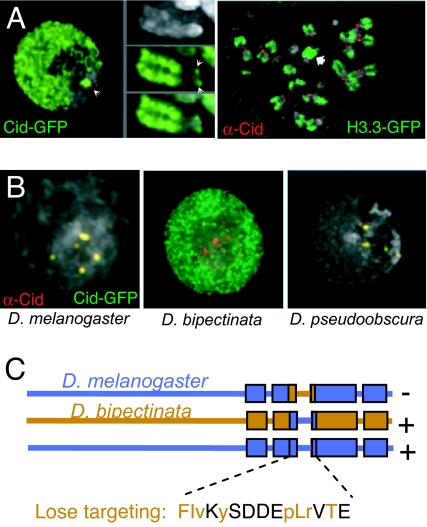
Overexpression of CenH3s results in their deposition outside of the centromere. Mislocalization was seen for overexpressed human CENP-A, which appeared to accumulate broadly throughout the nucleus (37). CENP-A is also overexpressed and mislocalized in some human cancers (38). Similarly, Drosophila Cid-GFP mislocalizes broadly within the nucleus when overexpressed, where the existence of a single large heterochromatic chromocenter reveals that mislocalization is strictly limited to euchromatin (Fig. 2A Left) (39). Mislocalization involves incorporation into euchromatin, as illustrated by the abundant presence of Cid-GFP on the arms of metaphase chromosomes. An important extension of these findings was the demonstration that mislocalized Cid could recruit kinetochore proteins and attach to microtubules, resulting in segregation defects (40). The segregation machinery appears to be sensitive to the levels of Cid produced in excess because in cells in which all chromosomes displayed prominent euchromatic localization of Cid-YFP, no chromosomes moved to the poles at anaphase, whereas chromosomes in cells with only a moderate excess of mislocalized Cid-YFP segregated normally (41). Although it is well established that CenH3s are necessary for kinetochore formation in many organisms, these results also provide strong evidence that some critical local concentration of Cid is sufficient for centromere function (40). Moreover, the competence of mislocalized Cid to incorporate throughout chromosome arms and direct kinetochore formation is further evidence that DNA sequence is not the key requirement for centromere identity in Drosophila.
Fig. 2.
Mislocalization of overexpressed and mutant Cid to euchromatin. (A) Overexpressed Cid-GFP localizes to both centromeres (arrowheads) and euchromatin in interphase cells and is incorporated into metaphase chromosomes in Drosophila Kc cells but not into the heterochromatic chromocenter (Left) (60). H3.3 is found in chromosome arms and rDNA (arrow) but is undetectable in heterochromatin and centromeres (Right). (B) GFP fusions with Cid from D. melanogaster and Drosophila pseudoobscura localize to D. melanogaster centromeres, whereas a GFP fusion with Cid from D. bipectinata shows a euchromatic distribution (80). (C) Swaps between segments of melanogaster (blue) and bipectinata (orange) Cid show that the 15-aa loop 1 segment alone is responsible for targeting (80). Single amino acid substitutions to glycine or alanine cause melanogaster Cid-GFP to display a euchromatic distribution (orange letters), where uppercase letters indicate mislocalization when expressed under both the Cid endogenous promoter and an induced heat shock promoter, and lowercase letters indicate when mislocalization occurred only with the induced heat shock promoter. [Reproduced with permission from ref. 39 (A Left), ref. 60 (A Right), and ref. 80 (B).]
In budding yeast, a mutant Cse4 protein that is resistant to proteolysis accumulates on chromosome arms, leading to segregation defects (42), and overexpressed Cse4 in wild-type cells shows evidence of mistargeting (21). Similar results were obtained in Drosophila, where growth in the presence of a proteasome inhibitor led to Cid accumulation on chromosome arms (41). Because nucleosome cores are especially resistant to proteolysis (43), these results are most easily explained by the incorporation and subsequent eviction of mislocalized CenH3. Promiscuous deposition and eviction of CenH3 appear to occur normally in certain situations: during the pachytene stage of Arabidopsis meiosis CenH3 accumulates at low levels on chromosome arms, followed at later stages by the appearance of spherical cytoplasmic particles (44) that resemble proteasome-rich aggresomes (45).
The ready eviction of CenH3 nucleosomes from chromosome arms is in striking contrast to its retention at centromeres (Fig. 2A Left). A possible explanation for this difference comes from the fact that centromeres are typically embedded in large blocks of pericentric heterochromatin, and heterochromatin appears to be inherently favorable for centromere function. For example, a distal ≈1-Mb block of Drosophila heterochromatic satellite DNA displays weak centromere activity, accumulates Cid and mediates segregation to poles at anaphase when released from linkage to the native centromere (46). Heterochromatin and centromeres are notably deficient in H3.3 (Fig. 2A Right) (39), which implies that histone turnover is especially low in these regions of the genome. The association of heterochromatin-associated protein 1 (HP1) with human neocentromeres (47) is consistent with low levels of histone turnover because in Drosophila, H3.3 is deficient in euchromatic genes that are rich in HP1 (48). Like human neocentromeres, rice centromere 8 is largely devoid of satellite DNA repeats, yet CenH3 chromatin also shows heterochromatic features, including high levels of H3K9me2 (49). Furthermore, the alternation of CENP-A with H2A.Z nucleosomal arrays at native mammalian centromeres might be another mechanism for reducing turnover because H2A.Z appears to stabilize H3-containing nucleosomes in HP1-rich heterochromatin (36, 50, 51). Therefore, CenH3s deposited in such regions might escape eviction, and indeed, heterologous CenH3s introduced into Drosophila and human cells accumulate specifically in pericentric heterochromatin (52). We suggest that this preferential accumulation in heterochromatin reflects the lack of histone turnover, which in euchromatin normally evicts histones during transcription and other active processes (53). Thus, the defining features of pericentric heterochromatin might be interpreted as adaptations to prevent CenH3 eviction, assuring an epigenetic mechanism by which kinetochores can assemble at the same location every cell cycle.
Chaperone-Mediated Deposition of CenH3 Nucleosomes
The correct wrapping of DNA around basic histone cores is energetically favorable, but it does not occur spontaneously under physiological ionic conditions. To facilitate this process in vivo, a variety of histone chaperones have evolved that promote DNA wrapping and prevent nonspecific aggregation in the context of high protein concentrations found in the nucleus (54). For example, anti-silencing factor 1 (Asf1) forms a trimolecular complex with H3 and H4, binding H3 within the H3/H3 dimerization surface such that it blocks dimerization in solution and releases upon deposition (55, 56). Other histone chaperones are found in complexes. The best studied of these chaperones is the chromatin assembly factor 1 (CAF-1) complex, which acts at the replication fork to assemble nucleosomes on newly synthesized DNA, tethered to the DNA polymerase processivity clamp (57). CAF-1 deposits H3/H4 as a dimer, not a tetramer (58), raising the possibility that successive incorporation of two H3/H4 dimers behind the replication fork is carried out by the successive action of CAF-1 and Asf1 (59).
Histone variants are distinguished from canonical histones by being deposited outside of replication. For example, canonical H3 is restricted to deposition during replication and is the substrate for CAF-1, but 3 aa on the H3.3 core that distinguish H3.3 from H3 allow replication-independent assembly (60). In human cells, the HirA complex deposits H3.3 in a replication-independent manner (58). CenH3s also are deposited outside of replication, but their low abundance relative to H3 and H3.3 and the small pools of soluble histones in cells relative to those found in chromatin have made the identification of the soluble CenH3 complex a challenging task (61). To overcome these limitations, we used a biotin-tagging system, which led to the identification of a trimolecular complex containing CenH3, H4, and RbAp48, an abundant chaperone protein that is also found in the CAF-1 and HirA complexes (62). The CenH3/H4-RbAp48 complex, whether purified or reconstituted, was able to assemble CenH3 nucleosomes in a standard supercoiling assay, which provided direct evidence that the soluble complex observed in vivo is sufficient for replication-independent nucleosome assembly. It is likely that the same complex assembles nucleosomes in many other lineages because RNAi knockdown of RbAp48 in human cells caused failure of CENP-A to localize (63). Moreover, Schizosaccharomyces pombe RbAp48 (Mis16) is required for centromere assembly (63). The absence of RbAp48 from purified human centromeric chromatin (64) suggests that it acts as a traditional protein chaperone [an “escort” (54)] as opposed to one that becomes tethered to the site of assembly.
The presence of RbAp48 as an equimolar component of the chaperone complexes specific for all three variant histone 3 forms (H3.1/H4, H3.3/H4, and CenH3/H4) is consistent with its role as a general chromatin assembler (65). RbAp48 is also a component of other chromatin-associated complexes, including a nucleosome remodeler (Drosophila NURF), a histone acetyltransferase (yeast HAT2), and a Polycomb group-silencing complex (Enhancer-of-zeste). RbAp48 is a WD-40 protein, characterized by seven β-propeller motifs organized in a ring, which could provide multiple surfaces for interactions with different components. The use of this abundant general histone chaperone for incorporation of CenH3/H4 (62) might account for promiscuous incorporation of CenH3 nucleosomes throughout euchromatin when overproduced (39–41).
It appears that the budding yeast counterpart of RbAp48 is not the only CenH3 chaperone because its ortholog (CAC3) is nonessential, and temperature-sensitive cse4− mutants show merely phenotypic enhancement in an RbAp48-null (cac3Δ) strain, not synthetic lethality (20). Recently, three studies described the properties of Scm3, a Cse4-binding protein (25, 66, 67) that had been originally identified as the product of a high-copy suppressor of Cse4 mutants, a phenotype consistent with a role in stabilizing soluble Cse4 (20). Scm3 is a 223-aa protein that has features of a histone chaperone, including a nuclear export signal and an essential heptad repeat domain that interacts with Cse4 (66). Scm3 resembles Asf1 and RbAp48 in that it forms a trimolecular complex with its dimeric substrate in solution (25). Like Asf1, Scm3 binds to the C-terminal portion of Cse4 (66), which is consistent with the possibility that Scm3 prevents Cse4 dimerization. However, unlike Asf1, Scm3 does not release from Cse4/H4 upon deposition but rather can be immunoprecipitated with chromatin in the immediate vicinity of the Cse4 nucleosome. Scm3 associates with Ndc10 (67), a component of the CBF3 complex, which is required to localize Cse4 to yeast centromeres (68). Although a model has been proposed in which two molecules of Scm3 are bound to a (Cse4/H4)2 tetrasome (25), the formation of a Cse4/Cse4 four-helix bundle at the presumed dimerization interface would seem to preclude retention of Scm3 at the same interface. Interestingly, fission yeast RbAp48 is also associated with centromeres (63), which might be another example of a protein chaperone playing a structural role at fungal centromeres. These biochemical studies appear to be most consistent with the assembly of a Cse4 hemisome at yeast centromeres such as what we have described for Drosophila.
Targeting CenH3s to Centromeres
The importance of faithful propagation of centromeres for segregation and genomic integrity means that there are likely to be multiple mechanisms that target CenH3 nucleosomes to centromeres. Unlike budding yeast, plants and animals lack specific sequences for localizing centromeres. This lack of sequence specificity is especially evident at human and barley neocentromeres (4, 69, 70) and native centromeres on rice chromosome 8, which is packaged in extensive arrays of CenH3 nucleosomes but is almost devoid of tandem repeats or other sequences found at most rice centromeres (49, 71). The promiscuous deposition and ready eviction of CenH3s from active regions are one way that high concentrations of CenH3 nucleosomes can be retained at centromeres. Another way would be to prevent the deposition of H3 nucleosomes at centromeres during replication. S phase expression of human CENP-A led to mislocalization throughout the nucleus (17), and several studies have shown that CenH3s from different species deposit from G2 through mitosis and the following G1 but not during S phase (72–77). These observations are consistent with the idea that CAF-1 leaves “holes” at centromeres that are later filled by CenH3s (78). An extension of this model is that targeting of CenH3 is a rather passive process, whereby CenH3/H4-RbAp48 deposits in holes (62). Further work is needed to establish the mechanistic basis for the specific exclusion of H3 nucleosomes from centromeres and for the longer spacing of CenH3 hemisomes at Drosophila centromeres.
The precise amino acid sequence of a CenH3 can also affect its localization to centromeres as opposed to being distributed throughout active chromatin, and there appear to be specific residues within the core of CenH3s that are necessary for preferential centromere targeting or retention (17, 79, 80). For example, Saccharomyces cerevisiae Cse4 can replace human CENP-A (81), yet the Cid from a close relative of Drosophila melanogaster, Drosophila bipectinata, even fails to localize to D. melanogaster centromeres correctly, instead accumulating throughout euchromatin (Fig. 2B) (80). The failure of D. bipectinata Cid to localize to D. melanogaster centromeres but rather to accumulate on chromosome arms provided us with an assay for precisely mapping the region responsible for targeting or retention (80). When D. bipectinata loop 1 together with several amino acids on either side was substituted for D. melanogaster loop 1, specific localization was lost, and conversely, when D. melanogaster loop 1 was substituted for D. bipectinata loop 1, centromere targeting was restored. Swapping of other regions of Cid had no effect on targeting, demonstrating that loop 1 is both necessary and sufficient for correct targeting of Cid. This conclusion appears to generalize because Cse4 loop 1 and CENP-A loop 1 are regions that do not appear to support viability (yeast) (79) or localization (human) (17) when substituted for their H3 counterparts. Lack of sufficiency in these earlier experiments might be explained by the presence of other differences between CenH3 and H3, which have evolved under very different constraints. For example, centromere targeting of an H3/CENP-A chimeric protein was achieved by using a 44-aa segment of CENP-A (10) that replaced both H3 loop 1 and the three H3 residues (Ser87, Val89, and Met90) that block replication-independent assembly (60).
CenH3 loop 1 is distinctive because it is longer than H3 loop 1 by 1–6 aa in all well studied species (9). To identify better the sequence constraints on loop 1, we mutated each of the 15 residues in the critical region of the D. melanogaster-targeting domain, which led to the identification of 8 residues near both ends of loop 1 that are required for targeting of melanogaster Cid and 7 that are not (Fig. 2C) (80). The fact that critical amino acids are distributed throughout most of loop 1 suggests that the overall conformation of loop 1 is important for Cid targeting.
Evolutionary studies have also implicated loop 1 in centromere specificity. In Drosophila, both the Cid core and tail are adaptively evolving, with the most frequent core residue differences lying in loop 1 (82). Similarly, Arabidopsis CenH3 is under positive selection in both the core and the tail, and adaptive evolution of the core is mostly accounted for by residues within loop 1 (83). Adaptive evolution implies an arms race, and we have proposed that the conflict is between centromere sequences attempting to “win” during female meiosis I by orienting favorably to the egg pole, resisted by kinetochore proteins encoded by the host that would favor parity between competing centromeres (84). As a result of this conflict, centromeric satellite repeats would expand to assemble or retain more CenH3 nucleosomes, only to encounter variant kinetochore proteins that would be selected if they restore meiotic parity.
A Possible Structural Basis for CenH3 Hemisome Formation
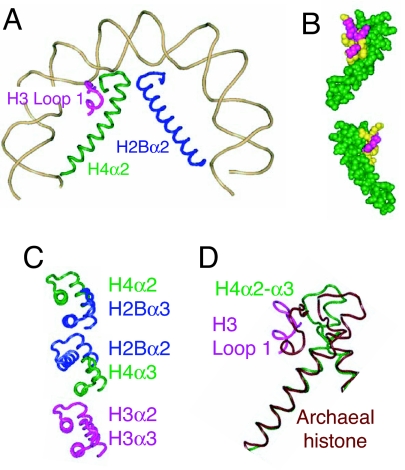
H3 loop 1 abuts H4α2 + loop 2 and extends to the surface of the octamer (Fig. 3A) (6). The extra amino acids present in CenH3 loop 1 seem most consistent with its further extension above the surface of the core. The proposed structural difference between the normal CenH3/H4/H2A/H2B tetramer and an octamer seems unlikely to involve binding of another protein to this extension because in vitro studies with pure components show similar DNA-packaging constraints (31). Rather, our attention is drawn to the interactions of H3 loop 1 with H4α2 and with H4 loop 2. α2 of all four core histones form pairs of struts that span the length of the H3/H4 and H2A/H2B heterodimers, providing the stiff superstructure that supports the tightly wrapped DNA gyres. Whereas the α2s of H3, H2A, and H2B are relatively straight, H4α2 is conspicuously bent (6) (Fig. 3B). Remarkably, mutated residues that result in mistargeting of Drosophila Cid (80) align with H3 residues that make direct contact with H4α2 + loop 2, interactions that cause H4α2 to bend tightly around the H3 loop 1 + H4 loop 2 subdomain (6). Thus, an attractive explanation for the failure of these loop 1 mutant proteins to target centromeres is that they alter the bending of H4α2, resulting in failure of the (CenH3/H4)2 tetramer to form.
Fig. 3.
A structural change proposed to underlie the transition from wild-type to mutant Cid. (A) Half-nucleosome showing the juxtaposition of H3 loop 1 (magenta), H4α2 (green), and H2Bα2 (blue) and the bending of H4α2 around the subdomain created by H3 loop 1 + H4 loop 2 [Protein Data Bank (PDB) ID code 1KX5] (6). (B) Space-fill model showing top and bottom views of the interactions between H3 loop 1 residues whose CenH3 counterparts are critical for targeting to centromeres (yellow) and H4α2 + loop 2 (green). (C) Alignment of α2 and α3 helices comprising the three four-helix bundles that hold together conventional octameric nucleosomes. The three-dimensional structure of the nucleosome shows that H2Bα2 and H4α3 (Middle) are misaligned relative to alignments of H4α2 with H2Bα3 (Top) and H3α2 with H3α3 (Bottom) (6). It is proposed that H2Bα2 and H4α3 become well aligned in CenH3 nucleosomes, thus strengthening the four-helix bundle in the middle of the CenH3 core. (D) Aligning the dimeric histone from M. kandleri (PDB ID code 1F1E) with H3/H4 at the N-terminal residues of H4α2 [from yeast Asf1-H3/H4 (PDB ID code 2HUE] shows that the corresponding helix in this tetrameric archaeal histone is straight, whereas H3/H4 bends tightly around the H3 loop1 + H4 loop 2 subdomain.
Straightening H4α2 could favor the formation of a CenH3/H4/H2A/H2B tetramer relative to an octamer by affecting the stability of four-helix bundles at both ends of the strut, which consist of C-terminal portions of α2 and α3 of H3, H4, and H2B (6). In octamers, the H3/H3 dimerization interface comprises a strong four-helix bundle at the dyad axis. The two H4/H2B four-helix bundles are weaker, probably because H2Bα2 is noticeably misaligned with H4α3 (Fig. 3C). We suggest that a straighter H4α2 will allow for better alignment of these two helices, thus strengthening the interface between CenH3/H4 and H2A/H2B dimers; as a result, the CenH3 hemisome would be more stable than a corresponding H3 half-nucleosome, which has not been detected in vitro or in vivo. At the same time, straightening of H4α2 would likely affect interactions at the other end of the strut. One possibility is a slight change in the orientation of the CenH3 interface with itself, resulting in a distortion of the four-helix bundle, and another is lengthening the distance between DNA gyres, resulting in an elongated nucleosome that might disfavor its tight wrapping around a CenH3/CenH3 four-helix bundle. In either case, octamer or homotypic tetramer formation would be disfavored. By this speculative scenario, the stability of CenH3 hemisomes that we observed in vivo would derive from both the strengthening of the interface between H4 and H2B and the weakening of the CenH3 homodimerization interface relative to the H3 octameric nucleosome. Straightening of H4α2 in CenH3-containing particles might also account for unusual properties of (CenH3/H4)2 and (CenH3/H4/H2A/H2B)2 nucleosomes assembled in vitro (10, 31, 85). These particles are reported to be smaller and conformationally rigid based on physical measurements and are unstable relative to H3-containing tetrasomes and nucleosomes. A consequence of straightening H4α2 is that its C-terminal end will move inward relative to the pseudodyad axis, resulting in more compact core particles that would require input of additional free energy to wrap the DNA more tightly. The relative instability of octameric CENP-A-containing nucleosomes has been attributed to the compactness of these particles (31), and we propose that the underlying cause is partial straightening of H4α2. Whether or not symmetrical CenH3 octamers exist in vivo is not known, although the reported inability to produce (CENP-A/H4)2 tetrasomes wrapping DNA in vitro (31) raises the question of what would stabilize folding intermediates during the multistep assembly of such a nucleosome.
Implications for Kinetochore Structure
Hemisomes represent a third structural form, the other two being the familiar octameric nucleosomes that package eukaryotic genomes and a variety of tetrameric nucleosomes that package many archaeal genomes. Unlike these other two forms, which show mirror-image symmetry around a pseudodyad axis, CenH3 hemisomes lack a homodimerization interface and so are expected to be inherently asymmetrical. This asymmetry, if maintained at mitosis, might serve a biological function. Whereas the fibers and higher-order structures formed by packaging histones have no preferred orientation, centromeres are inherently asymmetrical in the mitotic chromosome, whereby only the outer face is available for kinetochore formation and microtubule attachment, becoming the leading edge of the chromosome as it moves poleward at anaphase.
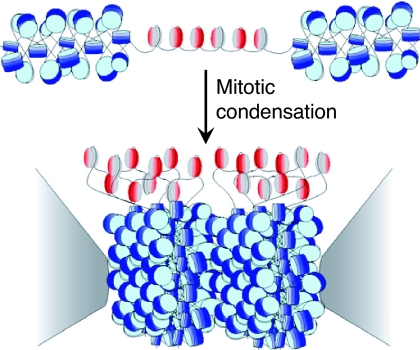
The strong anaphase pulling force exerted on the kinetochore has led to the widely held view that its chromatin foundation is rigid rather than flexible to withstand the presumed mitotic torque (10, 86). However, this concept is just the opposite of what is actually observed in vivo: a tetrameric nucleosome that does not cross-link to neighboring nucleosomes is separated by long linkers and is readily evicted (12, 41). How can these in vivo observations be reconciled with the intuitive notion that kinetochores should be strong and stiff? Perhaps the answer lies in the fact that kinetochores occupy only the outer surface of centromeric chromatin (87, 88), supported by a foundation of dense heterochromatin that is needed for proper cohesion (89, 90). At physiological concentrations, nucleosome core particles can form liquid crystalline stacks in vitro (91), and during chromosome condensation a regular densely packed structure is seen (92). If so, then a possible role of CenH3 nucleosomes would be to keep the surface accessible to kinetochore proteins, and the role of heterochromatin would be to provide the stiff foundation. A simple mechanism for forming an accessible surface within a three-dimensional liquid crystalline array of nucleosomes would be to “dope” with subunits that do not pack into the array, and asymmetric tetramers seem ideal for this role (Fig. 4). At a high enough local concentration along the chromosome, tetrameric nucleosomes would form a patch that resists condensation during mitosis, ending up on the surface of the liquid crystalline heterochromatin (92), at what becomes the primary constriction. It has long been thought that the repetitiveness of satellite DNA has evolved to phase nucleosomes for orderly packing (93), and the use of a reverse repeat of human α-satellite DNA for nucleosome crystallization studies supports this idea (6, 94). We propose that CenH3s have evolved to retain intrinsic tetramer flexibility and asymmetry so as to interrupt mitotic crystallization of heterochromatin octamers, thereby presenting a single accessible surface for kinetochore formation. Whereas packaging histones are tightly constrained by packing interactions and the need to package all of the sequences in the genome, the loose packing of centromeric nucleosomes would have allowed the CenH3 subunit to explore a broader mutational space to adapt to rapidly evolving centromeric satellite DNAs and to suppress the meiotic drive (95).
Fig. 4.
Model for the kinetochore. CenH3 hemisomes (red/gray disks) are separated by extended linker DNAs and so are decondensed relative to surrounding heterochromatin (blue disks). Asymmetric CenH3 nucleosomes assemble in random orientations [CenH3/H4 (red) and H2A/H2B (gray)]. Only one unit of a CenH3-rich block is shown. During mitotic condensation, heterochromatin packs tightly as a result of its homogeneity. Intervening blocks of CenH3 chromatin cannot pack into this crystal-like structure because of its smaller size, long linkers, and heterogeneity in its relative orientation, resulting in extruded loops of uncondensed CenH3 nucleosomes that serve as the foundation for kinetochore formation. The flanking gray cones represent pericentric regions flanking the primary constriction.
From Archaeal to CenH3 to Octameric Nucleosomes
The CenH3 hemisome might be an evolutionary link to more ancient forms of nucleosomes. All three major branches of Archaea include species that encode histone fold proteins, many of which are known to form tetrameric nucleosomes that function to package archaeal genomes (96). Although there is only limited sequence similarity between archaeal, H3/H4 and H2A/H2B dimers, their structural alignments within 2 Å provide compelling evidence that they arose from a common ancestor.
Conventional scenarios have assumed that the first eukaryal form was the (H3/H4)2 tetramer, which leads to the problem of the origin of H2A/H2B dimers (9). If H2A/H2B arose later in evolution than (H3/H4)2, we are left with the enigma that all extant eukaryotes [except for the late-branching dinoflagellates, which have lost their histones (97)] appear to encode H3, H4, H2A, and H2B forms, and yet a very long evolutionary period must have been necessary for H2A/H2B dimers to diverge so far from H3/H4 that there is no remaining sequence similarity. One possibility is that the present-day eukaryal octamer is a chimera derived from two different hypothetical ancestors.
An alternative scenario is that the four core histones evolved by neofunctionalization within an ancestral packaging tetramer to form a CenH3 heterotetramer. It is likely that CenH3 nucleosomes were present in the earliest eukaryotes because nearly all eukaryotic clades appear to encode a candidate CenH3, and mitosis is perhaps the most fundamental process that distinguishes eukaryotes from prokaryotes. Specialization of an asymmetric nucleosome for segregation and competition for a segregation advantage might have led to rapid divergence of the ancestral CenH3/H4 dimeric unit to yield completely distinct H2A/H2B and CenH3/H4 dimers similar to those found in present-day nucleosomes. CenH3 hemisomes might have evolved by direct descent from an archaeal–eukaryal common ancestor without ever having gone through a stable octameric intermediate form. Thus, the transition from hemisome to nucleosome might have involved only minor substitutions in loop 1, which would have altered contacts with H4α2 + loop 2. Emergence of symmetric octamers by mirror-image duplication of a CenH3 hemisome would have allowed cells to package their genomes more efficiently while retaining an ancestral hemisome dedicated to mitosis.
There are already precedents for differentiation within the archaeal tetramer. Although most archaeal nucleosomes are tetramers consisting of four single chains, at least two clades have independently evolved dimeric histone proteins consisting of an N-terminal core that structurally aligns with H3 and H2A and a C-terminal core that aligns with H4 and H2B (Fig. 3D) (96). Alignment of the Methanopyrus kandleri dimer with H3/H4 and H2A/H2B dimers shows that the archaeal dimer lacks the conspicuous bending of H4α2, providing strong support for our proposal that a straighter helix in this position is a characteristic of histone fold proteins that have evolved as tetramers. Small changes in loop 1 would then be sufficient for octamer formation. As more archaeal and protist genomes are sequenced, we anticipate the discovery of other tetrameric nucleosomes that could account for the remaining transitions in the evolution of archaeal and CenH3 nucleosomes.
Conclusion
Centromeres were described and their function deduced even before the rediscovery of Mendel's laws (98), and yet their basic properties have remained a subject of speculation. In part, this situation arises from the intractability of the highly repetitive sequences that most commonly studied centromeres inhabit and in part from the very low concentration of kinetochore components relative to bulk histones. Another factor is that the structure of the octameric nucleosome has been so important for our understanding of eukaryotic biology that the possibility of another very different eukaryal form has not been seriously considered. Our observation that Drosophila CenH3 assembles into stable hemisomes provides an attractive interpretation for many observations that were not satisfactorily explained when originally made. We hope that these insights will lead to a deeper understanding of centromere biology and nucleosome evolution.
Acknowledgments
We thank Harmit Malik, Paul Talbert, and Sue Biggins for sharing many insights on centromeric nucleosomes during the course of the studies described here. Y.D. was supported by National Science Foundation Grant DBI-0234960; T.F. was supported by National Institutes of Health (NIH) Grant F32-GM071230; D.V. was supported by NIH Grant R01-GM074108; S.H. was supported by the Howard Hughes Medical Institute.
Abbreviations
- Asf1
anti-silencing factor 1
- CAF-1
chromatin assembly factor 1
- CenH3
centromeric histone 3
- Cid
centromere identifier.
Footnotes
The authors declare no conflict of interest.
References
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Mol Biol Cell. New York: Garland; 1989. [Google Scholar]
- 2.Bloom K. Curr Opin Genet Dev. 2007;17:151–156. doi: 10.1016/j.gde.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Kamakaka RT, Biggins S. Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 4.Warburton PE. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 5.Thomas JO, Kornberg RD. Proc Natl Acad Sci USA. 1975;72:2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 7.Polizzi C, Clarke L. J Cell Biol. 1991;112:191–201. doi: 10.1083/jcb.112.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom KS, Carbon J. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- 9.Malik HS, Henikoff S. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 10.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 11.Bloom K, Sharma S, Dokholyan NV. Curr Biol. 2006;16:R276–R278. doi: 10.1016/j.cub.2006.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalal Y, Wang H, Lindsay S, Henikoff S. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavelle C, Prunell A. Cell Cycle. 2007;6:e1–e7. doi: 10.4161/cc.6.17.4631. [DOI] [PubMed] [Google Scholar]
- 14.Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, Caron C. J Cell Biol. 2007;176:283–294. doi: 10.1083/jcb.200604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowell JE, Kaiser F, Janzen CJ, Cross GA. J Cell Sci. 2005;118:5721–5730. doi: 10.1242/jcs.02688. [DOI] [PubMed] [Google Scholar]
- 16.Vafa O, Sullivan KF. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- 17.Shelby RD, Vafa O, Sullivan KF. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewish DR, Burgoyne LA. Biochem Biophys Res Commun. 1973;52:504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morey L, Barnes K, Chen Y, Fitzgerald-Hayes M, Baker RE. Eukaryot Cell. 2004;3:1533–1543. doi: 10.1128/EC.3.6.1533-1543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glowczewski L, Yang P, Kalashnikova T, Santisteban MS, Smith MM. Mol Cell Biol. 2000;20:5700–5711. doi: 10.1128/mcb.20.15.5700-5711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuyama S, Biggins S. Proc Natl Acad Sci USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espelin CW, Simons KT, Harrison SC, Sorger PK. Mol Biol Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 26.Westermann S, Cheeseman IM, Anderson S, Yates JR, III, Drubin DG, Barnes G. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto I, Winston F. EMBO J. 2000;19:1598–1612. doi: 10.1093/emboj/19.7.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meluh PB, Koshland D. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schittenhelm RB, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, Lehner CF. Chromosoma. 2007;116:385–402. doi: 10.1007/s00412-007-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Proc Natl Acad Sci USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conde ESN, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 32.Alilat M, Sivolob A, Revet B, Prunell A. J Mol Biol. 1999;291:815–841. doi: 10.1006/jmbi.1999.2988. [DOI] [PubMed] [Google Scholar]
- 33.Stein A, Bina-Stein M, Simpson RT. Proc Natl Acad Sci USA. 1977;74:2780–2784. doi: 10.1073/pnas.74.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mito Y, Henikoff J, Henikoff S. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 35.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 36.Jin C, Felsenfeld G. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 38.Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- 39.Ahmad K, Henikoff S. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno-Moreno O, Torras-Llort M, Azorin F. Nucleic Acids Res. 2006;34:6247–6255. doi: 10.1093/nar/gkl902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins KA, Furuyama S, Biggins S. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 43.Whitlock JP, Jr, Simpson RT. J Biol Chem. 1977;252:6516–6520. [PubMed] [Google Scholar]
- 44.Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gordon C. Curr Top Microbiol Immunol. 2002;268:175–184. doi: 10.1007/978-3-642-59414-4_7. [DOI] [PubMed] [Google Scholar]
- 46.Platero JS, Ahmad K, Henikoff S. Mol Cell. 1999;4:995–1004. doi: 10.1016/s1097-2765(00)80228-2. [DOI] [PubMed] [Google Scholar]
- 47.Saffery R, Irvine DV, Griffiths B, Kalitsis P, Wordeman L, Choo KH. Hum Mol Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- 48.de Wit E, Greil F, van Steensel B. PLoS Genet. 2007;3:e38. doi: 10.1371/journal.pgen.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 50.Fan JY, Rangasamy D, Luger K, Tremethick DJ. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 51.Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. Proc Natl Acad Sci USA. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henikoff S, Ahmad K, Platero JS, van Steensel B. Proc Natl Acad Sci USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Workman JL. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 54.Loyola A, Almouzni G. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 55.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Z, Shibahara KI, Stillman B. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 58.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 59.Annunziato AT. J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 60.Ahmad K, Henikoff S. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 61.Furuyama T, Henikoff S. Cell Cycle. 2006;5:1269–1274. doi: 10.4161/cc.5.12.2889. [DOI] [PubMed] [Google Scholar]
- 62.Furuyama T, Dalal Y, Henikoff S. Proc Natl Acad Sci USA. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 64.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, III, Cleveland DW. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 65.Hennig L, Bouveret R, Gruissem W. Trends Cell Biol. 2005;15:295–302. doi: 10.1016/j.tcb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 66.Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 68.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saffery R, Sumer H, Hassan S, Wong LH, Craig JM, Todokoro K, Ansderson M, Stafford A, Andy Choo KH. Mol Cell. 2003;12:509–516. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
- 70.Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR. Proc Natl Acad Sci USA. 2005;102:9842–9847. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan H, Jin W, Nagaki K, Tian S, Ouyang S, Buell CR, Talbert PB, Henikoff S, Jiang J. Plant Cell. 2005;17:3227–3238. doi: 10.1105/tpc.105.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maruyama S, Kuroiwa H, Miyagishima SY, Tanaka K, Kuroiwa T. Plant J. 2007;49:1122–1129. doi: 10.1111/j.1365-313X.2006.03024.x. [DOI] [PubMed] [Google Scholar]
- 73.Nagaki K, Kashihara K, Murata M. Plant Cell. 2005;17:1886–1893. doi: 10.1105/tpc.105.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shelby RD, Monier K, Sullivan KF. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jansen LE, Black BE, Foltz DR, Cleveland DW. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I. Plant Cell. 2006;18:2443–2451. doi: 10.1105/tpc.106.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schuh M, Lehner CF, Heidmann S. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 78.Sullivan KF. Curr Opin Genet Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- 79.Keith KC, Baker RE, Chen Y, Harris K, Stoler S, Fitzgerald-Hayes M. Mol Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vermaak D, Hayden HS, Henikoff S. Mol Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Mol Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malik HS, Henikoff S. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cooper JL, Henikoff S. Mol Biol Evol. 2004;21:1712–1718. doi: 10.1093/molbev/msh179. [DOI] [PubMed] [Google Scholar]
- 84.Henikoff S, Ahmad K, Malik HS. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 85.Black BE, Brock MA, Bedard S, Woods VL, Jr, Cleveland DW. Proc Natl Acad Sci USA. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Amor DJ, Kalitsis P, Sumer H, Choo KH. Trends Cell Biol. 2004;14:359–368. doi: 10.1016/j.tcb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 87.McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL. Chromosoma. 1998;107:366–375. doi: 10.1007/s004120050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blower MD, Sullivan BA, Karpen GH. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 90.Wines DR, Henikoff S. Genetics. 1992;131:683–691. doi: 10.1093/genetics/131.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leforestier A, Livolant F. Biophys J. 1997;73:1771–1776. doi: 10.1016/S0006-3495(97)78207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Konig P, Braunfeld MB, Sedat JW, Agard DA. Chromosoma. 2007;116:349–372. doi: 10.1007/s00412-007-0101-0. [DOI] [PubMed] [Google Scholar]
- 93.Vogt P. Hum Genet. 1990;84:301–336. doi: 10.1007/BF00196228. [DOI] [PubMed] [Google Scholar]
- 94.Harp JM, Uberbacher EC, Roberson AE, Palmer EL, Gewiess A, Bunick GJ. Acta Crystallogr D. 1996;52:283–288. doi: 10.1107/S0907444995009139. [DOI] [PubMed] [Google Scholar]
- 95.Dawe RK, Henikoff S. Trends Biochem Sci. 2006;31:662–669. doi: 10.1016/j.tibs.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 96.Sandman K, Reeve JN. Curr Opin Microbiol. 2006;9:520–525. doi: 10.1016/j.mib.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 97.Rizzo PJ. Cell Res. 2003;13:215–217. doi: 10.1038/sj.cr.7290166. [DOI] [PubMed] [Google Scholar]
- 98.Flemming W. Zellsubstanz, Kern und Zelltheilung. Leipzig, Germany: Vogel; 1882. [Google Scholar]