Abstract
γ-Irradiation-mediated ataxia telangiectasia mutated (ATM)-dependent dephosphorylation of serine 376 (S376) at the carboxyl terminus of human p53 results in the exposure of the 14-3-3 consensus-binding site, which includes serine 378 (S378). 14-3-3 binding potentiates p53's DNA-binding ability and causes G1 arrest. Moreover,endoplasmic reticulum stress-mediated S376 phosphorylation was shown to localize human p53 in the cytoplasm. Although many functions are conserved between human and mouse p53, the functional relevance of S376 and S378 mouse equivalents is not clear. We report here that γ-irradiation does not lead to 14-3-3 binding to mouse p53. Mouse p53 mutants, such as S373A/D (the equivalent of human S376), S375A/D (the equivalent of human S378), and combinatorial double mutants, were not impaired in their ability to transactivate p53-dependent target genes and were capable of inducing G1 arrest as efficiently as wild-type p53. Consistently, all mutant p53s were as potent as wild-type mouse p53 in inhibiting cellular colony formation. Furthermore, mouse S373A/D mutants were not defective in cytoplasmic localization in response to endoplasmic reticulum stress. Together, the data suggest that despite a high homology with human p53, neither phosphorylation status at S373 and S375 nor 14-3-3 binding may be a critical event for mouse p53 to be functional.
Keywords: 14-3-3 binding, mouse p53, S373, S375, phosphorylation
Introduction
p53 plays a central role in transmitting signals that regulate cellular survival, proliferation, differentiation, and DNA repair [1,2]. Optimal p53 function is achieved by several posttranslational modifications, especially on the amino and carboxyl termini of the protein, which include phosphorylation, acetylation, sumolyation, and neddylation [3,4]. These posttranslational modifications stabilize and recompartmentalize p53, resulting in both transcription-dependent and transcription-independent regulations of various cellular functions [5].
One such modification is the phosphorylation/dephosphorylation of serine 376 (S376) and serine 378 (S378) of human p53. It has been shown that γ-irradiation causes ataxia telangiectasia mutated (ATM)-dependent dephosphorylation at S376, generating a binding site for 14-3-3, which includes phosphorylated S378, thereby increasing the DNA-binding ability of p53 [6]. Subsequent studies showed that mutating these binding sites impaired the ability of p53 to induce G1 cell-cycle arrest [7]. However, S376 was shown to be phosphorylated by GSK-3β during endoplasmic reticulum (ER) stress response, which led to cyctoplasmic sequestration of human p53, thereby reducing the sensitivity of cells to other genotoxic stresses [8]. Moreover, phosphorylation of human p53 at S378 by the CDK7-cycH-p36 complex or by protein kinase C was shown to augment DNA-binding activity [9,10]. Hitherto, it has not been established whether such modifications on the mouse equivalents of S376 and S378 are required for efficient activation of mouse p53, although evidence points to the conservation of many posttranslational modifications between mouse and human p53 [11–13].
We have therefore investigated the functional relevance of S376 and S378 mouse equivalent sites in this study. The data presented here suggest that despite a high homology with human p53, neither phosphorylation status at S373 and S375 nor 14-3-3 binding may be a critical event for mouse p53 to be functional.
Materials and Methods
Cell Culture and Plasmids
The p53-null H1299 human lung cancer cell line, the human breast carcinoma cell line MCF-7, human diploid fibroblasts IMR90 and MRC5 (ATCC, Manassas, VA), and p53-/- mouse embryo fibroblasts (MEFs) were used in this study. All cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (Hyclone, Logan, UT) in the presence of 5% CO2 at 37°C, as described [14].
Transfections were carried out using Lipofectamine Plus reagent (Invitrogen, Carlsbad, CA) as per the manufacturer's description, generally using 5 µg of various p53 plasmids (unless otherwise stated). Fifteen hours after transfection, cells were γ-irradiated with 10 to 20 Gy and harvested 3 hours later for lysate preparation.
The S373 and S375 residues of mouse p53 were mutated to nonphosphorylatable alanine (A) residues or aspartic acid (D) that mimics phosphorylated residues by polymerase chain reaction (PCR)-based site-directed mutagenesis. PCR amplification was performed using pcDNA-based wild-type mouse p53 expression construct as template, primers carrying desired mutation, and Pfu polymerase (Promega, Madison, WI). PCR products were digested with DpnI restriction enzyme (New England Biolabs, Ipswich, MA) before transformation. Mutants were confirmed by restriction enzyme mapping and sequencing. Wild-type and mutant mouse p53 cDNA in pcDNA-based constructs were released by EcoRI (New England Biolaboratories) and subcloned into pBabe retroviral vector [15]. All plasmids were sequenced for verification. Green fluorescent protein (GFP)-encoding plasmid pRNAT-U6.1/Hygro (Genescript; GenScript Corp., Piscataway, NJ) was used for cotransfection to evaluate transfection efficiency in Western blot analysis. Human wild-type or phosphomutant p53 IND constructs were kindly provided by Dr. Thanos Halozenetis.
Colony Formation Assays
p53-/- MEFs were cotransfected with various pcDNA-based p53 constructs and pLoxPuro plasmid [16] at a 10:1 ratio. Cells were selected with 2 µg/ml puromycin (Sigma, St. Louis, MO) for 10 to 15 days and stained with crystal violet (Sigma).
Luciferase Assays
Various p53 plasmids (0.5 µg) and luciferase reporter plasmids (0.5 µg each) were cotransfected with 0.1 µg of plasmid encoding the β-galactosidase gene to evaluate transfection efficiency. The following pGL3 basic vector-based promoter reporter constructs were used in this study: p21-luc and mdm2-luc. Cells were generally collected 48 hours posttransfection, and luciferase assays were performed as described [17]. All experiments were performed in duplicate, and at least three independent times and representative results are shown.
Western Blot and Immunoprecipitation Analyses
Coimmunoprecipitation was performed using 1 mg of protein of whole-cell lysate with anti-14-3-3 antibody (K19)-conjugated agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA), as described previously [7]. Immunoprecipitates and 50 µg of whole-cell lysate were resolved by 10% SDS-PAGE and analyzed by Western blot analysis using the following antibodies: anti-p53 Pab240 and Pab421 (Oncogene, San Diego, CA), anti-DO-1 (Oncogene), anti-CM5 (Novocastra, Newcastle, England, UK), anti-14-3-3 K19 (Santa Cruz Biotechnology), anti-actin (Santa Cruz Biotechnology), and anti-GFP (Santa Cruz Biotechnology). Blots were detected using the ECL Western blot detection reagent (Amersham Biosciences, Buckinghamshire, England, UK).
RNA and Semiquantitative Reverse Transcription PCR
Total RNA was extracted from cultured cell lines using Trizol reagent (Invitrogen), as per the manufacturer's instructions, 24 hours after transfection. Total RNA was reverse-transcribed to first-strand cDNA, 1 µl of which was used as a template for quantitative PCR, subsequent to the optimization of the number of PCR cycles, annealing temperature, and extension time for each gene. The expression of endogenous p53 target genes (bax, noxa, mdm2, and p21) was determined by semiquantitative reverse transcription (RT) PCR and normalized against endogenous gaphd RNA level, as described previously [18].
Flow Cytometric Analyses
Asynchronous p53-/- MEFs were transfected with 5 µg of empty vector, wild-type p53 constructs, or phosphomutant p53 constructs, and were later γ-irradiated with 20 Gy. Simultaneous staining of intracellular p53 (with Pab246 primary antibody and fluorescein isothiocyanate-conjugated anti-mouse IgG antibody) and genomic DNA (by propidium iodide) was performed as described [19], and the cells were analyzed by flow cytometry with FACSCaliber (BD Biosciences, Franklin Lakes, NJ). p53-expressing cells, which emitted green fluorescence, were gated (except for vector-transfected controls), and their cell-cycle profile was determined by ModFIT cell-cycle analysis software (BD Biosciences). All experiments were performed in duplicate and at least twice independently.
Endoplasmic Stress Experiments
Cells were transfected with empty vector or p53-expressing plasmids. Fifteen hours after transfection, cells were treated with vehicle (DMSO), 10 µg/ml tunicamycin (TM), or 1 µM thapsigargin (TG) for 8 hours to induce ER stress as described [8]; fixed in 4% paraformaldehyde; and stained with anti-p53 Pab240 or Pab1620 antibodies (Oncogene), followed by Alexa488-conjugated anti-mouse IgG antibody (Molecular Probes, Eugene, OR) and propidium iodide. Fluorescence confocal microscopy was performed with an LSM 510 laser scanning confocal microscope (Zeiss, Jena, Germany).
Results
Mouse p53, Unlike Human p53, Does Not Bind to 14-3-3
Initial experiments were performed to evaluate the physical interaction of mouse p53 and 14-3-3 by coimmunoprecipitation assays. Mouse or human p53 cDNA was transiently transfected into either p53-null MEFs or human H1299 cells, respectively. Total cell lysates were used to immunoprecipitate 14-3-3, which was followed by immunoblotting with anti-p53 or anti-14-3-3 antibodies. Although human p53 was found to bind endogenous 14-3-3, mouse p53 was unable to pull down 14-3-3 in the same experiments, although 14-3-3 was abundantly expressed in mouse cell extracts (Figure 1A, left panel; compare lane 1 vs lane 2, and lane 3 vs lane 4). Both p53s were efficiently expressed in the cells, as determined by the immunoblotting of total cell lysates with anti-p53 antibodies (Figure 1A, left panel, lanes 6 and 8), ruling out lack of mouse p53 expression as a cause for the absence of 14-3-3 binding. It is noteworthy that the signal for mouse p53 was much weaker than that for human p53. As this could be due to the use of a mixture of Pab240 and DO-1 antibodies to detect both human and mouse p53 on the same blot, we reperformed this experiment using the antibodies separately and detected the blots individually. Similar to earlier results, human p53 was again found to bind endogenous 14-3-3, whereas mouse p53 was unable to pull down 14-3-3 in the same experiments, although 14-3-3 was abundantly expressed in mouse cell extracts (Figure 1A, right panel; compare lane 1 vs lane 2, and lane 3 vs lane 4). Both p53s were efficiently expressed in the cells, as determined by the immunoblotting of total cell lysates with anti-p53 antibodies (Figure 1A, right panel, lanes 5 and 6), ruling out lack of mouse p53 expression as a cause for the absence of 14-3-3 binding.
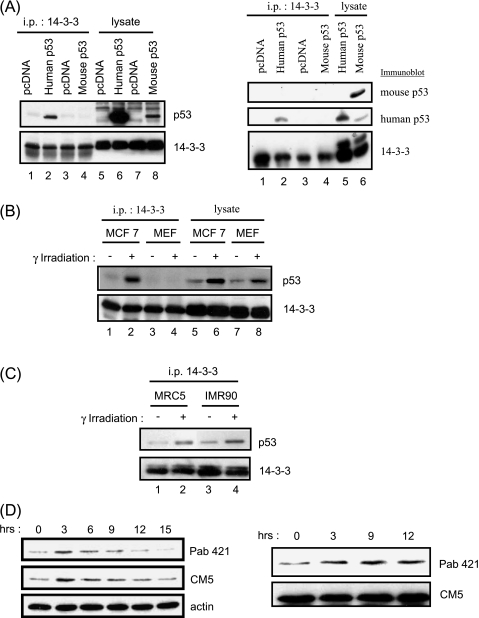
Figure 1.
Mouse p53 does not bind 14-3-3. (A) Empty pcDNA vector and wild-type human or mouse p53 were transfected into H1299 or p53-/- MEFs, respectively. Whole-cell lysates were immunoprecipitated with anti-14-3-3 antibody and analyzed by Western blot analysis using a mixture of Pab240 and DO-1 antibodies (left panel) or using these antibodies separately (right panel; Pab240 for mouse p53, and Pab421 for human p53) to detect p53 and K19 antibodies, which, in turn, detect 14-3-3. (B) Human MCF-7 and primary MEFs were γ-irradiated and collected after 3 hours, and 14-3-3 proteins were immunoprecipitated for Western blot analysis as described in (A), using Pab421 and Pab240 to detect p53. (C) Human diploid fibroblast cells IMR90 and MRC5 were γ-irradiated with 10 Gy, and the association of p53 with 14-3-3 was analyzed as described in (A), using DO-1 to detect human p53. All immunoprecipitation experiments have been repeated three times or more. (D) Western blot analysis of the phosphorylation status of mouse p53. Wild-type primary MEFs cells were γ-irradiated with 10 Gy and harvested at various time points as indicated. The phosphorylation state of p53 was analyzed with Pab421 antibody, the total amount of p53 was determined by CM5 antibody, and actin was used as loading control. Right panel: Blot with equal p53 protein loaded.
We further tested whether endogenous mouse p53 could bind to 14-3-3 after γ-irradiation. As shown in Figure 1B, whereas endogenous human p53 was coimmunoprecipitated with endogenous 14-3-3 on γ-irradiation in human MCF-7 cells, endogenous mouse p53 was not bound to 14-3-3 in mouse fibroblasts (Figure 1B, compare lanes 1 and 2 vs lanes 3 and 4). Nevertheless, p53 levels were induced by γ-irradiation in both cell types (Figure 1B, compare lane 5 vs lane 6, and lane 7 vs lane 8), indicating that mouse p53 was indeed induced by γ-irradiation and may be different from human p53 with respect to 14-3-3 binding.
Furthermore, the binding of p53 and 14-3-3 was also increased on γ-irradiation in the two human diploid fibroblastic lines IMR90 and MRC5, ruling out differences between mouse and human cell types as a cause for the lack of interaction in mouse fibroblasts (Figure 1C). Moreover, immunoreactivity to the Pab421 p53-specific antibody, which has a higher affinity for dephosphorylated S376 (mouse 373) residue, increased after γ-irradiation in primary MEFs (Figure 1D), as has been reported for human cells [6], suggesting that lack of 14-3-3 binding may not be due to the absence of change in the phosphorylation status of mouse p53 protein.
Characterization of Mouse Carboxyl-Terminal Phosphorylation Mutants
Because the 14-3-3 binding of human p53 through dephosphorylated S376 and phosphorylated S378 was shown to be required for p53 biologic activity [7], we investigated the significance of equivalent sites in mouse p53. To this end, mouse equivalents of human S375 and S378 were identified by comparing the protein sequence homology between the two species. The C-terminus of p53 is highly conserved, and S373 and S375 residues of mouse p53 were identified as human equivalents (Figure 2A). Then mouse p53 cDNA was cloned into two different expression vectors, driven by either the long terminal repeat of a retrovirus (pBabe) or the CMV promoter (pcDNA), and site-directed mutagenesis was performed to generate nonphosphorylatable alanine (A) mutants and pseudophosphorylated aspartic acid (D) mutants. The steady-state protein levels of all mutants generated were approximately equal in both pBabe-based and pcDNA-based constructs, suggesting that the phosphorylation state of these two residues does not affect the stability of the protein (Figure 2B). All subsequent experiments described were performed with p53 mutants based on both types of vectors, which showed similar and consistent results.
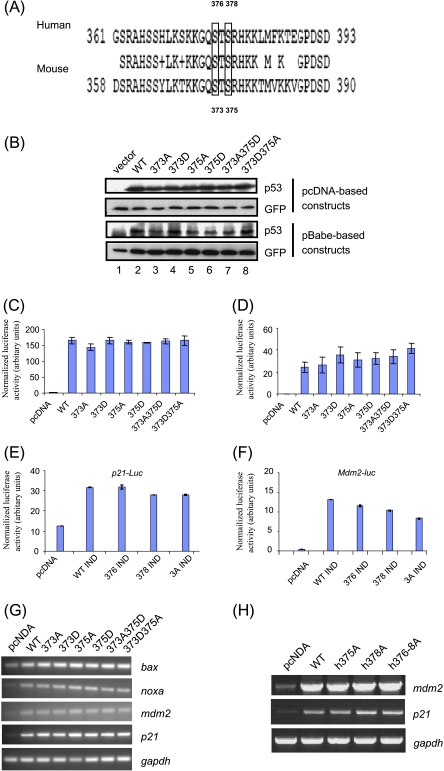
Figure 2.
Steady-state level and transactivation ability of mouse p53 phosphomutants. (A) The alignment of the C-termini of human (accession no. NP_000537) and mouse (accession no. NP_035770) p53 proteins was performed by the Web-based alignment program BLAST2 (National Center for Biotechnology Information). Conserved residues are shown in the middle row between human and mouse p53 sequences. Serine residues at positions 375 and 378 for human p53, and at positions 373 and 375 for mouse p53, are indicated by boxes. (B) The S373 and S375 residues of mouse p53 were mutated to nonphosphorylatable alanine (A) residues or aspartic acid (D) that mimics phosphorylated residues, and were expressed in pCDNA or pBabe vectors. p53-/- MEFs were cotransfected with 1 µg of empty vector, wild-type or mutant mouse p53, and 0.2 µg of GFP-encoding plasmids, and the steady-state level of p53 phosphomutants was determined by Western blot analysis. (C) The transcriptional activity of the mouse phosphomutants was determined by luciferase assays, using mdm2 promoter-luciferase construct. Luciferase activity was determined by chemiluminescence and was normalized against β-galactosidase activity. The experiment was performed thrice independently. Data from one of the experiments, performed in duplicate, are presented as mean ± SD. (D) Relative luciferase activity using the mdm2 promoter-luciferase construct was determined in mouse p53-transfected γ-irradiated (10 Gy) p53-/- MEFs, as described above. The experiment was performed twice independently. Data from one of the experiments, performed in duplicate, are presented as mean ± SD. (E and F) Relative luciferase activity using p21 (E) and mdm2 promoter-luciferase constructs (F) together with human wild-type or phosphomutant IND constructs in p53-null H1299 cells. Data from one of the experiments, performed in duplicate, are presented as mean ± SD. (G and H) The expression of endogenous p53 target genes (bax, noxa, mdm2, and p21) in vector-transfected and p53-transfected p53-/- MEFs (G) or human H1299 cells (H) was determined by semiquantitative RT-PCR and normalized against endogenous gapdh RNA level as described previously.
Using these constructs, we next evaluated the transactivation potential of all mouse phosphomutants by transiently transfecting them with the mdm2 promoter-luciferase reporter construct. The transactivation ability of all p53 phosphomutants was not significantly different from that of wild-type mouse p53 without and with γ-irradiation (Figure 2, C and D). Similar results were obtained with a synthetic multimerized p53-responsive element containing a luciferase reporter construct (PG13-luciferase; data not shown). Consistently, semiquantitative RT-PCR analysis indicted that all mouse phosphomutants were able to induce various endogenous p53 target genes, including bax, noxa, mdm2, and p21, to equal extents as wild-type p53 (Figure 2G). Concurrently, the transactivation ability of the corresponding human S376A, S378A, and S376-8A p53 mutants was determined by luciferase assay (Figure 2, E and F) and semiquantitative RT-PCR (Figure 2H). The results also showed that the transactivation ability of human p53 phosphomutants was comparable to that of the wild-type protein when overexpressed in H1299 cells, similar to previous observations indicating that both human and mouse p53 phosphomutants are transactivation-competent.
Mouse p53 Phosphomutants Are Not Defective in the Induction of Cell Cycle Arrest
Because the phosphorylation status of the S376 and S378 residues of human p53 was shown to be important for cell-cycle arrest induction on γ-irradiation, we tested the ability of various mouse p53 phosphomutants to induce cell-cycle arrest by transiently transfecting p53-/- MEFs with various p53 plasmids and by irradiating them. The transfected cells expressing p53 were monitored by flow cytometric analysis using an antibody that recognizes p53, and only p53+ cells were analyzed. Similar to wild-type mouse p53, all phosphomutants were capable of inducing G1 arrest without γ-irradiation, suggesting that all phosphomutants are functionally active (the G1 population of pcDNA versus WTp53-transfected cells: 40.81% vs 81.50%; all mutants were in a similar range) (Figure 3A, top panel). Consistent with previous findings, p53-null cells failed to arrest at G1 on γ-irradiation, which led to a transient accumulation of cells in G2/M phase (at 24 hours) (Figure 3A, middle panel). Subsequently, the cells re-entered the cell cycle, thereby resulting in an increase in polyploid 8N populations 48 hours after γ-irradiation (Figure 3A, bottom panel, leftmost). However, wild-type p53-expressing cells were arrested at both G1 and G2/M phases, and the cells that escaped G1 block were eventually arrested at G2/M phase 48 hours after γ-irradiation (Figure 3A). All phosphomutants analyzed were capable of inducing both G1 and G2/M cell-cycle arrests as efficiently as wild-type p53 (Figure 3A), suggesting that the phosphorylation state of the S373 and S375 residues of mouse p53 is not important for γ-irradiation-induced cell-cycle arrest.
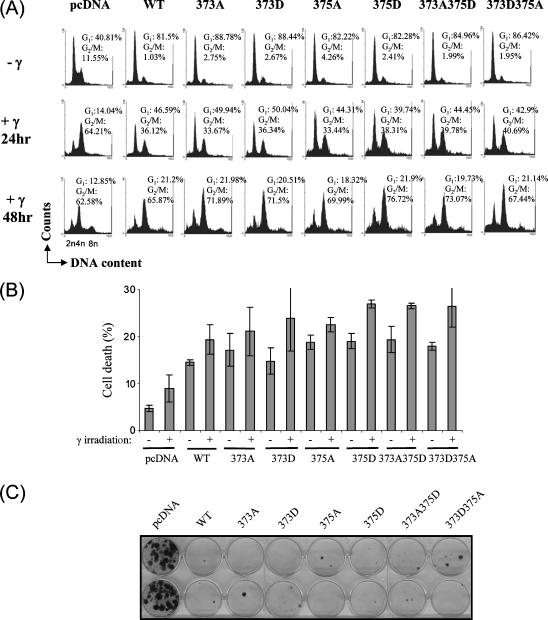
Figure 3.
Mouse p53 phosphomutants are not defective in inducing cell-cycle arrest. (A) Asynchronous p53-/- MEFs were transfected with 5 µg of empty vector, wild-type p53 constructs, or phosphomutant p53 constructs, and were γ-irradiated at 20 Gy. Simultaneous staining of intracellular p53 (with Pab246 primary antibody and fluorescein isothiocyanate-conjugated anti-mouse IgG antibody) and genomic DNA (by propidium iodide) was performed before analysis by flow cytometry. p53-expressing cells, which emitted green fluorescence, were gated (except for vector-transfected controls), and their cell-cycle profile was determined by ModFIT cell-cycle analysis software (BD Biosciences). (B) Cell death was analyzed by determining sub-G1 populations in cell-cycle analysis, as described in (A), by flow cytometry. Cells treated with and without 20 Gy γ-irradiation and harvested 24 hours after γ-irradiation were analyzed. The experiment was performed thrice independently. Data from three independent experiments are presented as mean ± SD. (C) p53-/- MEFs were cotransfected with pcDNA-based constructs, and cells were selected with 2 µg/ml puromycin (Sigma) for 10 to 15 days and stained with crystal violet (Sigma). Colony formation assay was performed four independent times, and representative data are shown.
Analysis of cell death, by measuring sub-G1 population in the cell-cycle profile, revealed that γ-irradiation caused a slight increase in cell death in pcDNA-transfected cells, and expression of wild-type p53 enhanced cell death in the absence and in the presence of γ-irradiation (Figure 3B, pcDNA versus WTp53-transfected cells: (-)γ-irradiation: 4.8 ± 0.71% vs 14.5 ± 0.57%; (+)γ-irradiation 8.95 ± 2.90% vs 19.4 ± 3.11%). Similar results were obtained with pBabe-based vectors (data not shown). All the phosphomutants tested were capable of inducing cell death with or without γ-irradiation as efficiently as wild-type p53, suggesting that mutations of phosphorylation sites had no significant impact on the mutants' ability to induce cell death. These findings were also confirmed in long-term survival (colony formation) assays in p53-/- MEFs. Similar to wild-type p53, all phosphomutant p53s were capable of significantly suppressing colony formation, compared to pcDNA-transfected cultures (Figure 3C). Together, the data suggest that the phosphorylation status of S373 and S375 may not be essential for the optimal function of mouse p53.
Mouse p53 Phosphomutants, Unlike Their Human Counterparts, Are Capable of Responding to ER Stress
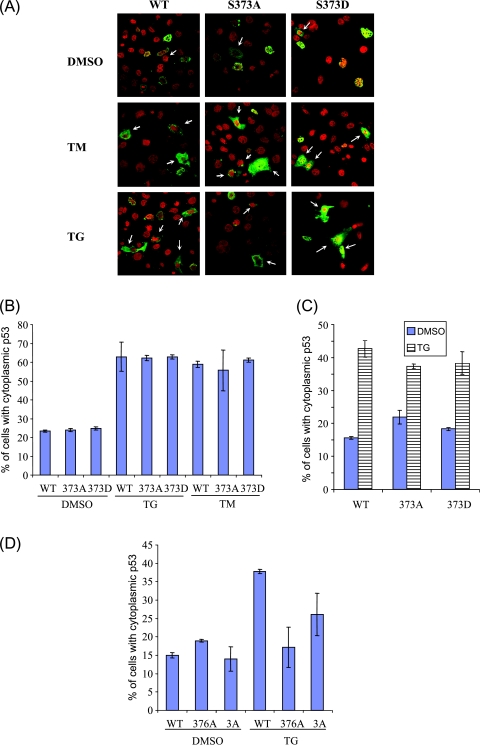
We finally evaluated the role of these phosphorylation sites in the cytoplasmic sequestration of p53 on induction of ER stress, which has been shown to be dependent on the phosphorylation state of the S376 residue of human p53 [8]. To this end, p53-/- MEFs were transfected with various S373 mouse mutant constructs and treated with ER-inducing drugs such as TM or TG, and the localization of p53 was determined by immunofluorescence using the Pab240 antibody. As shown in Figure 4A (upper panels), majority of the transfected p53 localized to the nucleus of the cells, regardless of phosphomutations (Figure 4B, WTp53 vs 373A vs 373D: 76.54 ± 0.61% vs 76.03 ± 0.88% vs 75.20 ± 0.83%). TM-mediated or TG-mediated ER stress resulted in a significant increase in the sequestration of wild-type mouse p53 in the cytoplasm (Figure 4A, arrows; B, untreated versus TM versus TG: 76.54 ± 0.61% vs 37.16 ± 7.75% vs 41.19 ± 1.68%). Both S373A and S373D phosphomutants were also found to translocate efficiently to the cytoplasm on ER stress (Figure 4, A and B). Similar results were obtained when analysis was performed using another anti-p53 antibody (Pab1620; Figure 4C). Moreover, the phosphorylation status of the S375 residue of mouse p53 was also found to be dispensable for cytoplasmic sequestration (data not shown), suggesting that the phosphorylation status of neither S373 nor S375 residue is important for p53 cytoplasmic localization on ER stress. Conversely and as published previously, S376A mutant human p53 failed to be exported from the nucleus on ER stress, in contrast to wild-type human p53, suggesting that the mechanisms of p53 nuclear export after ER stress may be different between mouse and human cells (Figure 4D).
Figure 4.
Mouse p53 phosphomutants, unlike their human counterparts, respond to ER stress as efficiently as wild-type p53. (A) p53-/- MEFs were transfected with empty vector, wild-type mouse p53, or phosphomutant mouse p53, and then treated with vehicle (DMSO), 10 µg/ml TM, or 1 µM TG for 8 hours to induce ER stress. Cells were fixed and stained with anti-p53 antibody and propidium iodide before fluorescence confocal microscopy. Representative images, in which green fluorescence represents p53 protein and red fluorescence represents genomic DNA, are shown. Arrows indicate cells in which p53 is cytoplasmic. (B) Quantitative analysis of cells having cytoplasmic p53. Approximately 500 p53-expressing cells were scored and classified into cells with or without nuclear p53 stain. Data from one of the experiments, performed in triplicate, are presented as mean ± SD. (C) Similar experiments were carried out as described in (A) and (B) and were analyzed using another anti-p53 antibody (Pab1620). (D) H1299 cells were transfected with empty vector, wild-type human p53, or phosphomutant human p53 (376A or 376-378A[3A]). ER stress was induced by 1 µM TG, as described in (A). Approximately 500 p53-expressing cells were scored and classified into cells with or without nuclear p53 stain. Data from one of the experiments, performed in triplicate, are presented as mean ± SD. All experiments were performed at least thrice independently.
Discussion
The results presented here suggest that mouse p53 is not similar to human p53 in some aspects, as the former is unable to bind 14-3-3, and, consistently, the serine residues that are thought to be required for the exposition of the 14-3-3-binding motif on human p53 do not seem to be functionally relevant in mouse p53. Although both human and mouse p53 are highly homologous in the carboxyl-terminal region of the protein, none of the mouse p53 phosphomutants (either A or D) was able to bind 14-3-3 (data not shown). These findings suggest that the phosphorylation status of S376 and S378 residues alone may not be sufficient for 14-3-3 binding in human p53 and that other unidentified factors may influence this phenomenon.
It is known that 14-3-3 isoforms bind to proteins in both a phosphorylation-dependent and a phosphorylation-independent manner. In phosphorylation-dependent binding, it recognizes RSXpSXP and RXXXpSXP epitopes, where pS represents phosphoserine (can be interchanged with phosphothreonin) and X represents any amino acid [20]. In the case of both human and mouse p53, the 14-3-3-binding site is believed to be KGQSTpSRH [6], in which arginine and proline residues critical for 14-3-3 binding are not found. Therefore, the site on p53 is not a perfect 14-3-3-binding motif and, hence, may be a low-affinity binding site. Moreover, a peptide containing two binding sites showed a 30-fold increased affinity for 14-3-3 than peptides having a single binding motif [20], consistent with the notion that 14-3-3 preferentially binds to some targets as a dimer [21]. However, the second putative 14-3-3-binding motif has not been identified in both human and mouse p53. Thus, it is not unconceivable that human p53 may only weakly bind to 14-3-3. Therefore, in addition to the above characteristics and given the substantial differences between human and mouse p53 in the amino acid sequence around this 14-3-3-binding site, it is possible that mouse p53 is not a good binding target for 14-3-3.
Moreover, the results prompt us to speculate that 14-3-3 binding to p53 may be an evolutionarily late event that occurred with only human p53 (perhaps to optimize its function) and was not essential for efficient functioning of mouse p53. In this respect, DNA damage-mediated interaction between human p53 and 14-3-3 was shown to stabilize p53, thereby facilitating oligomerization, blocking nuclear export, and increasing the transactivation ability of p53, in turn resulting in cell-cycle arrest [22,23]. However, all mouse phosphomutants were not altered in their steady-state levels or in their transactivation potential, further suggesting that modifications of these phosphorylation sites may not be crucial in regulating the function of the mouse protein. Furthermore, it is noteworthy that 14-3-3σ is a target of human p53 [22], suggesting the existence of a positive feedback loop between human p53 and 14-3-3 protein [23]. However, data suggesting the existence of this feedback loop in mouse cells are lacking, and mouse 14-3-3 appears to be unresponsive to p53 (M. Resnick et al., personal communication). Therefore, the data suggest that a positive feedback loop may not exist in the mouse system because we did not observe any binding of 14-3-3 to mouse p53 protein, thereby explaining the lack of any functional deficiencies of phosphomutant proteins. However, as both human and mouse p53 have extremely similar functions, it is possible that the differences between them are functionally irrelevant, or, alternatively, there has been convergent evolution where these two p53s perform similar functions through different mechanisms.
Consistent with the lack of 14-3-3 binding, mouse p53 phophomutants were not defective in cell-cycle arrest or apoptosis induction. In addition, phosphomutations had no effect on the subcellular shuttling of mouse p53, in contrast to human p53, on ER stress, which was shown to be dependent on the phosphorylation of the S315 and S375 residues of human p53 by GSK-3β in transient transfection assays [8]. However, unlike the absence of interaction between mouse p53 and 14-3-3, we observed the binding of mouse p53 with GSK-3β during ER stress in coimmunoprecipitation experiments (data not shown), similar to that observed with human p53, suggesting that this association may also be important for the shuttling of mouse p53 on ER stress. Nevertheless, the S373 residue of mouse p53 does not play a role in its localization of mouse p53 and raises the possibility that the S312 of mouse p53—which is the equivalent of S315, rather than S373, in human p53—may be essential for ER stress-mediated cytoplasmic shuttling.
This work was initiated to evaluate the functional significance of the two carboxyl-terminal phosphorylation sites in mouse p53, which has not been performed before. We found that γ-irradiation of mouse cells led to the induction of a band recognizable by Pab421, which recognizes human p53 that is dephosphorylated at the S376 residue, suggesting that dephosphorylation of mouse S373 may also occur on γ-irradiation. It should be noted that this epitope could also be regulated by other posttranslational modifications such as ubiquitination and acetylation, although we are not aware of any reports indicating that such modifications may affect recognition by the antibody. However, this event cannot be directly tested as the available anti-pS376 (human) antibody does not recognize mouse p53 (data not shown). This study was therefore undertaken to evaluate the significance of these posttranslational modifications in cultured cells using transient transfections before embarking on the generation of “knock-in” mouse models to address the in vivo significance of these phosphorylation sites. In this respect, it should be noted that most experiments aimed at defining the functional significance of human S376 and S378 were also performed by transient transfection experiments [8,7], suggesting that this system is sufficient to uncover any functional deficiencies using phosphomutant constructs in a short period of time. Being mindful of the high levels of proteins expressed in these transient systems, we had generated two sets of constructs driven by different promoter elements that result in either higher expression (pcDNA-based constructs) or expression levels similar to physiological settings (pBabe-based constructs), which gave similar results. Similar strategies have been employed in other studies aimed at understanding the physiological significance of various posttranslational modifications before the generation of knock-in mice, which is a time-consuming process. For example, the effects of phosphorylation at the S389 [11] and S18 [24] residues of mouse p53 were first investigated in cell culture models, and these findings were eventually confirmed with the use of knock-in models [13,25]. Hence, cell culture models, as described here, can provide useful clues for elucidating the significance of posttranslational modifications before embarking on the generation of modified mice.
In conclusion, we have tested the physiological relevance of the S373 and S375 residues of mouse p53 and have demonstrated that the phosphorylation status of these sites is not relevant to the efficient functioning of mouse p53. These data suggest that some of the posttranslational events regulating p53 function may have been acquired later in evolution, specifically for fine-tuning the regulation of essential properties in higher organisms.
Acknowledgements
We thank Thanos Halazonetis for his advise and kind help with the provision of human p53 IND mutant constructs.
Footnotes
This work was supported by grants from the National Medical Research Council and the Biomedical Research Council of Singapore to K.S.
References
- 1.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 4.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 5.Bargonetti J, Manfredi JJ. Multiple roles of the tumor suppressor p53. Curr Opin Oncol. 2002;14:86–91. doi: 10.1097/00001622-200201000-00015. [DOI] [PubMed] [Google Scholar]
- 6.Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 7.Stavridi ES, Chehab NH, Malikzay A, Halazonetis TD. Substitutions that compromise the ionizing radiation-induced association of p53 with 14-3-3 proteins also compromise the ability of p53 to induce cell cycle arrest. Cancer Res. 2001;61:7030–7033. [PubMed] [Google Scholar]
- 8.Qu L, Huang S, Baltzis D, Rivas-Estilla AM, Pluquet O, Hatzoglou M, Koumenis C, Taya Y, Yoshimura A, Koromilas AE. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes Dev. 2004;18:261–277. doi: 10.1101/gad.1165804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu H, Fisher RP, Bailey P, Levine AJ. The CDK7-cycH-p36 complex of transcription factor IIH phosphorylates p53, enhancing its sequence-specific DNA binding activity in vitro. Mol Cell Biol. 1997;17:5923–5934. doi: 10.1128/mcb.17.10.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takenaka I, Morin F, Seizinger BR, Kley N. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J Biol Chem. 1995;270:5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- 11.Hao M, Lowy AM, Kapoor M, Deffie A, Liu G, Lozano G. Mutation of phosphoserine 389 affects p53 function in vivo. J Biol Chem. 1996;271:29380–29385. doi: 10.1074/jbc.271.46.29380. [DOI] [PubMed] [Google Scholar]
- 12.MacPherson D, Kim J, Kim T, Rhee BK, Van Oostrom CT, DiTullio RA, Venere M, Halazonetis TD, Bronson R, De VA, et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J. 2004;23:3689–3699. doi: 10.1038/sj.emboj.7600363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sluss HK, Armata H, Gallant J, Jones SN. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol. 2004;24:976–984. doi: 10.1128/MCB.24.3.976-984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vikhanskaya F, Siddique MM, Kei LM, Broggini M, Sabapathy K. Evaluation of the combined effect of p53 codon 72 polymorphism and hotspot mutations in response to anticancer drugs. Clin Cancer Res. 2005;11:4348–4356. doi: 10.1158/1078-0432.CCR-04-1547. [DOI] [PubMed] [Google Scholar]
- 15.Passegue E, Wagner EF. JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J. 2000;19:2969–2979. doi: 10.1093/emboj/19.12.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arakawa H, Lodygin D, Buerstedde JM. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 2001;1:7. doi: 10.1186/1472-6750-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toh WH, Kyo S, Sabapathy K. Relief of p53-mediated telomerase suppression by p73. J Biol Chem. 2005;280:17329–17338. doi: 10.1074/jbc.M500044200. [DOI] [PubMed] [Google Scholar]
- 18.Lee MK, Hande MP, Sabapathy K. Ectopic mTERT expression in mouse embryonic stem cells does not affect differentiation but confers resistance to differentiation- and stress-induced p53-dependent apoptosis. J Cell Sci. 2005;118:819–829. doi: 10.1242/jcs.01673. [DOI] [PubMed] [Google Scholar]
- 19.Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 20.Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 21.Tzivion G, Luo ZJ, Avruch J. Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3-3 partners in vivo. J Biol Chem. 2000;275:29772–29778. doi: 10.1074/jbc.M001207200. [DOI] [PubMed] [Google Scholar]
- 22.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 23.Yang HY, Wen YY, Chen CH, Lozano G, Lee MH. 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Mol Cell Biol. 2003;23:7096–7107. doi: 10.1128/MCB.23.20.7096-7107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayr GA, Reed M, Wang P, Wang Y, Schweds JF, Tegtmeyer P. Serine phosphorylation in the NH2 terminus of p53 facilitates transactivation. Cancer Res. 1995;55:2410–2417. [PubMed] [Google Scholar]
- 25.Bruins W, Zwart E, Attardi LD, Iwakuma T, Hoogervorst EM, Beems RB, Miranda B, Van Oostrom CT, van den BJ, van den Aardweg GJ, et al. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol Cell Biol. 2004;24:8884–8894. doi: 10.1128/MCB.24.20.8884-8894.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]