Abstract
Cori’s disease is a glycogen storage disorder characterized by a deficiency in the glycogen debranching enzyme, amylo-1,6-glucosidase,4-α-glucanotransferase (AGL). Here, we demonstrate that the G1448R genetic variant of AGL is unable to bind to glycogen and displays decreased stability that is rescued by proteasomal inhibition. AGL G1448R is more highly ubiquitinated than its wild-type counterpart and forms aggresomes upon proteasome impairment. Furthermore, the E3 ubiquitin ligase Malin interacts with and promotes the ubiquitination of AGL. Malin is known to be mutated in Lafora disease, an autosomal recessive disorder clinically characterized by the accumulation of polyglucosan bodies resembling poorly branched glycogen. Transfection studies in HepG2 cells demonstrate that AGL is cytoplasmic whereas Malin is predominately nuclear. However, after depletion of glycogen stores for 4 h, ∼90% of transfected cells exhibit partial nuclear staining for AGL. Furthermore, stimulation of cells with agents that elevate cAMP increases Malin levels and Malin/AGL complex formation. Refeeding mice for 2 h after an overnight fast causes a reduction in hepatic AGL levels by 48%. Taken together, these results indicate that binding to glycogen crucially regulates the stability of AGL and, further, that its ubiquitination may play an important role in the pathophysiology of both Lafora and Cori’s disease.
Keywords: Glycogen debranching enzyme, Lafora disease, Cori’s disease, ubiquitination, Malin
Lafora disease (OMIM 254780) is an autosomal recessive form of epilepsy with onset in late childhood or adolescence (Chan et al. 2005; Ganesh et al. 2006). Primary symptoms include seizures, ataxia, myoclonus, and the progressive development of severe dementia. Most inflicted individuals die by 25 years of age. Currently, there is no known long-term treatment. Clinically, Lafora disease is characterized by the presence of inclusion bodies (Lafora bodies) in most organs, including the brain, heart, liver, muscle, and skin. These inclusions consist of high levels of polyglucosans that resemble abnormally branched glycogen (Yokoi et al. 1968), suggesting that in Lafora disease, enzymes involved in glycogen metabolism are dysregulated.
Positional cloning studies have characterized two major loci implicated in Lafora disease, EPM2A (encoding Laforin) (Minassian et al. 1998; Serratosa et al. 1999) and EPM2B (encoding Malin) (Chan et al. 2003). Laforin is a dual specificity phosphatase consisting of a C-terminal catalytic domain and an N-terminal carbohydrate-binding domain (CBD) that localizes the protein to glycogen, as well as polyglucosan bodies (Ganesh et al. 2000, 2004; Wang et al. 2002; Chan et al. 2004). Laforin is conserved in all vertebrates and a subclass of protists (Gentry et al. 2007). Mice deficient for Laforin or those that overexpress the catalytically inactive form (C266S) progressively develop Lafora bodies and the disease symptoms with age (Ganesh et al. 2002a; Chan et al. 2004). Malin is a RING finger-containing E3 ubiquitin ligase that interacts with Laforin, promoting its ubiquitination and degradation (Gentry et al. 2005; Lohi et al. 2005). However, since loss-of-function mutations in either Laforin or Malin paradoxically cause Lafora disease, the significance of this interaction remains uncertain.
The levels of Laforin protein in cells seem to correlate with the amount of glycogen (Wang et al. 2007). However, how Laforin participates in glycogen metabolism remains unclear. Laforin has been shown to bind PTG/R5, a protein phosphatase 1 (PP1) glycogen targeting subunit (Fernandez-Sanchez et al. 2003). PTG acts as a scaffolding molecule that enhances the ability of PP1 to dephosphorylate and thus activate glycogen synthase (Printen et al. 1997; Brady and Saltiel 2001). Laforin has also been shown to dephosphorylate and activate the kinase GSK3β (Lohi et al. 2005; Wang et al. 2006). One direct target of GSK3β is glycogen synthase, and its phosphorylation by GSK3β leads to inactivation, implying that Laforin could directly affect glycogen metabolism through this pathway. However, recent evidence suggests that alterations in the GSK3β/glycogen synthase pathway are not primarily responsible for the formation of Lafora bodies (Wang et al. 2007; Worby et al. 2006), and precisely how Laforin affects glycogen metabolism remains unknown.
The presence of polyglucosan bodies in Lafora disease could also implicate a disruption in the controls that regulate the glycogen branching/debranching system, a process that is poorly understood (Roach 2002). Glycogen branching is catalyzed by glucan (1,4-α-),branching enzyme 1 (GBE1). The relative activities of glycogen synthase:GBE1 are likely to play a role in the overall degree of branching. In mice that overexpress glycogen synthase, glycogen is less branched and polyglucosan bodies develop (Raben et al. 2001; Pederson et al. 2003). Conversely, the absence of GBE1 activity observed in type IV glycogen storage disease results in poorly branched glycogen and polyglucosan bodies similar to those seen in Lafora disease (Moses and Parvari 2002).
During glycogenolysis, the activity of glycogen phosphorylase is stalled when the enzyme encounters a branch point in glycogen four glucose residues away. To allow further glycogen degradation, the removal of this branch must be carried out by the glycogen debranching enzyme, amylo-1,6-glucosidase,4-α-glucanotransferase (AGL) (Shen and Chen 2002). Cori’s disease (glycogen storage disease type III; OMIM 232,400) is a human autosomal recessive disorder resulting from a deficiency in AGL activity. Most AGL mutations identified to date lead to premature truncation and loss of activity. However, one missense variant, G1448R, is located within the putative CBD (Okubo et al. 1999). How this mutation affects AGL function remains unclear.
In this study, we show that the E3 ubiquitin ligase Malin promotes the ubiquitination of AGL in a highly regulated process. The G1448R mutant found in Cori’s disease disrupts the CBD of AGL, causing it to undergo proteasomal degradation. Glycogenolytic agents known to activate cAMP signaling increase the levels of Malin and the formation of Malin/AGL complexes. In overnight fasted mice, refeeding causes a reduction in AGL levels. These results suggest that the regulation of AGL levels may play an important role in both Lafora and Cori’s disease.
Results
Disrupting the CBD of AGL leads to increased degradation
The glycogen debranching enzyme AGL is well conserved from humans to Saccharomyces cerevisiae, with 38% identity (52% similarity) at the protein level (Shen and Chen 2002). Mutations in AGL that lead to premature truncation and loss of activity occur in Cori’s disease, or glycogen storage disease type III. However, one missense variant (G1448R) associated with the disease is located within the putative CBD (Okubo et al. 1999). To test the possibility that this mutant would affect the ability of AGL to bind to glycogen, AGL mutants with a truncation at the C terminus within the CBD domain (AGL ΔCBD) or the G1448R mutation were generated (Fig. 1A). We overexpressed these mutants as Flag-tagged AGL proteins in COS cells, and subjected them to pull-down assays using amylose resin to evaluate carbohydrate binding (Fig. 1B). Whereas wild-type AGL readily bound to amylose, both the ΔCBD and G1448R mutants did not. We further confirmed these results in whole cells by performing high-speed ultracentrifugation to isolate glycogen-enriched fractions (Fig. 1C). Wild-type AGL was found only in the glycogen-enriched (high-speed pellet) fraction. In contrast, both mutants localized predominantly to the cytosolic (high-speed supernatant) portion. These results demonstrate that both AGL mutants are deficient in binding carbohydrate.
Figure 1.
Disrupting the CBD of AGL leads to decreased protein stability. (A) Schematic of wild-type and mutant AGL proteins. AGL possesses transferase, glucosidase, and CBDs. Truncations and a G1448R mutant were made. (B) AGL mutants are unable to bind amylose resin. The indicated constructs were transfected into COS cells, and lysates were subjected to a pull-down assay using amylose resin beads. (C) AGL mutants do not fractionate into the glycogen-enriched fraction. Transfected COS cells were subjected to differential centrifugation, and the proportion of AGL proteins in each fraction was determined by immunoblotting. (D) AGL carbohydrate-binding mutants exhibit decreased protein stability. Cells were transfected with Flag-tagged wild-type, ΔCBD, or G1448R AGL. The next day, cells were treated with cycloheximide and labeled with 35S methionine for 2 h. Cell lysates were immunoprecipitated with anti-Flag beads and subjected to SDS-PAGE. Gels were transferred to nitrocellulose membranes and processed for autoradiography. (E) Proteasomal inhibition increases the levels of mutant AGL proteins. Transfected cells were treated with 100 μg/mL cycloheximide with or without 10 μM MG-132 for 6 h. Cell lysates were then analyzed by immunoblotting with the indicated antibodies. (F) Mutant AGL proteins exhibit increased ubiquitination. Cells were transfected as in B, in addition with HA-UB. Cells were lysed in denaturing immunoprecipitation buffer, and lysates were immunoprecipitated with anti-Flag antibodies. Immunoprecipitates were immunoblotted with the indicated antibodies.
During the course of our studies, we noticed that the AGL ΔCBD and G1448R mutants were relatively unstable. Pulse chase analysis revealed that the wild-type protein had a half-life of at least 9 h, whereas the mutants that disrupt glycogen binding exhibited a significantly shorter half-life of <3 h (Fig. 1D). Treatment of cells with the proteasome inhibitor MG-132 had no effect on wild-type AGL levels, consistent with its observed long half-life (Fig. 1E). In contrast, the levels of AGL ΔCBD or G1448R were significantly increased upon proteasomal inhibition.
To further confirm the involvement of the proteasome, we assessed the levels of ubiquitination of the AGL proteins. Flag-tagged wild-type and mutant AGL proteins were expressed in COS cells, along with HA-tagged ubiquitin. The cells were lysed in 1% SDS, boiled, and diluted, and AGL proteins were immunoprecipitated with anti-Flag antibodies. Immunoprecipitates were then analyzed for covalently linked ubiquitin by immunoblot analysis with anti-HA antibodies. Both the ΔCBD and G1448R AGL mutants exhibited increased ubiquitination (Fig. 1F). This was not due to the differential expression of HA-ubiquitin (HA-UB), as analysis of total cell lysates (TCL) demonstrated similar ubiquitination in all transfected lanes. Taken together, these results suggest that the CBD of AGL is crucial to maintaining its stability and protection from ubiquitin-mediated degradation.
The AGL G1448R mutant forms aggresomes upon proteasome impairment
The ubiquitination of mutant AGL proteins may facilitate their removal and prevent the accumulation of toxic or aggregated products. In this regard, proteasomal inhibition has been shown to lead to the formation of inclusion bodies containing aggregates termed “aggresomes.” These structures form specifically at the centrosome in a microtubule-dependent manner and consist of the aggregated proteins as well as chaperones and components of the proteasome (Kopito 2000). Aggresome formation may be pathogenic in many neurodegenerative disorders, such as Parkinson’s and Huntington’s disease, leading to cellular degeneration or dysfunction.
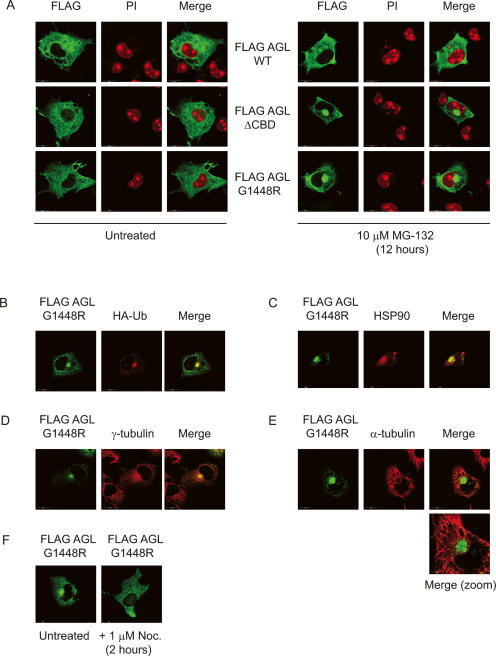
We asked whether mutant forms of AGL would undergo a similar process. COS cells were transfected with Flag-tagged wild-type AGL, or the ΔCBD or G1448R mutants, and confocal immunofluorescence microscopy was performed in the presence or absence of MG-132. In the absence of proteasomal inhibition, all the AGL proteins exhibited a cytoplasmic staining pattern (Fig. 2A). However, treatment with 10 μM MG-132 for 12 h dramatically induced perinuclear aggregate formation in ∼90% of the cells transfected with AGL mutant proteins. In contrast, wild-type AGL was less efficient in producing these aggregates, in both size and number (<50% in transfected cells).
Figure 2.
The AGL (G1448R) mutant forms aggresomes. (A) Disrupting the CBD of AGL induces inclusion body formation upon proteasomal impairment. Flag-tagged wild-type (WT), ΔCBD, or G1448R AGL proteins were immunostained with Flag antibodies (green), and nuclei were visualized with propidium iodide (PI, red). Localization of AGL in COS cells growing in culture in the absence (left panels) or presence (right panels) of proteasome inhibitor. Cells were transfected with AGL constructs for 12 h, and then 10 μM MG-132 was included during the media change and cells were allowed to continue for an additional 12 h before immunofluorescence analysis. (B–E) The AGL (G1448R)-containing inclusions possess aggresome-like properties. Flag-tagged AGL G1448R was transfected in COS cells, and 12 h later, 10 μM MG-132 was included during the media change. Cells were allowed to grow for 12 more hours before immunofluorescence analysis with the indicated antibodies. Costaining reveals that Flag-AGL G1448R inclusions (green) colocalize with HA-tagged ubiquitin (red) (B), the molecular chaperone HSP90 (red) (C), and the centrosomal marker γ-tubulin (red) (D). (E) Costaining with α-tubulin (red) reveals the G1448R inclusion is “caged” by a microtubule network. (F) Disruption of the microtubule network by 1 μM nocodazole for 2 h results in loss of G1448R inclusion formation.
We also found that the G1448R-containing aggregates contained both ubiquitin and the molecular chaperone HSP90 (Fig. 2B,C). Costaining with the centrosomal marker γ-tubulin demonstrates that the inclusion bodies colocalize with the centrosome (Fig. 2D). Furthermore, the aggregates seem to be “caged” by a microtubular network, as revealed by immunostaining with α-tubulin (Fig. 2E). Finally, the microtubule-disrupting drug nocodazole disrupted the formation of the inclusion bodies (Fig. 2F). Similar results were obtained using the ΔCBD mutant (data not shown). Taken together, these results demonstrate that the AGL G1448R Cori’s disease mutant forms aggresome-like bodies upon proteasome impairment.
Subcellular localization of Malin, AGL, and Laforin under glycogenolytic conditions
Recently it has been shown that upon proteasome impairment, the E3 ubiquitin ligase Malin relocalizes within cells to aggresomes (Mittal et al. 2007). In COS cells, Malin is found in both nuclear and endoplasmic reticulum (ER) compartments (Chan et al. 2003). In our hands, low-level overexpression of Malin in COS cells results in nuclear localization, whereas at high levels, Malin assumes a perinuclear ER-like staining (Fig. 3A). When overexpressed in HepG2 cells, myc-tagged Malin was localized predominately in the nucleus, as indicated by immunofluorescence staining (Fig. 3B). In contrast to Malin, overexpressed HA-AGL exhibits cytoplasmic staining (Fig. 3C). Surprisingly, we found that under glycogenolytic conditions (glucose deprivation, Forskolin [FSK]/3-isobutyl-1-methylxanthine [IBMX] treatment), HA-AGL partially translocated to the nucleus (Fig. 3C). Complete depletion of glycogen levels resulted in up to 90% of transfected cells displaying AGL staining in the nucleus (Fig. 3D,E). Under these conditions, Malin-myc staining was slightly diffuse and partly cytoplasmic, although still predominately nuclear (data not shown). This suggests that upon release from glycogen, AGL is directed toward the nuclear compartment. In this regard, treatment of cells with a glycogen phosphorylase inhibitor prevented nuclear staining of AGL (Fig. 3F), concomitant with the blockade of glycogenolysis (Fig. 3G).
Figure 3.
Localization of AGL, Laforin, and Malin under glycogenolytic conditions. (A) Malin is a predominately nuclear protein. COS cells were transfected with Malin-myc for 12 h (low expression) or 24 h (high expression) and processed for immunofluorescence analysis. Cells were stained with anti-myc antibodies (for Malin) and propidium iodide (PI). (B) Localization of Malin-myc in HepG2 cells. (C) AGL and Laforin exhibit partial nuclear staining upon treatment with glycogenolytic agents. HepG2 cells were transfected with the indicated constructs, treated with 50 μM FSK/500 μM IBMX in glucose-free media or left in regular media for 2 h, and then processed for immunofluorescence analysis. (D) Quantification of AGL nuclear staining with different treatments. Cells were scored as positive when nuclear staining was at least as strong as cytoplasmic staining. Error bars represent SEM. (E) Corresponding glycogen levels during the treatments. Levels were normalized to untreated cells. Error bars represent SEM. (F) Inhibition of glycogen phosphorylase prevents nuclear staining of AGL. Cells expressing HA-AGL were preincubated with or without 1 μM phosphorylase inhibitor for 1 h, before treatment with the glycogenolytic agents. New inhibitor was added when medium was changed. Cells were scored as in D. (G) Corresponding glycogen levels during the treatments. Levels were normalized to untreated cells. Error bars, SEM.
Malin has been previously demonstrated to promote the degradation of Laforin (Gentry et al. 2005; Lohi et al. 2005). We therefore asked whether glycogenolytic signals also trigger the nuclear localization of Laforin. Under basal conditions, Laforin exhibited a cytoplasmic localization similar to AGL (Fig. 3C). Treatment with glycogenolytic agents caused a significant nuclear localization of Laforin. Similar results were also seen with the C266S mutant of Laforin (data not shown). It is interesting to note that a splice variant of Laforin has been shown to localize in both the cytoplasm and nucleus (Ganesh et al. 2002b). It is possible that differential localization allows Laforin to target different substrates. Alternatively, nuclear localization of Laforin may target it for Malin-mediated proteasomal degradation.
cAMP signaling increases levels of Malin and Malin/AGL complexes
We next asked whether Malin and AGL could interact. We expressed Flag-AGL together with the wild-type or a catalytically inactive mutant (C26S) of Malin-myc (Gentry et al. 2005) in cells, and performed coimmunoprecipitation experiments. Flag-AGL was found in immunoprecipitates of both wild type and C26S Malin-myc, but not with control beads (Fig. 4A).
Figure 4.
Agents that elevate cAMP increase Malin levels and Malin/AGL complexes. (A) Malin interacts with AGL. 293T cells were grown to 50%–75% confluence in 6-cm dishes prior to transfection with Fugene 6. Flag-AGL was transfected with vector, wild type, or C26S Malin-myc. Cell lysates were immunoprecipitated with anti-myc antibodies and probed for the presence of Flag-AGL. (B) Overexpression of Malin promotes AGL ubiquitination. Cells were transfected as in A, in addition with HA-UB. Cells were lysed in denaturing buffer, and lysates were immunoprecipitated with anti-Flag antibodies. Immunoprecipitates were immunoblotted with the indicated antibodies. (C,D) Increased Malin/AGL coimmunoprecipitation during treatment with FSK but not glucose deprivation alone. Flag-AGL and C26S Malin-myc were cotransfected into 293T cells for 16 h, followed by treatment with the indicated reagents. Lysates were subjected to immunoprecipitations with anti-myc antibodies and analyzed for the presence of Flag-AGL. (E) FSK, but not glucose deprivation, causes an increase in Malin levels. Malin-myc was transfected into 293T cells and treated with the indicated reagents. Lysates were analyzed with the indicated antibodies.
Interestingly, Flag-AGL interacted with the mutant C26S Malin-myc better than with the wild-type protein, suggesting that Malin may mediate the ubiquitination of AGL and then release it. To address this question, we expressed Flag-AGL, HA-UB, and Malin-myc wild-type or its C26S mutant in 293T cells, and performed denaturing immunoprecipitations (Fig. 4B). Probing Flag immunoprecipitates with HA antibodies revealed that expression of wild-type Malin significantly increased AGL ubiquitination, but was without effect using the C26S mutant.
To determine whether carbohydrate binding influenced this interaction, we examined the interaction between AGL and Malin under glycogenolytic conditions. For these experiments, we used the C26S mutant of Malin, so that the overall levels of AGL would not be affected. When cells were glucose-deprived and treated with FSK, the association of Malin with AGL markedly increased, as determined by coimmunoprecipitation (Fig. 4C). This was due to the unexpected increase in Malin levels under these conditions. Interestingly, the increase in Malin/AGL complex formation was not observed upon glucose deprivation alone (Fig. 4D), suggesting that release from glycogen is insufficient for AGL and Malin to interact. In addition to FSK, both 8-Bromo-cAMP as well as IBMX, produced an increase in Malin levels (data not shown). Furthermore, the presence of glucose had no impact, suggesting that an elevation in cAMP levels is sufficient for this effect (Fig. 4E).
Regulation of AGL levels during fasting/refeeding
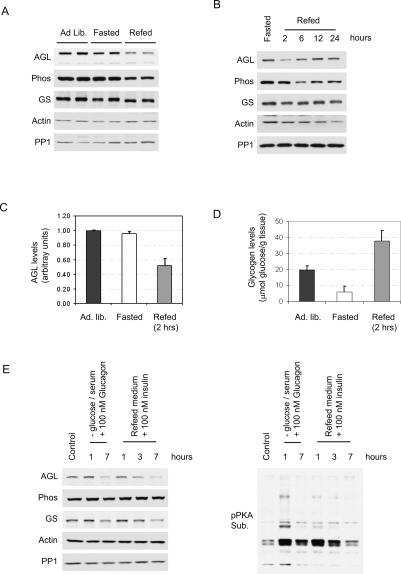
Consistent with a role for Malin in promoting AGL ubiquitination, we observed that coexpression of AGL with wild type but not C26S Malin could cause a decrease in AGL levels (data not shown). Since the Malin/AGL interaction is affected by glycogenolytic agents, we sought to determine whether AGL levels are regulated in vivo during conditions such as fasting or refeeding. Fasting mice overnight (24 h) produced no appreciable difference in endogenous AGL levels (Fig. 5A). However, refeeding mice for 2 h led to a 48 ± 9% decrease in AGL levels that was restored upon continued refeeding (Fig. 5B,C). Glycogen phosphorylase levels were also decreased over this period, although glycogen synthase levels remained unaltered. During these 2 h of refeeding, glycogen was repleted beyond normal levels (Fig. 5D), probably as a result of overcompensation (Mamedova et al. 2003).
Figure 5.
Regulation of AGL levels in liver and primary hepatocytes during fasting–refeeding. (A) AGL levels in mouse liver decrease during the refeeding after an overnight fast. Three-month-old female C57Bl/6 mice were either allowed to eat freely (Ad. lib.), fasted for 24 h, or refed for 2 h after fasting. Liver lysates were obtained and processed for immunoblotting with the indicated antibodies. Each lane represents one mouse. (B) AGL levels during the time course of refeeding. Fasted mice were allowed to be refed for the indicated times, and then liver lysates were processed as above. (C) Quantification of AGL levels. Three mice were analyzed in each case. Error bars represent SEM. (D) Liver glycogen levels during fasting and refeeding. Glycogen levels are expressed as micromolar glucose released from amyloglucosidase digestion normalized to the weight of the liver. Three mice were analyzed in each case. Error bars represent SEM. (E) AGL levels in primary mouse hepatocytes during a starvation/refeeding cycle. Starvation was induced by glucose/serum deprivation and addition of 100 nM glucagon for either 1 or 7 h. After starvation for 1 h, refeeding was initiated by replacing the media with regular growth medium supplemented with 100 nM insulin. Lysates were then obtained and processed for immunoblotting with the indicated antibodies.
To further evaluate this phenomenon in primary hepatocytes, starvation was mimicked by glucose deprivation and treatment with 100 nM glucagon. After 1 h of glucose deprivation, refeeding was then initiated by changing the media to include glucose as well as 100 nM insulin. Treatment of cells with glucose/insulin caused a time-dependent decrease in AGL levels, as did prolonged (7 h) starvation (Fig. 5E). One possible explanation for the discrepancy between liver and primary hepatocytes could be the nonphysiological concentrations of the hormones used. Indeed, the decrease in AGL levels correlated with a down-regulation in PKA signaling (phospho-PKA blot), suggesting that the degradation of AGL requires an additional signal related to the desensitization of cAMP levels.
AGL colocalizes with polyglucosans
One major clinical feature of Lafora disease is the presence of polyglucosan inclusions termed “Lafora bodies” (Chan et al. 2005; Ganesh et al. 2006). Overexpression of glycogen synthase in mice leads to rapid polyglucosan formation (Raben et al. 2001; Pederson et al. 2003). Likewise, expression of glycogen synthase in HepG2 cells caused the appearance of polyglucosans that was surrounded by glycogen synthase itself (Fig. 6A). Thus we were interested to determine if AGL also localizes to these structures.
Figure 6.
AGL colocalizes with polyglucosans formed by overexpression of glycogen synthase. (A) Expression of myc-tagged glycogen synthase (myc-GS) causes the formation of inclusions that stain positive for periodic acid Schiff (PAS). HepG2 cells were transfected with myc-GS for 16 h, fixed in formalin, and processed for immunofluorescence microscopy using anti-myc antibodies (green) or PAS stain. Black arrows point to positive PAS stain. (B) Expression of myc-GS causes wild type but not the ΔCBD mutant of AGL to aggregate around the PAS-stain-positive inclusions. HepG2 cells were transfected for 16 h with myc-GS together with either HA-tagged wild-type AGL (HA-AGL) or HA-AGL ΔCBD. Cells were fixed in formalin and processed for immunofluorescence microscopy using anti-HA (green) and anti-myc (red) antibodies. White arrows indicate colocalization of HA-AGL and myc-GS.
When AGL was coexpressed with glycogen synthase, the localization of AGL switched from a diffuse cytoplasmic pattern (Fig. 3C) to one concentrated with glycogen synthase (Fig. 6B). The ability of AGL to localize to these polyglucosans is likely through its CBD domain, as AGL ΔCBD was unable to do so.
Discussion
The glycogen debranching enzyme, AGL, is involved in the breakdown of glycogen by primarily removing branch points that impede the action of glycogen phosphorylase. However, how AGL itself is regulated is poorly understood. In this report, we identify ubiquitination as a novel means of regulating AGL levels, and uncover some of the signals that regulate this process.
In humans, Cori’s disease is characterized by a deficiency in the activity of AGL. However, most mutations identified to date result in premature truncations, providing little by way of insight into its regulation. One exception is the G1448R mutation (Okubo et al. 1999) that is localized in the putative CBD. We demonstrate here that this mutant cannot bind to amylose or efficiently partition into a high-speed glycogen-enriched fraction. Interestingly, the G1448R mutant is unstable due to enhanced ubiquitination that is rescued by proteasomal inhibition. These results suggest that binding to glycogen may be involved in regulating AGL stability.
In addition to its reduced stability, we also observed that AGL G1448R forms aggresomes upon proteasome inhibition. Two proteins that are mutated in Lafora disease, Laforin and Malin, can also relocalize to aggresomes (Mittal et al. 2007). Lafora disease is characterized by the presence of inclusion bodies resembling poorly branched glycogen (Chan et al. 2005; Ganesh et al. 2006). We demonstrated that AGL and Malin directly interact, and further that this interaction increased upon treatment with cAMP-elevating agents, due in part to an increase in Malin levels. This suggests that a PKA-dependent pathway is involved in regulating Malin levels.
Because Malin is a ubiquitin ligase, these data suggest that AGL levels might also be regulated by fluctuations in glycogen stores. Indeed, fasted mice that were refed for 2 h exhibited a decrease in AGL levels in the liver. It is interesting to note that the glycogen levels had been restored by this time. We thus speculate that the suppression of AGL levels may be important in the repletion of glycogen stores and/or its proper branching.
In primary hepatocytes either refeeding or prolonged starvation caused a decrease in AGL levels. Thus release from glycogen alone is not sufficient for AGL degradation, but may act in a permissive role. Such compartmentalization of ubiquitination in the nucleus followed by cytoplasmic export for degradation has been shown for other proteins (Groulx and Lee 2002).
Taken together, these data lead to a model for the regulation of AGL by ubiquitination (Fig. 7). Glycogenolytic signals trigger the release of AGL from glycogen, allowing it to translocate to the nucleus. This permits AGL to interact with Malin, which then promotes its ubiquitination. Signals that elevate cAMP levels may further enhance this process by increasing the levels of Malin. Subsequent degradation of AGL probably involves an additional signal that might be related to the down-regulation of cAMP/PKA signaling. Thus, individuals with loss-of-function mutations in Malin could accumulate AGL inappropriately during instances of low glycogen stores, and lead to a decrease in the overall level of glycogen branching. Indeed, Lafora bodies have been shown to possess fewer α-1,6-glycosidic branches (Yokoi et al. 1968).
Figure 7.
Regulation of AGL during glycogenolysis. AGL binds to glycogen and is involved in the debranching process. Signals that induce glycogenolysis trigger the release of AGL and allow it to enter the nucleus. There, Malin promotes its ubiquitination. Signals that elevate cAMP levels may further enhance this process by increasing the levels of Malin. Subsequent degradation of AGL probably involves an additional signal that might be related to the down-regulation of cAMP/PKA signaling. In Lafora disease, loss of Malin function could potentially lead to increased AGL levels and/or activity. Conversely, in Cori’s disease, the G1448R mutant is unable to bind glycogen and undergoes rapid proteasomal-mediated degradation.
Lafora bodies are intracellular polyglucosan inclusions. These structures are found in several other glycogen-related disorders, such as Andersen disease (glycogen storage disease type IV) and adult polyglucosan body disease (APGD) (Moses and Parvari 2002; Chan et al. 2005; Ganesh et al. 2006). However, the symptoms and target organs can differ substantially between each disease. Whereas Lafora disease and APGD can present severe neurological problems, Andersen disease typically affects the heart and liver. The onset of disease is also quite different. Individuals affected by Andersen disease typically exhibit symptoms soon after birth. In contrast, Lafora disease progresses after 10 years of age.
Recently, an interesting case of Lafora disease involving two siblings was reported (Gomez-Garre et al. 2007). Although both individuals exhibited hepatic polyglucosan bodies, only one of the siblings developed severe liver cirrhosis and failure, suggesting the possible involvement of modifier genes. Thus, although disruptions of different genes can lead to polyglucosan formation, the tissue expression pattern and function plays an important role in manifestation of the phenotype.
In mice, overexpression of glycogen synthase leads to rapid polyglucosan formation and has been suggested as a model of Lafora disease (Raben et al. 2001; Pederson et al. 2003). In HepG2 cells, both AGL and glycogen synthase accumulate around polyglucosans (Fig. 6). Whether AGL assists glycogen synthase in this process is currently unknown. It is also possible that polyglucosans contain glycogen-related proteins as a result of carbohydrate–protein aggregation. Perhaps one function of Malin is to remove these proteins so that the cell can find a way to dispose of the excess carbohydrate. Further studies will be required to clarify these possibilities.
Materials and methods
Reagents
Anti-AGL antibodies were purchased from Abgent. Antibodies against AKT and phospho-PKA substrate were obtained from Cell Signaling Technology. Anti-glycogen synthase antibodies were purchased from Chemicon and USBiological. Protein A/G agarose and antibodies against Ubiquitin, PP1, HA, and myc were from Santa Cruz Biotechnology. Antibodies against γ-tubulin, α-tubulin, Flag, and anti-Flag (M2) agarose beads were from Sigma. Anti-phosphorylase antibodies were described previously (Crosson et al. 2003). The proteasomal inhibitor MG-132 was purchased from EMD Biosciences. N-Ethylmaleimide (NEM), FSK, and IBMX were obtained from Sigma. The glycogen phosphorylase inhibitor (Klabunde et al. 2005) was purchased from EMD Biosciences.
Plasmids
Constructs for HA-UB, Flag-Laforin, and myc-Malin were described previously (Wang et al. 2002; Gentry et al. 2005). The AGL cDNA was obtained by RT–PCR from mouse liver RNA. A CBD mutant was generated by deletion of the last 116 amino acids of AGL (where the glycogen-binding domain is located). AGL and the ΔCBD and G1448R mutants were cloned into various mammalian expression vectors by PCR. All constructs were verified by sequencing.
Cell culture and transfection
HepG2 hepatoblastoma, 293T, and COS cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS; Invitrogen). COS and 293T cells were transfected with Fugene 6 (Roche) according to the manufacturer’s instructions. HepG2 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions
Preparation of cell lysates and immunoblotting
For isolation of glycogen-enriched fractions, cells were scraped in 50 mM Tris (pH 7.5) and 150 mM NaCl supplemented with Complete EDTA-free protease inhibitor. Cell extracts were further sonicated (30% maximal setting, 10 pulses), and then debris was pelleted at 3,000g for 10 min. The supernatant was then successively pelleted at 14,000g for 10 min and then at 100,000g for 1 h to obtain the cytosol (supernatant) and glycogen-enriched fraction (pellet). The pellet was solubilized in lysis buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1% Triton X-100, Complete EDTA-free protease inhibitor) by rotating end-over-end for 15 min. The lysates were then cleared one more time at 14,000g to remove unwanted debris.
For isolation of TCL, cells were washed twice in ice-cold phosphate-buffered saline (PBS) and then scraped in lysis buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM Na3VO4, 10 mM NaF, 5 mM NEM, 10 μM MG-132, Complete EDTA-free protease inhibitor). The lysates were rocked end-over-end for 15 min at 4°C and then clarified by centrifugation at 14,000g for 10 min. For mouse liver, tissue fragments were dounce-homogenized in the above lysis buffer and processed similarly.
For denaturing immunoprecipitations, cells were lysed in denaturing buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1% Triton, 1% SDS, 1 mM Na3VO4, 10 mM NaF, 5 mM NEM, 10 μM MG-132) and boiled for 10 min. Lysates were diluted 1:10 with the same buffer lacking SDS and incubated with the appropriate antibody/Protein A/G agarose beads for 2 h rotating at 4°C. Immunoprecipitates were extensively washed in lysis buffer (without SDS), and proteins were eluted at 95°C in SDS loading buffer, separated by SDS PAGE, and transferred to nitrocellulose.
Confocal fluorescence microscopy
Immunofluorescence studies were performed as described previously (Zhang et al. 2006). Cells were grown on glass coverslips in six-well or 12-well dishes. Following the fixation with 10% formalin for 20 min, cells were permeabilized with 0.5% Triton X-100 for 5 min and then blocked with 1% bovine serum albumin, 1% ovalbumin, and 2% goat serum for 1 h. For some experiments, methanol fixation at −20°C for 10 min was performed. Coverslips were incubated with primary and Alexa Fluor secondary antibodies in blocking solution and mounted on glass slides with Vectashield (Vector Laboratories). Cells were imaged using confocal fluorescence microscope (Olympus IX SLA). Images were then imported into Photoshop (Adobe Systems, Inc.) for processing.
Glycogen determination
Determination of glycogen in liver was performed as described previously (Suzuki et al. 2001). HepG2 cells were grown to near confluence in six-well plates. Cells were washed twice in ice-cold PBS and then snap-frozen in liquid nitrogen until further use. Cells were scraped in 50 mM sodium acetate (pH 4.8), and lysis was achieved by sonication. Lysates were cleared by centrifugation at 6,000g for 10 min. An aliquot was saved for protein determination and immunoblot analysis. The assay was initiated by addition of amyloglucosidase (Sigma) and incubation for 2 h at 37°C. The glucose released was then measured using the Auto CII glucose kit.
Acknowledgments
We thank Xiaowei Chen and Dr. Dave Bridges for helpful discussions. We also thank Dr. Chang Liu with technical help in hepatocyte isolations. This work was supported by a grant from the National Institutes of Health (2R01DK060597-06). A.C. was supported in part by a post-doctoral fellowship from the Canadian Institutes of Health Research and a Phyllis Locke grant from the Canadian Diabetes Association.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1553207
References
- Brady M.J., Saltiel A.R., Saltiel A.R. The role of protein phosphatase-1 in insulin action. Recent Prog. Horm. Res. 2001;56:157–173. doi: 10.1210/rp.56.1.157. [DOI] [PubMed] [Google Scholar]
- Chan E.M., Young E.J., Ianzano L., Munteanu I., Zhao X., Christopoulos C.C., Avanzini G., Elia M., Ackerley C.A., Jovic N.J., Young E.J., Ianzano L., Munteanu I., Zhao X., Christopoulos C.C., Avanzini G., Elia M., Ackerley C.A., Jovic N.J., Ianzano L., Munteanu I., Zhao X., Christopoulos C.C., Avanzini G., Elia M., Ackerley C.A., Jovic N.J., Munteanu I., Zhao X., Christopoulos C.C., Avanzini G., Elia M., Ackerley C.A., Jovic N.J., Zhao X., Christopoulos C.C., Avanzini G., Elia M., Ackerley C.A., Jovic N.J., Christopoulos C.C., Avanzini G., Elia M., Ackerley C.A., Jovic N.J., Avanzini G., Elia M., Ackerley C.A., Jovic N.J., Elia M., Ackerley C.A., Jovic N.J., Ackerley C.A., Jovic N.J., Jovic N.J., et al. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat. Genet. 2003;35:125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- Chan E.M., Ackerley C.A., Lohi H., Ianzano L., Cortez M.A., Shannon P., Scherer S.W., Minassian B.A., Ackerley C.A., Lohi H., Ianzano L., Cortez M.A., Shannon P., Scherer S.W., Minassian B.A., Lohi H., Ianzano L., Cortez M.A., Shannon P., Scherer S.W., Minassian B.A., Ianzano L., Cortez M.A., Shannon P., Scherer S.W., Minassian B.A., Cortez M.A., Shannon P., Scherer S.W., Minassian B.A., Shannon P., Scherer S.W., Minassian B.A., Scherer S.W., Minassian B.A., Minassian B.A. Laforin preferentially binds the neurotoxic starch-like polyglucosans, which form in its absence in progressive myoclonus epilepsy. Hum. Mol. Genet. 2004;13:1117–1129. doi: 10.1093/hmg/ddh130. [DOI] [PubMed] [Google Scholar]
- Chan E.M., Andrade D.M., Franceschetti S., Minassian B., Andrade D.M., Franceschetti S., Minassian B., Franceschetti S., Minassian B., Minassian B. Progressive myoclonus epilepsies: EPM1, EPM2A, EPM2B. Adv. Neurol. 2005;95:47–57. [PubMed] [Google Scholar]
- Crosson S.M., Khan A., Printen J., Pessin J.E., Saltiel A.R., Khan A., Printen J., Pessin J.E., Saltiel A.R., Printen J., Pessin J.E., Saltiel A.R., Pessin J.E., Saltiel A.R., Saltiel A.R. PTG gene deletion causes impaired glycogen synthesis and developmental insulin resistance. J. Clin. Invest. 2003;111:1423–1432. doi: 10.1172/JCI17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Sanchez M.E., Criado-Garcia O., Heath K.E., Garcia-Fojeda B., Medrano-Fernandez I., Gomez-Garre P., Sanz P., Serratosa J.M., de Rodriguez Cordoba S., Criado-Garcia O., Heath K.E., Garcia-Fojeda B., Medrano-Fernandez I., Gomez-Garre P., Sanz P., Serratosa J.M., de Rodriguez Cordoba S., Heath K.E., Garcia-Fojeda B., Medrano-Fernandez I., Gomez-Garre P., Sanz P., Serratosa J.M., de Rodriguez Cordoba S., Garcia-Fojeda B., Medrano-Fernandez I., Gomez-Garre P., Sanz P., Serratosa J.M., de Rodriguez Cordoba S., Medrano-Fernandez I., Gomez-Garre P., Sanz P., Serratosa J.M., de Rodriguez Cordoba S., Gomez-Garre P., Sanz P., Serratosa J.M., de Rodriguez Cordoba S., Sanz P., Serratosa J.M., de Rodriguez Cordoba S., Serratosa J.M., de Rodriguez Cordoba S., de Rodriguez Cordoba S. Laforin, the dual-phosphatase responsible for Lafora disease, interacts with R5 (PTG), a regulatory subunit of protein phosphatase-1 that enhances glycogen accumulation. Hum. Mol. Genet. 2003;12:3161–3171. doi: 10.1093/hmg/ddg340. [DOI] [PubMed] [Google Scholar]
- Ganesh S., Agarwala K.L., Ueda K., Akagi T., Shoda K., Usui T., Hashikawa T., Osada H., Delgado-Escueta A.V., Yamakawa K., Agarwala K.L., Ueda K., Akagi T., Shoda K., Usui T., Hashikawa T., Osada H., Delgado-Escueta A.V., Yamakawa K., Ueda K., Akagi T., Shoda K., Usui T., Hashikawa T., Osada H., Delgado-Escueta A.V., Yamakawa K., Akagi T., Shoda K., Usui T., Hashikawa T., Osada H., Delgado-Escueta A.V., Yamakawa K., Shoda K., Usui T., Hashikawa T., Osada H., Delgado-Escueta A.V., Yamakawa K., Usui T., Hashikawa T., Osada H., Delgado-Escueta A.V., Yamakawa K., Hashikawa T., Osada H., Delgado-Escueta A.V., Yamakawa K., Osada H., Delgado-Escueta A.V., Yamakawa K., Delgado-Escueta A.V., Yamakawa K., Yamakawa K. Laforin, defective in the progressive myoclonus epilepsy of Lafora type, is a dual-specificity phosphatase associated with polyribosomes. Hum. Mol. Genet. 2000;9:2251–2261. doi: 10.1093/oxfordjournals.hmg.a018916. [DOI] [PubMed] [Google Scholar]
- Ganesh S., Delgado-Escueta A.V., Sakamoto T., Avila M.R., Machado-Salas J., Hoshii Y., Akagi T., Gomi H., Suzuki T., Amano K., Delgado-Escueta A.V., Sakamoto T., Avila M.R., Machado-Salas J., Hoshii Y., Akagi T., Gomi H., Suzuki T., Amano K., Sakamoto T., Avila M.R., Machado-Salas J., Hoshii Y., Akagi T., Gomi H., Suzuki T., Amano K., Avila M.R., Machado-Salas J., Hoshii Y., Akagi T., Gomi H., Suzuki T., Amano K., Machado-Salas J., Hoshii Y., Akagi T., Gomi H., Suzuki T., Amano K., Hoshii Y., Akagi T., Gomi H., Suzuki T., Amano K., Akagi T., Gomi H., Suzuki T., Amano K., Gomi H., Suzuki T., Amano K., Suzuki T., Amano K., Amano K., et al. Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum. Mol. Genet. 2002a;11:1251–1262. doi: 10.1093/hmg/11.11.1251. [DOI] [PubMed] [Google Scholar]
- Ganesh S., Suzuki T., Yamakawa K., Suzuki T., Yamakawa K., Yamakawa K. Alternative splicing modulates subcellular localization of Laforin. Biochem. Biophys. Res. Commun. 2002b;291:1134–1137. doi: 10.1006/bbrc.2002.6590. [DOI] [PubMed] [Google Scholar]
- Ganesh S., Tsurutani N., Suzuki T., Hoshii Y., Ishihara T., Delgado-Escueta A.V., Yamakawa K., Tsurutani N., Suzuki T., Hoshii Y., Ishihara T., Delgado-Escueta A.V., Yamakawa K., Suzuki T., Hoshii Y., Ishihara T., Delgado-Escueta A.V., Yamakawa K., Hoshii Y., Ishihara T., Delgado-Escueta A.V., Yamakawa K., Ishihara T., Delgado-Escueta A.V., Yamakawa K., Delgado-Escueta A.V., Yamakawa K., Yamakawa K. The carbohydrate-binding domain of Lafora disease protein targets Lafora polyglucosan bodies. Biochem. Biophys. Res. Commun. 2004;313:1101–1109. doi: 10.1016/j.bbrc.2003.12.043. [DOI] [PubMed] [Google Scholar]
- Ganesh S., Puri R., Singh S., Mittal S., Dubey D., Puri R., Singh S., Mittal S., Dubey D., Singh S., Mittal S., Dubey D., Mittal S., Dubey D., Dubey D. Recent advances in the molecular basis of Lafora’s progressive myoclonus epilepsy. J. Hum. Genet. 2006;51:1–8. doi: 10.1007/s10038-005-0321-1. [DOI] [PubMed] [Google Scholar]
- Gentry M.S., Worby C.A., Dixon J.E., Worby C.A., Dixon J.E., Dixon J.E. Insights into Lafora disease: Malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of Laforin. Proc. Natl. Acad. Sci. 2005;102:8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry M.S., Dowen R.H., Worby C.A., Mattoo S., Ecker J.R., Dixon J.E., Dowen R.H., Worby C.A., Mattoo S., Ecker J.R., Dixon J.E., Worby C.A., Mattoo S., Ecker J.R., Dixon J.E., Mattoo S., Ecker J.R., Dixon J.E., Ecker J.R., Dixon J.E., Dixon J.E. The phosphatase Laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J. Cell Biol. 2007;178:477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Garre P., Gutierrez-Delicado E., Gomez-Abad C., Morales-Corraliza J., Villanueva V.E., de Rodriguez Cordoba S., Larrauri J., Gutierrez M., Berciano J., Serratosa J.M., Gutierrez-Delicado E., Gomez-Abad C., Morales-Corraliza J., Villanueva V.E., de Rodriguez Cordoba S., Larrauri J., Gutierrez M., Berciano J., Serratosa J.M., Gomez-Abad C., Morales-Corraliza J., Villanueva V.E., de Rodriguez Cordoba S., Larrauri J., Gutierrez M., Berciano J., Serratosa J.M., Morales-Corraliza J., Villanueva V.E., de Rodriguez Cordoba S., Larrauri J., Gutierrez M., Berciano J., Serratosa J.M., Villanueva V.E., de Rodriguez Cordoba S., Larrauri J., Gutierrez M., Berciano J., Serratosa J.M., de Rodriguez Cordoba S., Larrauri J., Gutierrez M., Berciano J., Serratosa J.M., Larrauri J., Gutierrez M., Berciano J., Serratosa J.M., Gutierrez M., Berciano J., Serratosa J.M., Berciano J., Serratosa J.M., Serratosa J.M. Hepatic disease as the first manifestation of progressive myoclonus epilepsy of Lafora. Neurology. 2007;68:1369–1373. doi: 10.1212/01.wnl.0000260061.37559.67. [DOI] [PubMed] [Google Scholar]
- Groulx I., Lee S., Lee S. Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor suppressor protein. Mol. Cell. Biol. 2002;22:5319–5336. doi: 10.1128/MCB.22.15.5319-5336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde T., Wendt K.U., Kadereit D., Brachvogel V., Burger H.J., Herling A.W., Oikonomakos N.G., Kosmopoulou M.N., Schmoll D., Sarubbi E., Wendt K.U., Kadereit D., Brachvogel V., Burger H.J., Herling A.W., Oikonomakos N.G., Kosmopoulou M.N., Schmoll D., Sarubbi E., Kadereit D., Brachvogel V., Burger H.J., Herling A.W., Oikonomakos N.G., Kosmopoulou M.N., Schmoll D., Sarubbi E., Brachvogel V., Burger H.J., Herling A.W., Oikonomakos N.G., Kosmopoulou M.N., Schmoll D., Sarubbi E., Burger H.J., Herling A.W., Oikonomakos N.G., Kosmopoulou M.N., Schmoll D., Sarubbi E., Herling A.W., Oikonomakos N.G., Kosmopoulou M.N., Schmoll D., Sarubbi E., Oikonomakos N.G., Kosmopoulou M.N., Schmoll D., Sarubbi E., Kosmopoulou M.N., Schmoll D., Sarubbi E., Schmoll D., Sarubbi E., Sarubbi E., et al. Acyl ureas as human liver glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. J. Med. Chem. 2005;48:6178–6193. doi: 10.1021/jm049034y. [DOI] [PubMed] [Google Scholar]
- Kopito R.R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Lohi H., Ianzano L., Zhao X.C., Chan E.M., Turnbull J., Scherer S.W., Ackerley C.A., Minassian B.A., Ianzano L., Zhao X.C., Chan E.M., Turnbull J., Scherer S.W., Ackerley C.A., Minassian B.A., Zhao X.C., Chan E.M., Turnbull J., Scherer S.W., Ackerley C.A., Minassian B.A., Chan E.M., Turnbull J., Scherer S.W., Ackerley C.A., Minassian B.A., Turnbull J., Scherer S.W., Ackerley C.A., Minassian B.A., Scherer S.W., Ackerley C.A., Minassian B.A., Ackerley C.A., Minassian B.A., Minassian B.A. Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum. Mol. Genet. 2005;14:2727–2736. doi: 10.1093/hmg/ddi306. [DOI] [PubMed] [Google Scholar]
- Mamedova L.K., Shneyvays V., Katz A., Shainberg A., Shneyvays V., Katz A., Shainberg A., Katz A., Shainberg A., Shainberg A. Mechanism of glycogen supercompensation in rat skeletal muscle cultures. Mol. Cell. Biochem. 2003;250:11–19. doi: 10.1023/a:1024980710799. [DOI] [PubMed] [Google Scholar]
- Minassian B.A., Lee J.R., Herbrick J.A., Huizenga J., Soder S., Mungall A.J., Dunham I., Gardner R., Fong C.Y., Carpenter S., Lee J.R., Herbrick J.A., Huizenga J., Soder S., Mungall A.J., Dunham I., Gardner R., Fong C.Y., Carpenter S., Herbrick J.A., Huizenga J., Soder S., Mungall A.J., Dunham I., Gardner R., Fong C.Y., Carpenter S., Huizenga J., Soder S., Mungall A.J., Dunham I., Gardner R., Fong C.Y., Carpenter S., Soder S., Mungall A.J., Dunham I., Gardner R., Fong C.Y., Carpenter S., Mungall A.J., Dunham I., Gardner R., Fong C.Y., Carpenter S., Dunham I., Gardner R., Fong C.Y., Carpenter S., Gardner R., Fong C.Y., Carpenter S., Fong C.Y., Carpenter S., Carpenter S., et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat. Genet. 1998;20:171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- Mittal S., Dubey D., Yamakawa K., Ganesh S., Dubey D., Yamakawa K., Ganesh S., Yamakawa K., Ganesh S., Ganesh S. Lafora disease proteins Malin and Laforin are recruited to aggresomes in response to proteasomal impairment. Hum. Mol. Genet. 2007;16:753–762. doi: 10.1093/hmg/ddm006. [DOI] [PubMed] [Google Scholar]
- Moses S.W., Parvari R., Parvari R. The variable presentations of glycogen storage disease type IV: A review of clinical, enzymatic and molecular studies. Curr. Mol. Med. 2002;2:177–188. doi: 10.2174/1566524024605815. [DOI] [PubMed] [Google Scholar]
- Okubo M., Kanda F., Horinishi A., Takahashi K., Okuda S., Chihara K., Murase T., Kanda F., Horinishi A., Takahashi K., Okuda S., Chihara K., Murase T., Horinishi A., Takahashi K., Okuda S., Chihara K., Murase T., Takahashi K., Okuda S., Chihara K., Murase T., Okuda S., Chihara K., Murase T., Chihara K., Murase T., Murase T. Glycogen storage disease type IIIa: First report of a causative missense mutation (G1448R) of the glycogen debranching enzyme gene found in a homozygous patient. Hum. Mutat. 1999;14:542–543. doi: 10.1002/(SICI)1098-1004(199912)14:6<542::AID-HUMU15>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Pederson B.A., Csitkovits A.G., Simon R., Schroeder J.M., Wang W., Skurat A.V., Roach P.J., Csitkovits A.G., Simon R., Schroeder J.M., Wang W., Skurat A.V., Roach P.J., Simon R., Schroeder J.M., Wang W., Skurat A.V., Roach P.J., Schroeder J.M., Wang W., Skurat A.V., Roach P.J., Wang W., Skurat A.V., Roach P.J., Skurat A.V., Roach P.J., Roach P.J. Overexpression of glycogen synthase in mouse muscle results in less branched glycogen. Biochem. Biophys. Res. Commun. 2003;305:826–830. doi: 10.1016/s0006-291x(03)00862-3. [DOI] [PubMed] [Google Scholar]
- Printen J.A., Brady M.J., Saltiel A.R., Brady M.J., Saltiel A.R., Saltiel A.R. PTG, a protein phosphatase 1-binding protein with a role in glycogen metabolism. Science. 1997;275:1475–1478. doi: 10.1126/science.275.5305.1475. [DOI] [PubMed] [Google Scholar]
- Raben N., Danon M., Lu N., Lee E., Shliselfeld L., Skurat A.V., Roach P.J., Lawrence J.C., Musumeci O., Shanske S., Danon M., Lu N., Lee E., Shliselfeld L., Skurat A.V., Roach P.J., Lawrence J.C., Musumeci O., Shanske S., Lu N., Lee E., Shliselfeld L., Skurat A.V., Roach P.J., Lawrence J.C., Musumeci O., Shanske S., Lee E., Shliselfeld L., Skurat A.V., Roach P.J., Lawrence J.C., Musumeci O., Shanske S., Shliselfeld L., Skurat A.V., Roach P.J., Lawrence J.C., Musumeci O., Shanske S., Skurat A.V., Roach P.J., Lawrence J.C., Musumeci O., Shanske S., Roach P.J., Lawrence J.C., Musumeci O., Shanske S., Lawrence J.C., Musumeci O., Shanske S., Musumeci O., Shanske S., Shanske S., et al. Surprises of genetic engineering: A possible model of polyglucosan body disease. Neurology. 2001;56:1739–1745. doi: 10.1212/wnl.56.12.1739. [DOI] [PubMed] [Google Scholar]
- Roach P.J. Glycogen and its metabolism. Curr. Mol. Med. 2002;2:101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- Serratosa J.M., Gomez-Garre P., Gallardo M.E., Anta B., de Bernabe D.B., Lindhout D., Augustijn P.B., Tassinari C.A., Malafosse R.M., Topcu M., Gomez-Garre P., Gallardo M.E., Anta B., de Bernabe D.B., Lindhout D., Augustijn P.B., Tassinari C.A., Malafosse R.M., Topcu M., Gallardo M.E., Anta B., de Bernabe D.B., Lindhout D., Augustijn P.B., Tassinari C.A., Malafosse R.M., Topcu M., Anta B., de Bernabe D.B., Lindhout D., Augustijn P.B., Tassinari C.A., Malafosse R.M., Topcu M., de Bernabe D.B., Lindhout D., Augustijn P.B., Tassinari C.A., Malafosse R.M., Topcu M., Lindhout D., Augustijn P.B., Tassinari C.A., Malafosse R.M., Topcu M., Augustijn P.B., Tassinari C.A., Malafosse R.M., Topcu M., Tassinari C.A., Malafosse R.M., Topcu M., Malafosse R.M., Topcu M., Topcu M., et al. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2) Hum. Mol. Genet. 1999;8:345–352. doi: 10.1093/hmg/8.2.345. [DOI] [PubMed] [Google Scholar]
- Shen J.J., Chen Y.T., Chen Y.T. Molecular characterization of glycogen storage disease type III. Curr. Mol. Med. 2002;2:167–175. doi: 10.2174/1566524024605752. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Lanner C., Kim J.H., Vilardo P.G., Zhang H., Yang J., Cooper L.D., Steele M., Kennedy A., Bock C.B., Lanner C., Kim J.H., Vilardo P.G., Zhang H., Yang J., Cooper L.D., Steele M., Kennedy A., Bock C.B., Kim J.H., Vilardo P.G., Zhang H., Yang J., Cooper L.D., Steele M., Kennedy A., Bock C.B., Vilardo P.G., Zhang H., Yang J., Cooper L.D., Steele M., Kennedy A., Bock C.B., Zhang H., Yang J., Cooper L.D., Steele M., Kennedy A., Bock C.B., Yang J., Cooper L.D., Steele M., Kennedy A., Bock C.B., Cooper L.D., Steele M., Kennedy A., Bock C.B., Steele M., Kennedy A., Bock C.B., Kennedy A., Bock C.B., Bock C.B., et al. Insulin control of glycogen metabolism in knockout mice lacking the muscle-specific protein phosphatase PP1G/RGL. Mol. Cell. Biol. 2001;21:2683–2694. doi: 10.1128/MCB.21.8.2683-2694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Stuckey J.A., Wishart M.J., Dixon J.E., Stuckey J.A., Wishart M.J., Dixon J.E., Wishart M.J., Dixon J.E., Dixon J.E. A unique carbohydrate binding domain targets the lafora disease phosphatase to glycogen. J. Biol. Chem. 2002;277:2377–2380. doi: 10.1074/jbc.C100686200. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu Y., Wu C., Zhang H., Zheng X., Zheng Z., Geiger T.L., Nuovo G.J., Zheng P., Liu Y., Wu C., Zhang H., Zheng X., Zheng Z., Geiger T.L., Nuovo G.J., Zheng P., Wu C., Zhang H., Zheng X., Zheng Z., Geiger T.L., Nuovo G.J., Zheng P., Zhang H., Zheng X., Zheng Z., Geiger T.L., Nuovo G.J., Zheng P., Zheng X., Zheng Z., Geiger T.L., Nuovo G.J., Zheng P., Zheng Z., Geiger T.L., Nuovo G.J., Zheng P., Geiger T.L., Nuovo G.J., Zheng P., Nuovo G.J., Zheng P., Zheng P. Epm2a suppresses tumor growth in an immunocompromised host by inhibiting Wnt signaling. Cancer Cell. 2006;10:179–190. doi: 10.1016/j.ccr.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Wang W., Lohi H., Skurat A.V., Depaoli-Roach A.A., Minassian B.A., Roach P.J., Lohi H., Skurat A.V., Depaoli-Roach A.A., Minassian B.A., Roach P.J., Skurat A.V., Depaoli-Roach A.A., Minassian B.A., Roach P.J., Depaoli-Roach A.A., Minassian B.A., Roach P.J., Minassian B.A., Roach P.J., Roach P.J. Glycogen metabolism in tissues from a mouse model of Lafora disease. Arch. Biochem. Biophys. 2007;457:264–269. doi: 10.1016/j.abb.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby C.A., Gentry M.S., Dixon J.E., Gentry M.S., Dixon J.E., Dixon J.E. Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J. Biol. Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi S., Austin J., Witmer F., Sakai M., Austin J., Witmer F., Sakai M., Witmer F., Sakai M., Sakai M. Studies in myoclonus epilepsy (Lafora body form). I. Isolation and preliminary characterization of Lafora bodies in two cases. Arch. Neurol. 1968;19:15–33. doi: 10.1001/archneur.1968.00480010033002. [DOI] [PubMed] [Google Scholar]
- Zhang M., Liu J., Cheng A., Deyoung S.M., Chen X., Dold L.H., Saltiel A.R., Liu J., Cheng A., Deyoung S.M., Chen X., Dold L.H., Saltiel A.R., Cheng A., Deyoung S.M., Chen X., Dold L.H., Saltiel A.R., Deyoung S.M., Chen X., Dold L.H., Saltiel A.R., Chen X., Dold L.H., Saltiel A.R., Dold L.H., Saltiel A.R., Saltiel A.R. CAP interacts with cytoskeletal proteins and regulates adhesion-mediated ERK activation and motility. EMBO J. 2006;25:5284–5293. doi: 10.1038/sj.emboj.7601406. [DOI] [PMC free article] [PubMed] [Google Scholar]