Abstract
The levels of oxidatively damaged, carbonylated, proteins increase with the replicative age of yeast mother cells. We show here that such carbonylated proteins are associated with Hsp104p-containing protein aggregates and that these aggregates, like oxidized proteins, are retained in the progenitor cell during cytokinesis by a Sir2p-dependent process. Deletion of HSP104 resulted in a breakdown of damage asymmetry, and overproduction of Hsp104p partially restored damage retention in sir2Δ cells, suggesting that functional chaperones associated with protein aggregates are required for the establishment of damage asymmetry and that these functions are limited in sir2Δ cells. In line with this, Hsp104p and several Hsp70s displayed elevated damaged in sir2Δ cells, and protein aggregates were rescued at a slower rate in this mutant. Moreover, overproduction of Hsp104p suppressed the accelerated aging of cells lacking Sir2p, and drugs inhibiting damage segregation further demonstrated that spatial quality control is required to rejuvenate the progeny.
Keywords: Hsp104, Sir2, aging, protein aggregates, protein oxidation, yeast
Mitosis in the yeast Saccharomyces cerevisiae is distinctively asymmetrical and encompasses mother cell-specific aging (Kennedy et al. 1994). That is, during progressive divisions the cell undergoes age-related changes, including an increased generation time, increase in size, decline in mating ability, accumulation of extrachromosomal rDNA circles (ERCs) (Sinclair and Guarente 1997), and a switch to a hyperrecombinational state (McMurray and Gottschling 2003). Thus, with each division the mother cell becomes phenotypically older and will, on average, enter terminal senescence after having completed 20–30 divisions. However, mother cells (if not too close to their terminal state) (Kennedy et al. 1994) generate offspring exhibiting a full replicative potential, and the acquired aging phenotype is thus somehow expunged in the progeny during the process of asymmetric division (Jazwinski 2004). This argues for the existence of a senescence factor that accumulates in the mother cells but is either prevented from transmission to the offspring or somehow eradicated in newborn cells (Egilmez and Jazwinski 1989). ERCs (Sinclair and Guarente 1997) and dysfunctional mitochondria (Lai et al. 2002) have been suggested to be such senescence factors, but conclusive evidence for one primary culprit is lacking.
Mother cell-specific aging in yeast is similar to senescence in mammals, worms, flies, and bacteria characterized by a progressive accumulation of oxidized proteins (Stadtman 1992; Dukan and Nyström 1998; Aguilaniu et al. 2003; Reverter-Branchat et al. 2004), of which protein carbonyls are among the best studied. Carbonyl derivatives are formed by a direct and irreversible metal-catalyzed oxidative (MCO) attack on specific amino acid side chains (Stadtman 1992; Requena et al. 2003). This modification usually impairs or totally abolishes the activity of the targeted protein, and heavily carbonylated proteins, oxidized in vitro, tend to form high-molecular-weight aggregates (Bota and Davies 2002). Such aggregates may become cytotoxic and are associated with a large number of age-related disorders in humans, including Parkinson’s disease, Alzheimer’s disease, and diabetes (Grune et al. 2004). To what extent carbonylated proteins form aggregates in vivo is not known, however.
The development of an in situ detection of protein carbonyls in single yeast cells revealed a distinctively asymmetrical distribution of carbonylated proteins during cytokinesis, with most being detected in the mother cell compartment. A screen for mutants with a more symmetrical distribution of carbonylated proteins identified the sirtuin, Sir2p, as an essential component in maintaining damage asymmetry during mitosis (Aguilaniu et al. 2003). Sir2p, a conserved NAD-dependent histone deacetylase, is a key regulator of aging in a variety of organisms, including yeast, worms, and flies (Guarente 2000; Sinclair 2002; Tissenbaum and Guarente 2001; Rogina and Helfand 2004). In yeast, the role of Sir2p in regulating longevity has been linked to recombination at rDNA loci and the accumulation of ERCs (Sinclair and Guarente 1997), whereas its effects on aging in nematodes are suggested to be intimately connected to defense against damage caused by reactive oxygen species (ROS) (Hekimi and Guarente 2003).
This study demonstrates that damage asymmetry is the result of a specific retention mechanism acting on nearly all species of carbonylation-damaged proteins in old progenitor cells. This damage retention is accomplished by a spatial protein quality control (PQC) system involving Hsp104 and the actin cytoskeleton. The spatial PQC suffers from elevated damage and its activity is reduced in cells lacking Sir2p. Hsp104 overproduction ameliorates damage retention and suppresses the reduced life span of sir2Δ cells. Furthermore, a transient collapse of damage retention is shown to impair the full rejuvenation of the progeny.
Results
Cells lacking Sir2p exhibit elevated levels of oxidatively damaged molecular chaperones
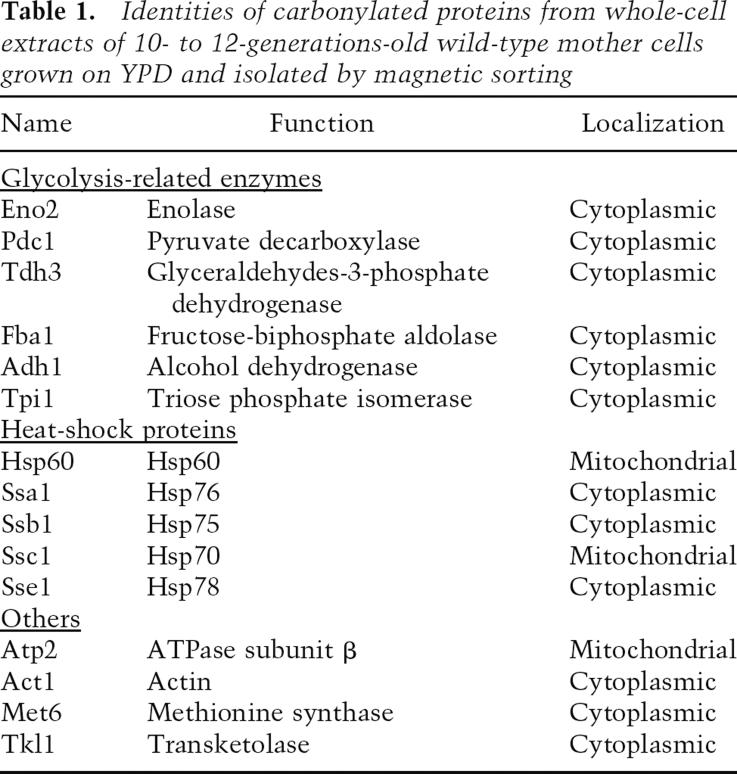
The possibility that unequal rates of proteolysis in mother and daughter cells are the cause for damage asymmetry has been refuted previously on the grounds that the rate of degradation of carbonylated proteins in the mother and daughter cell compartment is similar and that the absence of Sir2p did not affect this degradation (Aguilaniu et al. 2003). Therefore, we approached the possibility that a general or specific barrier between the mother and daughter cell may impede inheritance of carbonylated proteins into the daughter cell compartment. To distinguish between general and specific retention processes giving rise to damage asymmetry, we first identified which of the many yeast proteins were targets for age-dependent carbonylation (Table 1) using a proteomic approach (Dukan and Nyström 1998). As seen in Table 1, the functions of proteins becoming increasingly oxidized upon replicative aging exhibit a bias toward involvement in central carbon metabolism and protein management (see also Reverter-Branchat et al. 2004). The proteins contributing most to the carbonylation load of wild-type cells are Eno2p, Pdc1p, and Tdh3p (Fig. 1A), which is partly due to their abundance. However, the pattern of carbonylation does not simply reflect protein abundance since other highly abundant proteins did not exhibit any detectible carbonylation damage (e.g., Hxk2p, Zwf1p). The pattern of carbonylation is somewhat different in cells lacking Sir2p. Total carbonylation is twofold higher in old sir2Δ cells (Fig. 1B), mostly as a result of a two- to threefold higher carbonylation of several chaperones, including Ssa1p, Ssb1p, Ssc1p, Sse1p, and Hsp60 (Fig. 1A). By normalizing the carbonylation load of each protein to its relative abundance, it became apparent that molecular chaperones are preferential targets of elevated damage in sir2Δ cells (Fig. 1C).
Table 1.
Identities of carbonylated proteins from whole-cell extracts of 10- to 12-generations-old wild-type mother cells grown on YPD and isolated by magnetic sorting
Figure 1.
Cellular chaperones are preferential targets of age-dependent protein oxidation in the short-lived sir2Δ mutant. (A) Oxidation signal of single proteins from 10- to 12-generations-old wild-type (black bars) and sir2Δ (gray bars) cells. The specific oxidation signal was derived by comparing the carbonylation intensity of single proteins with the total carbonylation signal from all proteins. Protein identities are found in Table 1; arrows point to chaperones. (B) Comparison of total carbonylation (normalized to protein concentration) of the whole proteome from aged wild-type and sir2Δ cells. (C) Oxidation index of single proteins from aged wild-type (black bars) and sir2Δ (gray bars) cells. The specific oxidation index was calculated by normalizing the carbonylation intensity (carbonylation signal in the specific protein per total carbonylation signal) of single proteins to their abundance (Coomassie-derived signal in specific protein per total Coomassie signal on the gel). An index >1 indicates a protein whose oxidation level exceeds that of the average carbonylated protein. Values shown are the average of at least four different 2D gels and two independent sorting events; error bars in A–C represent the standard deviation of the mean.
Sir2p-dependent establishment of damage asymmetry is the result of a specific retention of oxidized proteins
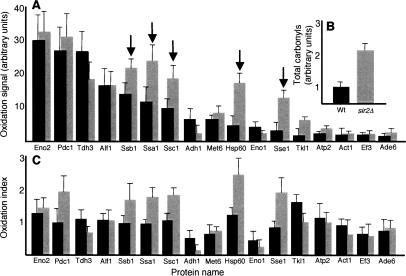
We next determined whether the entire pool of a protein, or only its carbonylated subpopulation, differed between the mother and daughter cells. This approach demonstrated an unequal distribution of only the oxidized form of a protein (see Eno2p, Tdh3p, and Pdc1p as an example in Fig. 2A), not the protein per se (Fig. 2A–C). A similar pattern was observed for all species of carbonylated proteins, ranging widely in size, isoelectric point, cellular localization, or function; moreover, they all required Sir2p for their asymmetrical distribution (Fig. 2A–C). Knowing that ERCs accumulate in cells lacking Sir2p but can be suppressed by deletion of FOB1, we next looked at whether ERC accumulation was involved in the failure of sir2Δ mutants to establish damage asymmetry. Damage segregation was normal in fob1Δ cells, whereas sir2Δ, fob1Δ double mutants displayed the same failure to segregate damaged proteins as sir2Δ single mutants (data not shown). Thus, the failure of sir2Δ cells to establish damage asymmetry cannot be linked to the accumulation of ERC.
Figure 2.
Damage asymmetry is the result of a specific Sir2p-mediated retention of oxidized proteins in the mother cell during cytokinesis. (A) 2D PAGE carbonylation (oxyblot) and abundance (Coomassie) of the three major carbonylated proteins—Eno2p, Pdc1p, and Thd3p—in wild-type (top rows) and sir2Δ (bottom rows) mother and daughter cells. The mother cells are 10–12 generations old. (B) Bud/mother oxidation ratio in aged wild type (black bars) and sir2Δ (gray bars). The ratio was calculated for each of the identified proteins, by comparing the specific oxidation signals (see Fig. 1) of daughter cells and their 10- to 12-generations-old mothers. (C) Bud/mother protein ratio in aged wild type (black bars) and sir2Δ (gray bars). The ratio was determined by comparing the specific abundance of daughter and mother proteins as seen by Coomassie staining. In both, B and C, the values shown are the average of at least four different 2D gels and two independent sorting events. Budding wild-type (D,F) and sir2Δ (E,G) cells subjected to FRAP analysis. Signal recovery of Eno2p-GFP (D,E) and Tdh3p-GFP (F,G), expressed as the percentage of the signal prior to bleaching, gives a measure of the rate of de novo protein synthesis between mothers (filled squares) and their daughters (empty squares). (H) Controls were carried out by blocking protein synthesis with cyclohexamide, prior to bleaching. Representative curves are shown in each case.
Since an apparent asymmetry of damaged proteins could be established by a general retention of all proteins paralleled by a higher rate of de novo synthesis in the daughter cell, we used GFP-tagged proteins and fluorescence recovery after photobleaching (FRAP) to determine the rate of synthesis of specific proteins in the mother and daughter cells. Analysis of the rate of signal recovery of Eno2p-GFP (Fig. 2D,E) and Tdh3p-GFP (Fig. 2F,G), two main targets of carbonylation, demonstrated that the rate of specific protein synthesis was not higher in the daughter cells and, furthermore, was not affected by the absence of Sir2p (Fig. 2D–G). The inclusion of the protein synthesis inhibitor cyclohexamide in the FRAP analysis confirmed that the fluorescence recovery reported on de novo protein synthesis rather than diffusion (Fig. 2H). Taken together, the results suggest that the asymmetrical distribution of carbonylated proteins during cytokinesis is governed by a Sir2p-dependent retention mechanism that specifically affects the damaged subpopulation of the proteins while undamaged proteins may diffuse freely between mother and daughter cell compartments. This notion is consistent with previous results on the free diffusion of a cytoplasmic GFP from mother to daughter (Dobbelaere and Barral 2004).
Carbonylated proteins are associated with Hsp104-containing aggregates, which are asymmetrically distributed during cytokinesis
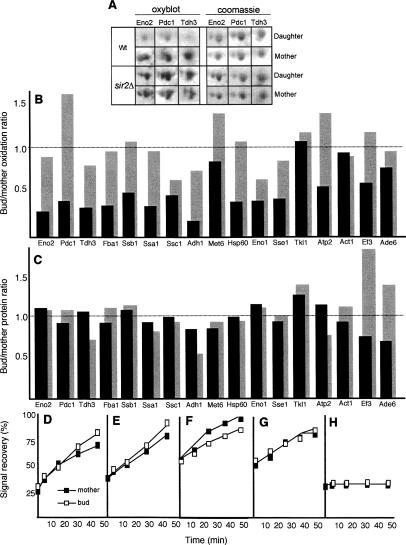
In view of the facts that the carbonylated proteins do not appear to behave as freely diffusible, soluble, proteins and that the carbonyl signal becomes speckled as the cells age (Fig. 3A,B), we asked whether carbonylated proteins formed aggregates in vivo. To approach this, we used an Hsp104-GFP construct as an in situ marker for protein aggregation. Hsp104p is a stress tolerance factor with remodeling activity that acts on trapped, aggregated proteins and together with conventional chaperones—e.g., Hsp70 and Hsp40—promotes refolding of the intermediates it produces (Glover and Lindquist 1998; Weibezahn et al. 2005). Upon heat shock, Hsp104p accumulates at the periphery of electron-dense aggregates, which can thus be visualized as intense cytosolic Hsp104-GFP-containing foci (Lum et al. 2004), but remains diffuse in the absence of aggregates. We induced protein aggregation by hydrogen peroxide or tBOOH (tert-butylhydroperoxide) exposure (Fig. 3C–E) and heat (data not shown) to confirm that the Hsp104-GFP construct could be used to detect aggregation in situ. As shown in Figure 3C–E, a mild oxidative stress resulted in small Hsp104-containing aggregates, whereas larger aggregates developed upon prolonged exposure to oxidative stress. While unstressed young cells displayed no protein aggregates (Fig. 3C), these were found to accumulate during replicative aging (Fig. 3F), reminiscent of the accumulation of carbonylated proteins. Costaining of Hsp104-GFP (using antibodies against GFP) and protein carbonyls demonstrated that protein carbonyls and Hsp104p-containing aggregates to a large extent colocalized in both tBOOH- or hydrogen peroxide-stressed cells and old untreated cells (Fig. 3G,H). Costaining with DAPI revealed that Hsp104p-containing aggregates formed both in the cytoplasm and in the nucleus, but only the former appeared to coincide with protein carbonyl-rich sites (data not shown). Thus, the protein carbonylation signal detected in situ in aging yeast cells is to a large extent localized to cytosolic protein aggregates.
Figure 3.
Carbonyl-damaged proteins form aggregates associated with Hsp104p in vivo in aged yeast cells. Protein carbonylation signal in young wild-type cells (A) and in aged (10–12 generations old) mother cells (B). Bright-field pictures (top) and carbonyl immunodetection (bottom) are shown. (C–F) Hsp104-GFP distribution in young cells subjected to no stress (C), mild H2O2 stress (D), and severe H2O2 stress (E), and in unstressed 10- to 12-generations-old mother cells (F). The corresponding bright-field pictures are also shown; 95% of cells in the population displayed the hereby shown phenotype. Intense cytosolic Hsp104-GFP foci correspond to sites of electron-dense aggregates (Lum et al. 2004). Foci of carbonylated proteins and Hsp104p in H2O2 stressed cells (G) and in unstressed 12-generations-old cells (H). From left to right are shown a bright-field image, a carbonyl image, a Hsp104 foci, and a merge between carbonyls and Hsp104 foci. Bars, 5 μm.
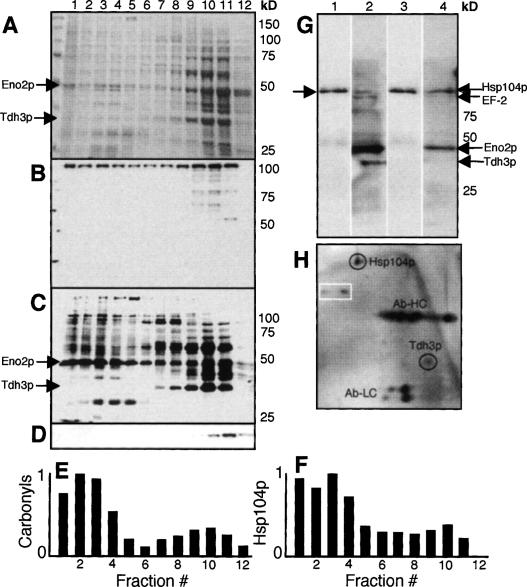
To confirm that some carbonylated proteins are associated with aggregates, we first performed gradient centrifugation analysis of protein extracts (Pappenberger et al. 2006). As can be seen in Figure 4, a large number of carbonylated proteins, including full-length Eno2p (confirmed by mass spectrometry), are found in the high-molecular-weight fractions (fractions 1–4 in Fig. 4A–F). Normalizing to total protein concentrations in each fraction demonstrates that carbonylated proteins and the Hsp104p chaperone are enriched in these high-molecular-weight fractions. Since the CCT chaperone, which is 950 kDa, comes down in fractions 6 and 7, many carbonylated proteins reside in structures >950 kDa (Fig. 4E,F). Next, we found that TapTag purification of Hsp104 also pulled down carbonylated proteins, including Eno2p, Tdh3p, and EG-2 (Fig. 4G). In addition, immunoprecipitation pull-down of Hsp104 demonstrated that Tdh3p, one of the major carbonylated proteins in the cytoplasm (Fig. 1), copurifies with Hsp104 (Fig. 4H). This, together with Hsp104p/carbonyl colocalization analysis by microscopy (Fig. 3G,H), demonstrates that carbonylated proteins are associated with Hsp104-containing aggregates.
Figure 4.
Carbonylated proteins are enriched in high-molecular aggregates and copurify with Hsp104p. Proteins were size-fractionated by sucrose gradient centrifugation, and total protein patterns (A), abundance of Hsp104p (B), carbonylated proteins (C), and actin (D) were analyzed in each fraction. Total protein was determined by Coomassie brilliant blue staining, while Hsp104p, actin, and carbonylated proteins were determined by immunodetection as described in Materials and Methods. The abundance of carbonylated proteins (E) and Hsp104p (F) in each size fraction normalized to total protein in the fractions. (G) Hsp104p (lanes 1,3) and carbonylated proteins (lanes 2,4) from oxidatively stressed (lanes 1,2) and unstressed (lanes 3,4) cells copurified by pull-down of Hsp104-TapTag as described in Materials and Methods. (H) 2D gel showing copurification of one of the major carbonylated proteins, Tdh3p, upon immunoprecipitation of Hsp104p. The identity of Tdh3p was confirmed by mass spectrometry. The heavy (Ab-HC) and light (Ab-LC) chains of the antibody are depicted on the 2D gel. The spots in the white box mark nonspecific proteins coming down also in the control (bead only).
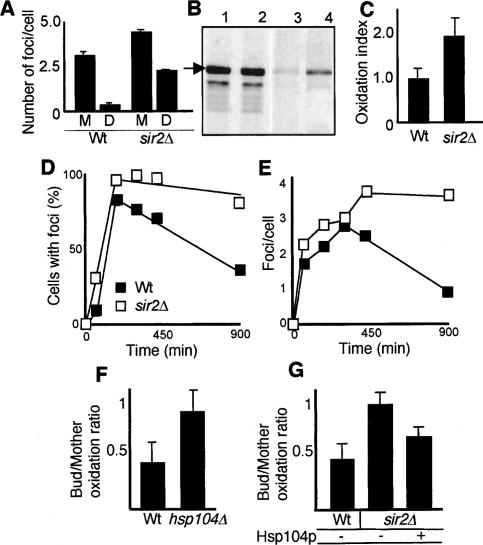
The Hsp104p-containing aggregates, like carbonylated proteins, were distributed unevenly in mother and daughter cells during cytokinesis. Old mother cells displayed on average five times more intense Hsp104-GFP cytosolic foci than their daughters (Fig. 5A). Similar to carbonylated proteins, segregation of Hsp104-GFP aggregates was less pronounced in cells lacking Sir2p (Fig. 5A). This reduction in aggregate asymmetry during cytokinesis of sir2Δ cells was independent of the overall number of detectable Hsp104-containing aggregates.
Figure 5.
Hsp104p-dependent PQC is required for damage segregation and is diminished in cells lacking Sir2p. (A) Distribution of Hsp104-GFP foci in mothers (M) and daughters (D) during cytokinesis of 10–12-generations-old wild-type (n = 55) and sir2Δ mutant (n = 74) cells. (B) Hsp104p levels (lanes 1,2) and Hsp104 carbonylation (lanes 3,4) in wild-type (lanes 1,3) and sir2Δ (lanes 2,4) cells. The arrow indicates Hsp104p. (C) Carbonyl levels normalized to total levels of Hsp104p in wild-type and sir2Δ mutant cells. (D) Fraction of cells in the population displaying Hsp104p foci before and during exposure to 1 mM H2O2 for 120 min, and after removal of the oxidant. Between 200 and 400 cells were scored at each time point and were defined as having either a diffuse cytosolic, nonaggregated distribution of Hsp104-GFP or, conversely, a discrete, aggregated one (see also Lum et al. 2004). Results are shown for both wild type (black squares) and sir2Δ (open squares). (E) The average number of Hsp104-GFP foci per cell before, during, and after oxidative stress (as in C); between 58 and 198 cells were analyzed at each time point. (F) Retention of protein damage in 12-generations-old wild-type and hsp104Δ mother cells, as determined by in situ carbonylation. (G) Effects of overproducing Hsp104p on damage asymmetry in sir2Δ cells. Shown are values for the wild type carrying the vector control (n = 44), sir2Δ carrying the vector control (n = 53), and sir2Δ overexpressing HSP104 (n = 43). Error bars correspond to the standard error (A–C) or the standard deviation of the mean (F–G).
Hsp104p overproduction retards aging and re-establishes damage retention in cells lacking Sir2p
The elevated damage to molecular chaperones in sir2Δ cells (Fig. 1A,B) may form the basis for the failure of this mutant to retain protein carbonyls in the mother cell, since this process may rely on chaperone-dependent binding and management of protein aggregates (Figs. 3, 4, 5). The Hsp104 protein was not always visible on our two-dimensional (2D) gels, and we therefore purified Hsp104p by an immunoprecipitation procedure to determine whether this chaperone also displayed elevated damage in sir2Δ cells. Indeed, sir2Δ mutant cells suffered from an almost twofold higher carbonylation load on their Hsp104 chaperone (Fig. 5B,C), suggesting that these cells may exhibit diminished Hsp104 functions. Thus, we next analyzed the formation and disappearance of Hsp104-GFP foci, as a measure of the in vivo capacity of cells to break up protein aggregates formed upon oxidative stress (Fig. 5D,E) and a transient heat shock (data not shown). The time-course experiment demonstrated that cells lacking Sir2p were notably slower than wild type in recovering and reinitiating growth after aggregation-inducing stress. In line with this, sir2Δ cells displayed a retarded rate of aggregate breakup after relief from the stress (Fig. 5D,E). The same results were obtained when heat was used as the aggregation-inducing agent. This was evident both by measuring the fraction of cells displaying Hsp104-GFP-containing foci (Fig. 5D) and the average number of such foci per cell (Fig. 5E) in the population. In addition, the number of Hsp104-foci generated upon stress was higher in sir2Δ cells (Fig. 5D,E). Similar to unstressed cells, wild-type mother cells were markedly more efficient than sir2Δ mothers in retaining the aggregates generated during the stress (data not shown). Taken together, the data suggest that Hsp104p-dependent activities are diminished in cells lacking Sir2p.
To test if Hsp104p-dependent functions were indeed important for segregation of damaged proteins, we measured in situ carbonylation of proteins in cells lacking and overexpressing HSP104. Mother cells (10–12 generations old) deficient for Hsp104p displayed an almost complete lack of carbonyl retention during cytokinesis (Fig. 5F). Moreover, overproduction of Hsp104p in cells lacking Sir2p partially restored segregation of carbonylated proteins such that the mother cells now retained some of the damage (Fig. 5G). In contrast, Hsp104p overproduction in wild-type cells did not further affect damage segregation (data not shown), implying Hsp104p functions to be limited in sir2Δ cells but not wild-type cells.
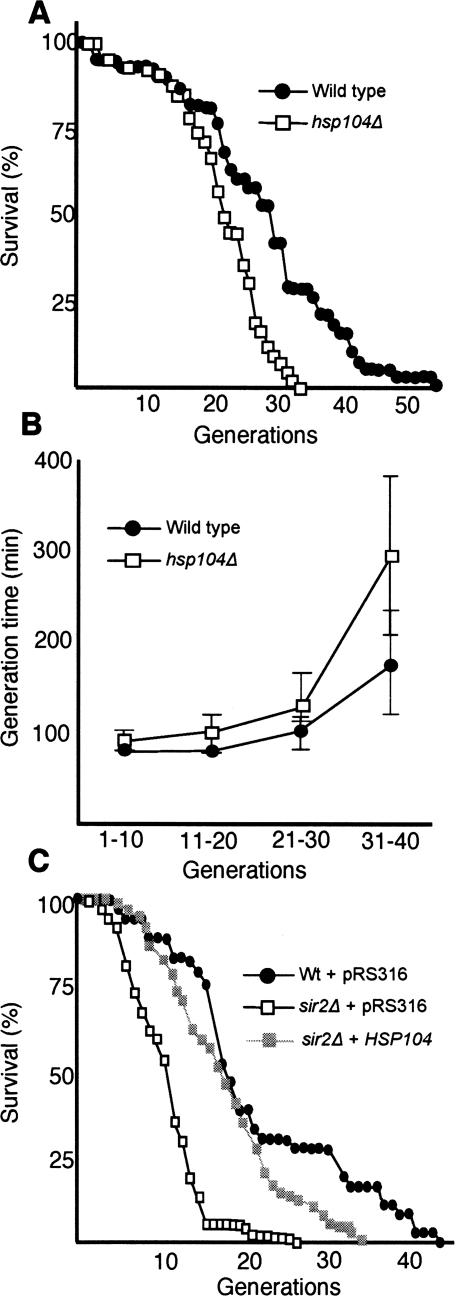
We next asked whether limited Hsp104p functions are, in fact, partly responsible for the accelerated aging of sir2Δ cells. Hsp104p is clearly required for cells to maintain a full replicative potential since hsp104Δ cells aged more rapidly than wild type (Fig. 6A). In addition, hsp104Δ cells exhibited an accelerated loss of fitness (the generation time becomes progressively longer as a function of the replicative age of the cells) (Fig. 6B), particularly so in the last third of the life span, when aggregates become more abundant. However, overproduction of Hsp104p in wild-type cells had no effect on replicative life span (data not shown), suggesting that longevity is not limited by Hsp104p-dependent segregation or remodeling of protein aggregates. In contrast, overproduction of Hsp104p in sir2Δ cells markedly increased the mean life span to almost the same value as wild-type cells carrying the vector control (Fig. 6C). The maximal life span of sir2Δ cells was also extended by Hsp104p overproduction, albeit to a more modest extent (Fig. 6C).
Figure 6.
Hsp104 is required for yeast longevity, and its overproduction suppresses the reduced life span of Sir2p-deficient cells. (A) Replicative life span of wild-type (black circles) and the hsp104Δ mutant (open squares) on YPD. The standard error for the wild type was ±0.4 (n = 78) and for the hsp104Δ mutant was ±0.1 (n = 81). (B) Generation times of wild type (black circles) and the hsp104Δ mutant (open squares) as a function of their replicative age. Error bars correspond to the standard deviation of the mean. (C) Replicative life span of control wild-type (black circles) and sir2Δ cells harboring the empty vector (open squares), and sir2Δ cells overproducing Hsp104p (gray squares). The standard error for wild type was ±0.2 (n = 72), for sir2Δ cells was ±0.7 (n = 74), and for sir2Δ cells overproducing Hsp104p was ±0.2 (n = 74). Cells were grown on selective YNB-Uracil medium. All life-span measurements were performed in duplicate or triplicate.
Damage segregation determines the life span of the progeny
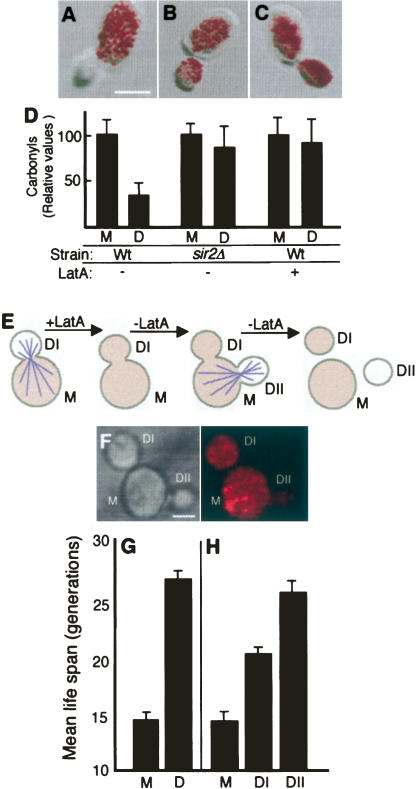
Damage segregation has been proposed to ensure the fitness of the progeny after completion of cytokinesis (Aguilaniu et al. 2003). The data presented here showing that ectopic overproduction of Hsp104p re-establishes damage asymmetry and, in parallel, extends the replicative life span of sir2Δ cells suggest that damage segregation may play a role also in determining the replicative life span of the progeny. We approached this notion by performing an experiment in which damage asymmetry was transiently abolished using Latrunculin-A (Lat-A). Lat-A, a compound that binds to G-actin preventing its polymerization into filaments and thus causing a rapid depolymerization of the actin cytoskeleton, has previously been suggested to affect the development of damage asymmetries based on the fact that daughter cells isolated after completion of cytokinesis exhibited elevated levels of carbonylated proteins upon Lat-A treatment (Aguilaniu et al. 2003). We found that Lat-A abolished damage segregation during cytokinesis, similar to a sir2Δ mutation, by in situ imaging of protein carbonyls in dividing cells (Fig. 7A–D). Thus, when Lat-A is added to cells engaged in cytokinesis, the daughter cell receives a full load of oxidatively damaged proteins (see Fig. 7C,D). This is consistent with data demonstrating that cytoskeletal structures provide a scaffold for protein aggregates (Ganusova et al. 2006).
Figure 7.
Transient depolymerization of the actin cytoskeleton eliminates damage asymmetry and limits the replicative potential of the daughter cell. Overlays of transmission images and in situ detection of carbonyls in untreated wild-type cells (A) and sir2Δ (B) and wild-type (C) cells treated with 200 μM Lat-A. (D) Quantification of carbonyls in mothers (M) and daughters (D) of cells treated as in A–C. (E) Schematic representation of the experiment in which a transient depolymerization of the actin cytoskeleton is achieved by a short-term Lat-A treatment. After removal of the drug, the mother (M) gives rise to two daughters nearly simultaneously: a first one affected by the treatment (DI), and a second one (DII) generated at a new bud site once the cytoskeleton had oriented itself again. Damaged proteins (shaded color) are distributed equally between mother and daughter DI, and this damage symmetry persists after cell separation. As the actin cytoskeleton is reoriented, damaged proteins are largely prevented from being inherited by daughter DII. (F) In situ carbonylation of cells exposed to a transient Lat-A treatment, as described above. Bar, 5 μm. (G) Mean replicative life span of 10–12-generations-old mothers and their daughters treated with DMSO only. The standard error (error bars in the figure) for mother cells (M) was ±0.4 and for daughter cells (D) was ±0.8. (D) Mean replicative life span of age-matched mothers (M) and their “twin” daughters, DI and DII, after treatment with Lat-A. Error bars correspond to the standard error of three independent measurements. The standard error for mother cells was ±1.0, for DI daughter cells was ±0.6, and for DII daughter cells was ±1.2.
If Lat-A, after causing a breakdown of damage asymmetry, is subsequently removed from the medium, a substantial number of mother cells initiate a new cell cycle and select a new bud site at which a new daughter cell is produced concomitantly with the completion and separation of the bud derived from the previous landmark. The result is two daughter cells produced almost simultaneously—one of which inherits a high load of oxidative damage (DI), whereas the other displays a normal, low degree of damage (DII) (Fig. 7E,F). We performed transient Lat-A exposure experiments on mother cells that had already produced 10 daughters, such that the progenitor cells had accumulated age-related damage and the life span of mother and daughter could be clearly distinguished; the mean replicative life span of these mother cells is 14 generations, whereas their offspring can produce, on average, 27 daughter cells (Fig. 7G). Transient exposure to Lat-A did not affect the life span of the mother cells (Fig. 7H), demonstrating that the Lat-A treatment did not cause general negative effects on longevity. However, the daughter DI produced under transiently depolarizing conditions exhibited a 20% shorter life span (Fig. 7H). In contrast, the DII daughter displayed a full replicative potential (Fig. 7H) typical of daughters produced from untreated mother cells (Fig. 7G). When young mothers (having produced zero to one daughter cell) were isolated and used as progenitor cells, treatment with Lat-A did not significantly affect the replicative potential of the progeny, and the replicative life span of mothers and daughters was indistinguishable (data not shown). This is consistent with young mothers exhibiting very low levels of damage (Aguilaniu et al. 2003), and Lat-A treatment is therefore not expected to have notable effects on the life span of the progeny.
Discussion
Aging of organisms, ranging from yeast to humans (Stadtman 1992), is accompanied by the accumulation of oxidative damage to nucleic acids and proteins. Carbonylation is one example of an oxidative modification on proteins that has attracted special attention in aging research because of its irreversible nature and association with a large variety of age-related disorders (Stadtman 1992). The detrimental effects of this modification are underscored also by the fact that its accumulation is associated with the physiological age/performance, or life expectancy, of an organism rather than with its chronological age. For example, carbonyl levels are higher in crawlers (low life expectancy) than fliers in a cohort of houseflies of the same chronological age (Yan and Sohal 2000). In the case of chronologically aging Escherichia coli cells, the cells displaying low carbonyl levels remain reproductively competent, whereas cells with a high carbonyl load become genetically dead (Desnues et al. 2003). Interestingly, the plant Arabidopsis thaliana appears to have evolved mechanisms that allow carbonylation to drop abruptly prior to the vegetative-to-reproductive transition (Johansson et al. 2004), whereas in mice, carbonylated proteins are eliminated upon early embryonic development of the blastocysts (Hernebring et al. 2006). In addition, as demonstrated here, the asymmetrically dividing yeast S. cerevisiae has evolved a Sir2p-dependent system that segregates carbonylated proteins during mitotic cytokinesis, ensuring low levels of oxidative damage in the progeny.
We demonstrate in this study that such segregation encompasses a damage-retention mechanism that affects almost all protein species found to be carbonylated in the cell. The protein carbonylation signal is dispersed throughout the cytoplasm in young cells but becomes increasingly localized in foci in aging cells (Fig. 3A,B), suggesting in vivo aggregation of carbonylated proteins. Such aggregates are poor substrates for the proteasome (Bota and Davies 2002; Grune et al. 2004), whereas carbonylated proteins that remain soluble are rapidly degraded (Davies 1987). It has been demonstrated previously that the carbonylated proteins observed by in situ immunodetection in old yeast cells were extremely stable (Aguilaniu et al. 2003), which suggests that these proteins exist in an aggregated form inaccessible to proteolytic attack.
The Hsp104p chaperone, in concert with Hsp40, Hsp70, and Hsp26, denotes a PQC system that binds to and reactivates proteins that have been denatured and allowed to aggregate (Glover and Lindquist 1998; Lum et al. 2004; Haslbeck et al. 2005; Weibezahn et al. 2005). Hsp104p is also critically involved in formation, elimination, and inheritance of yeast prions (Shorter and Lindquist 2004). We show here that Hsp104p-containing aggregates accumulate in aging cells and colocalize with sites of intense carbonylation and that many carbonylated proteins, including the highly damaged Eno2p, are enriched in high-molecular-weight fractions of >950 kDa. In addition, Hsp104 pull-down experiments demonstrated that carbonylated proteins copurify with Hsp104p (Fig. 4). Thus, carbonylated proteins may not only form aggregates in vitro but also coalesce into larger aggregates in vivo under nonstress conditions upon cellular aging. Additional proof of the correlation between oxidized proteins and Hsp104p aggregates comes from the observation that, like carbonylated proteins, Hsp104p-containing aggregates are segregated during cytokinesis by a Sir2p-dependent process (Fig. 5A). In addition, Hsp104 deficiency results in a failure to segregate carbonylated proteins in otherwise wild-type cells (Fig. 5F). Moreover, the failure of sir2Δ cells to segregate carbonylation-damaged proteins appears linked to a diminished Hsp104p activity, since overproduction of Hsp104p partially restored segregation of damage in cells lacking Sir2p but had no effect on segregation in wild-type cells. Consistently, sir2Δ cells were significantly slower than wild type in rescuing protein aggregates after exposure to oxidative agents (Fig. 5D,E) or heat. The ability to segregate aggregates and recover proteins from aggregates appears essential not only for the cells’ fitness under adverse conditions, but also throughout a cell’s life span. Indeed, a diminished function of the Hsp104p system is, in part, responsible for the accelerated aging of sir2Δ cells, as ectopic overproduction of Hsp104p extended the mean life span of sir2Δ cells to a value close to that of the wild type (Fig. 6C). However, it is not presently clear whether this extension is due to restoring damage asymmetry or a more global effect on recovering aggregation management in the sir2Δ cells.
The elevated carbonylation of chaperones, including Hsp104p and several Hsp70s, in cells lacking Sir2p raises the question of how a nuclear-localized deacetylase affects protein oxidation in the cytoplasm. It is possible that the deletion of SIR2 affects the abundance and/or activity of cytoplasmic proteins involved in oxidative defenses and/or protein management. For example, the lack of SIR2 has been reported to cause a redistribution of the cytosolic Sir2p homolog Hst2p to the nucleus (Perrod et al. 2001), and this may limit the deacetylation of possible targets in the cytosol involved in stress defense and protein homeostasis. In addition, it has been reported recently that sir2Δ cells tend to display aberrant actin cable formation and a reduced actin cable-dependent localization of actin patch proteins (Erjavec and Nyström 2007). Since reduced actin dynamics increase ROS production (Gourlay et al. 2004), this could have an impact on protein carbonylation, but it is not clear why chaperones would be specific targets for such damage.
Damage segregation has previously been suggested to affect the fitness of the offspring (Aguilaniu et al. 2003), and we show here that it also affects the replicative potential of the daughter cell. Controlled and transient depolymerization of the actin cytoskeleton during mitosis generates a high number of “twin” daughters, only one of which has been produced under conditions of failed segregation (see Fig. 7). Analysis of the replicative life span of such “twins” unequivocally showed that segregation was required for the daughter cell to attain a full replicative potential. It may be argued that the life span of the mother and daughter cells produced under failed segregation should be the same, considering that the oxidative damage (on a per-unit protein level) is the same in these cells. However, this can be explained by the size difference between the mother and daughter cells and the fact that the latter has a higher potential to dilute damage during the cell cycle leading up to the next division. It should be noted that the “twin” experiment does not imply the segregation of carbonylated proteins as the sole factor required for the daughter cell to attain a full replicative potential, since many other types of damage may be subjected to retention by the actin cytoskeleton.
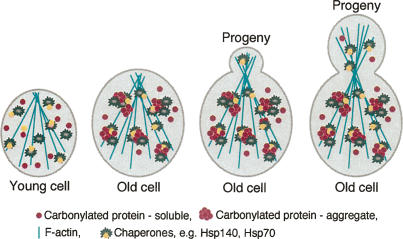
The fact that carbonylated proteins reside in Hsp104p-containing aggregates and that an intact actin cytoskeleton (Fig. 7; Aguilaniu et al. 2003) is required for the segregation of carbonylated proteins points to a plausible mechanism for damage retention. It has been shown that the actin cytoskeleton provides a scaffold for large aggregates—e.g., prion Sup35-derived aggregates—and that this resembles the mammalian aggresomes (Ganusova et al. 2006). Hsp104p is thought to mediate the interaction between such aggregates and the cytoskeletal components, thus helping the cell to localize misfolded prion proteins and prevent them from causing damage (Ganusova et al. 2006). Likewise, Hsp104p may provide a bridge between carbonylated aggregates and the cytoskeleton that not only prevents proteotoxicity but reduces the inheritance of these aggregates to the progeny. The Hsp70 chaperones are other possible candidates assisting in the retention of damage because of their fondness for hydrophobic, carbonylated proteins (Fredriksson et al. 2005). Thus, the Hsp70 chaperones may work in concert with Hsp104p on trapped, aggregated proteins tethered on the actin cytoskeleton (Fig. 8).
Figure 8.
Model for the proposed segregation of damaged aggregated proteins during yeast cytokinesis. Actin cables align themselves along the bud–mother axis during bud emergence and persist as such throughout cytokinesis. This allows for a vectorial transport of cargo between mother and bud that is mediated by adaptor proteins. In addition, it has been shown that the actin cytoskeleton provides a scaffold for large aggregates—e.g., prion Sup35-derived aggregates—and that this resembles the mammalian aggresomes (Ganusova et al. 2006). Hsp104p is implicated in the interaction between prion protein aggregates and the cytoskeletal components, thus helping the cell to localize misfolded prion proteins and prevent them from causing damage (Ganusova et al. 2006). Likewise, we suggest that Hsp104p may provide a bridge between carbonylated aggregates and the cytoskeleton that not only prevents proteotoxicity but reduces the inheritance of these aggregates to the progeny.
The exact role of Sir2 in promoting replicative longevity in yeast and somatic longevity in higher organisms (Tissenbaum and Guarente 2001; Rogina and Helfand 2004) is not known. Sir2p has been argued to promote replicative longevity in yeast by repressing the accumulation of ERCs (Sinclair and Guarente 1997). Such a mechanism cannot explain its life-span-promoting effects in higher organisms, which instead have been linked to Sir2p-dependent effects on stress management, especially oxidative stress (Hekimi and Guarente 2003). The work presented here puts the spotlight on a novel role for Sir2p in PQC of carbonylated proteins in Hsp104p-containing aggregates. A study on cultured cells reports on another histone deacetylase (HDAC6) working as an adaptor between polyubiquitinylated misfolded proteins and the dynein motors on microtubules (Kawaguchi et al. 2003). In the absence of HDAC6, or of its deacetylating activity, misfolded proteins will remain as cytoplasmic aggregates instead of being transported to aggresomes, where they can be processed. Sir2p could play a similar mediatory role in yeast, where the actin cytoskeleton, rather than microtubules, provides the scaffold on which chaperones and their cargo of damaged proteins may be trapped (Fig. 8). Possibly, segregation of damaged/aggregated proteins and accumulation of ERCs, both subjected to control by Sir2p, are two aging factors in mitotic yeast that both contribute to replicative deterioration. This dual role of Sir2p could explain why sir2 deletions have a negative effect on the yeast replicative life span (Kaeberlein et al. 1999), as well as the segregation of damaged proteins, in cells lacking Fob1p, in which formation of ERCs is blocked.
Materials and methods
Strains
All S. cerevisiae strains used in this study are derivatives of BY4741 (MATa; his3Δ1; leu2Δ; met15Δ; ura3Δ), here referred to as wild type, and have the gene of interest replaced by the KanMX4 cassette (EUROSCARF). C-terminally GFP-tagged and TAP-tagged strains were from Invitrogen (Huh et al. 2003). The mutants sir2Δ ENO2-GFP, sir2Δ TDH3-GFP, sir2Δ HSP104-GFP, and sir2Δ fob1Δ were constructed by replacing the SIR2 gene with a URA3 marker and were verified thereafter by PCR. HSP104-overexpressing wild-type and sir2Δ strains were generated by transforming the cells with a pRS316-HSP104 plasmid (Addgene). Control strains were generated by transforming with the pRS316 plasmid alone, and functionality of the HSP104 plasmid was checked by successful complementation of an hsp104Δ mutant. All strains were grown at 30°C on YPD (1% yeast extract, 2% peptone, 2% glucose), except for HSP104-overexpressing (and corresponding control) strains, which were grown on Yeast Nitrogen Base lacking Uracil.
Separation of mother and daughter cells
Old mother cells were obtained either by magnetic sorting (Smeal et al. 1996) or by centrifugal elutriation (Woldringh et al. 1995). In the first case, exponentially growing cells were harvested, labeled with 1 mg/mL Sulfo-NHS-LC Biotin (Pierce), washed, and allowed to grow overnight. They were then incubated with 2.5 × 107 Streptavidin-coated Magnetic Beads per milliliter (Pierce), and unlabeled cells were removed from the population by magnetic sorting. The cells were grown for the desired number of generations, sorted, and then allowed to go through one division. At this point, old mother cells (pellet) and their corresponding daughters (supernatant) were isolated again by magnetic sorting.
For elutriation, a J-20 XP centrifuge, equipped with a JE 5.0 rotor (Beckman-Coulter), was used. To enrich the culture for old cells, two successive rounds of elutriation were performed, allowing for overnight growth in between. Cells were subsequently resuspended in PBS and loaded in a 40-mL separation chamber at 32 mL/min and 1800–2000 rpm; old cells were elutriated at 90 mL/min and 600–800 rpm. The efficiency of every sorting was confirmed by Calcofluor White (Sigma) staining and bud scar counting (Smeal et al. 1996).
Protein and protein damage analysis
Protein extracts were prepared as described previously (Aguilaniu et al. 2001), and protein concentration was determined by the Bradford method (Pierce). Derivatization for detection of protein carbonyls was performed by mixing 1 vol of protein extract with 1 vol of 10% SDS and 2 vol of 20 mM DNPH (2,4-dinitrophenylhydrazine) in 10% trifluoroacetic acid. The sample was incubated for 20 min and then neutralized by adding 1.5 vol of 2 M tris base in 30% glycerol (Levine et al. 1994). Samples were precipitated with 10% TCA and washed with cold acetone, prior to resuspension in loading buffer containing 6 M urea and 0.7 M β-mercaptoethanol. They were then applied onto 2D PAGE consisting of capillary IEF (Bio-Rad), with a 3.5–10 pH range of ampholines (Amersham) in the first dimension and 7.5% SDS-PAGE in the second dimension (O’Farrell 1975). Gels were either fixed with Coomassie Blue in acetic acid or transferred to a PVDF membrane using a semidry blotting module (Bio-Rad) and probed with anti-DNP HRP-conjugated antibodies (Dako). The signal was revealed upon addition of ECL+ substrate (Amersham) using a Fuji chemiluminescence camera (Fuji). Quantification was done with the Image Gauge software (Fuji), and correct loading was confirmed by staining the membranes with Coomassie. Spots were cut out of the corresponding gels and identified by MALDI-MS, using a Mascot search tool (http://www.matrixscience.com). To complement for a poor 2D PAGE separation in the basic range (pH 8–10), samples were analyzed also by nonequilibrium gel electrophoresis (O’Farrell 1975); however, no additional targets could be found. Each strain was sorted independently at least three times, and each sample was run at least four times on a 2D PAGE.
Quantification of Hsp104p levels was performed by Western blot using an anti-Hsp104 rabbit polyclonal antibody (MBL) and an HRP-conjugated secondary antibody thereafter.
Biochemical analysis of protein aggregates
Sucrose gradient fractionation (Pappenberger et al. 2006) was performed by applying crude protein extracts to 10%–40% (w/v) sucrose gradients containing 15% glycerol and centrifuging them at 85,000g for 24 h at 4°C using a Beckman ultracentrifuge equipped with an SW55TI rotor. This allowed for the recovery of 12 fractions, which prior to loading on 9% SDS-PAGE gel were DNP-derivatized (Chemicon Oxyblot kit) for 20 min, after which the reaction was stopped by adding 10 μL of neutralizing solution (Chemicon Oxyblot kit), and probed with anti-GFP (Roche) and anti-DNP (Chemicon) antibodies.
Immunoprecipitation of Hsp104-GFP was carried out by mixing 0.5 mg of crude extract in breaking buffer (50 mM HEPES at pH 7.2, 90 mM KCl, 0.5% [w/v] Igepal) with 6–8 μg of anti-GFP antibody (Roche). The mixture was incubated on ice for 2–3 h, then a 40-μL slurry of 1:1 Protein G-Sepharose 4B beads (Sigma) in PBS was added and incubated on a rotary wheel for 3 h at 4°C. The beads were then washed with 3× 0.5 mL of breaking buffer followed by a 10-min incubation on a rotary wheel at 4°C with wash buffer (50 mM Tris-HCl at pH 7.5, 0.25 M NaCl, 0.1% NP-40, 0.05% [w/v] deoxycholate) in order to reduce any nonspecific binding. Twenty microliters of the recovered beads were DNP-derivatized (Chemicon Oxyblot kit) for 20 min, after which the reaction was stopped by adding 16 μL of neutralizing solution (Chemicon Oxyblot kit) and examined for the extent of Hsp104 carbonylation. Samples were loaded on 9% SDS-PAGE gels followed by blotting onto nitrocellulose membranes and probing with anti-DNP (Chemicon) antibodies using an Odyssey IR scanner (LI-COR Biosciences). Controls were performed by incubating the beads with buffer only. For 2D PAGE analysis, dried beads were then mixed with 40 μL of IEF buffer and separated by IEF with a 3–10 pH range of ampholines and loaded on 9% SDS-PAGE followed by silver staining to identify proteins coprecipitating with Hsp104p.
Tandem affinity purification was performed on Hsp104-TAP-tagged cells grown in YPD to 1.5 × 107 cells per milliliter and incubated without and with 1 mM H2O2 in YPD for 3 h. The cells were then harvested, washed twice with cold water, resuspended in IPP150-A buffer (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.1% NP-40, 1 mM Pefablock), and broken with an equal volume of glass beads. Crude protein extracts were applied onto 500 μL of IgG-Sepharose beads (Amersham) and incubated on a rotary wheel overnight at 4°C. The unbound fraction was eluted with IPP50-A buffer, and the IgG-A-bound fraction was collected upon cutting with 10 U of TEV protease (Invitrogen) for 3 h at 10°C. This was then derivatized (see above) and resolved on 11.5% SDS-PAGE. The gels were either stained with Coomassie or blotted onto nitrocellulose membranes and probed with anti-CBP (Upstate Biotechnology) or anti-DNP (Molecular Probes) antibodies, using an Odyssey IR scanner (LI-COR Biosciences). The specificity of the carbonylation pattern was compared with that of a Hst2- and Hsp26-TAP-tagged controls. Protein identifications were performed by MS–MS analysis (see above).
FRAP
FRAP (Reits and Neefjes 2001) was performed using a Radiance 2000 confocal system (Bio-Rad), equipped with a Nikon TE300 inverted microscope. Whole Eno2- and Tdh3-GFP-tagged cells were zoomed in and bleached for 1 sec, using a 488-nm Ar laser at a 100% output. Single-plane images and stacks of 0.50-μm Z-scans were recorded before and after the bleaching using the same laser at a 2% output. To diminish unintentional bleaching, frames were acquired at intervals of 5–10 min, and cells were followed for at least one generation. Control experiments were performed in which cells were incubated for 30 min with 100 μg/mL cyclohexamide prior to bleaching. Quantification was performed using NIH ImageJ software (NIH). All experiments were performed in YPD on at least 20 different cells for each strain.
In situ immunodetection and fluorescence imaging
In situ detection of carbonyls was carried out and analyzed as described previously (Aguilaniu et al. 2003), while costaining of protein aggregates was performed by introducing some modifications to the abovementioned protocol. In brief, Hsp104-GFP-tagged cells were incubated simultaneously with anti-DNP (Zymo Research) and anti-GFP (Roche) antibodies. These were then visualized with TexasRed- and Cy5-conjugated secondary antibodies (Jackson Immuno Research), respectively. Cells were imaged with a confocal microscope (see above), and three-dimensional images were reconstructed from 0.20-μm Z-stacks using ImageJ software (NIH). Approximately 60–70 cells were analyzed for each sample.
Hsp104-GFP localization in live wild-type and sir2Δ cells was done using a Leica DMRXA fluorescence microscope. Exponentially growing cells were subjected to stress with 0.7 mM tBOOH or H2O2 for 2–3 h at 30°C in YPD, washed twice, resuspended in fresh YPD, and allowed to recover thereafter. Aliquots were taken at different time points to check for absorbance at 660 nm and Hsp104-GFP localization in 200–400 cells per time point. Experiments with each strain were carried out twice, and controls were performed by subjecting the cells to heat shock for 30 min at 42°C, followed by a period of recovery (Lum et al. 2004). Costaining for nuclei was carried out by fixing the cells with cold 70% ethanol for 15 min and incubating them with 100 ng/mL DAPI for 5 min.
Life-span analysis
Cells from an exponentially growing culture were placed on a Petri plate with a micromanipulator (Singer Instruments) and allowed to bud once. Mother cells were discarded, and buds were used as starting virgin cells. Thereafter, buds were removed after every division, and the number of times each mother cell divided was recorded (Kennedy et al. 1994). Experiments were performed independently at least twice for each strain, with 72–80 virgin cells per plate.
Lat-A treatment and recovery analysis
Ten- to twelve-generations-old cells were harvested and resuspended in YPD containing 200 μM Lat-A dissolved in DMSO. Cells were incubated for 15 min at 30°C with gentle shaking and then washed twice with ice-cold water. Control samples were treated with an equivalent amount of DMSO only. At this point, cells were resuspended in 10 mL of YPD and either used for the determination of protein carbonyls in situ or for assessing their replicative life span. In the first case, the cells were incubated for 60–90 min at 30°C with gentle shaking in order to allow for recovery and then treated as described above (Aguilaniu et al. 2003).
In the second case, 10 μL of suspension were spread on a YPD plate, and 30–40 dividing cells were positioned at different coordinates with the help of a manipulator (Singer Instruments). Mother cells were allowed to complete the ongoing division, and their daughters (DI) were placed elsewhere on the same plate. The same was repeated with the daughters (DII) arising from the second division. Replicative life span of both sets of daughter cells and their mothers, plus controls, was performed as described elsewhere in this section (Kennedy et al. 1994). For every cell sample, replicative life span of mothers and daughters was done in triplicate.
Disruption and reappearance of actin were monitored upon fixation of the cells with 3.7% formaldehyde and incubation with 0.05 U/μL Fluorescein-conjugated Phalloidin (Molecular Probes), as suggested by the manufacturer.
Acknowledgments
This work was supported by grants from the Swedish Natural Research Council and an award from the Göran Gustafsson Foundation for Scientific Research in Molecular Biology. Part of the work of T.N. is supported by the European Commission (contract 518230, acronym: Proteomage).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.439307
References
- Aguilaniu H., Gustafsson L., Rigoulet M., Nyström T., Gustafsson L., Rigoulet M., Nyström T., Rigoulet M., Nyström T., Nyström T. Protein oxidation in G0 cells of Saccharomyces cerevisiae depends on the state rather than rate of respiration and is enhanced in pos9 but not yap1 mutants. J. Biol. Chem. 2001;276:35396–35404. doi: 10.1074/jbc.M101796200. [DOI] [PubMed] [Google Scholar]
- Aguilaniu H., Gustafsson L., Rigoulet M., Nyström T., Gustafsson L., Rigoulet M., Nyström T., Rigoulet M., Nyström T., Nyström T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- Bota D.A., Davies K.J., Davies K.J. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- Davies K.J. Protein damage and degradation by oxygen radicals. I. General aspects. J. Biol. Chem. 1987;262:9895–9901. [PubMed] [Google Scholar]
- Desnues B., Cuny C., Gregori G., Dukan S., Aguilaniu H., Nyström T., Cuny C., Gregori G., Dukan S., Aguilaniu H., Nyström T., Gregori G., Dukan S., Aguilaniu H., Nyström T., Dukan S., Aguilaniu H., Nyström T., Aguilaniu H., Nyström T., Nyström T. Differential oxidative damage and expression of stress defence regulons in culturable and non-culturable cells of Escherichia coli cells. EMBO Rep. 2003;4:400–405. doi: 10.1038/sj.embor.embor799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Barral Y., Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;350:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- Dukan S., Nyström T., Nyström T. Bacterial senescence: Stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes & Dev. 1998;12:3431–3441. doi: 10.1101/gad.12.21.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egilmez N.K., Jazwinski S.M., Jazwinski S.M. Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J. Bacteriol. 1989;171:37–42. doi: 10.1128/jb.171.1.37-42.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N., Nyström T., Nyström T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2007;104:10877–10881. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson Å., Ballesteros M., Dukan S., Nyström T., Ballesteros M., Dukan S., Nyström T., Dukan S., Nyström T., Nyström T. Defense against protein carbonylation by DnaK/DnaJ and proteases of the heat shock regulon. J. Bacteriol. 2005;187:4207–4213. doi: 10.1128/JB.187.12.4207-4213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusova E.E., Ozolins L.N., Bhaga S., Newman G.P., Wegrzyn R.D., Sherman M.Y., Chernoff Y.O., Ozolins L.N., Bhaga S., Newman G.P., Wegrzyn R.D., Sherman M.Y., Chernoff Y.O., Bhaga S., Newman G.P., Wegrzyn R.D., Sherman M.Y., Chernoff Y.O., Newman G.P., Wegrzyn R.D., Sherman M.Y., Chernoff Y.O., Wegrzyn R.D., Sherman M.Y., Chernoff Y.O., Sherman M.Y., Chernoff Y.O., Chernoff Y.O. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., Lindquist S., Lindquist S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Gourlay C.W., Carpp L.N., Timpson P., Winder S.J., Ayscough K.R., Carpp L.N., Timpson P., Winder S.J., Ayscough K.R., Timpson P., Winder S.J., Ayscough K.R., Winder S.J., Ayscough K.R., Ayscough K.R. A role for the actin cytoskeleton in cell death and aging in yeast. J. Cell Biol. 2004;164:803–809. doi: 10.1083/jcb.200310148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T., Jung T., Merker K., Davies K.J.A., Jung T., Merker K., Davies K.J.A., Merker K., Davies K.J.A., Davies K.J.A. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid and ‘aggresomes’ during oxidative stress, aging, and disease. Int. J. Biochem. Cell Biol. 2004;36:2519–2530. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes & Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- Haslbeck M., Miess A., Stromer T., Walter S., Buchner J., Miess A., Stromer T., Walter S., Buchner J., Stromer T., Walter S., Buchner J., Walter S., Buchner J., Buchner J. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J. Biol. Chem. 2005;280:23861–23868. doi: 10.1074/jbc.M502697200. [DOI] [PubMed] [Google Scholar]
- Hekimi S., Guarente L., Guarente L. Genetics and the specificity of the aging process. Science. 2003;299:1351–1354. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- Hernebring M., Brolen G., Aguilaniu H., Semb H., Nyström T., Brolen G., Aguilaniu H., Semb H., Nyström T., Aguilaniu H., Semb H., Nyström T., Semb H., Nyström T., Nyström T. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc. Natl. Acad. Sci. 2006;103:7700–7705. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.-K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Howson R.W., Weissman J.S., O’Shea E.K., Weissman J.S., O’Shea E.K., O’Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jazwinski S.M. Yeast replicative life span—The mitochondrial connection. FEMS Yeast Res. 2004;5:119–125. doi: 10.1016/j.femsyr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Johansson E., Olsson O., Nyström T., Olsson O., Nyström T., Nyström T. Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J. Biol. Chem. 2004;279:22204–22208. doi: 10.1074/jbc.M402652200. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McVey M., Guarente L., McVey M., Guarente L., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., Yao T.-P., Kovacs J.J., McLaurin A., Vance J.M., Ito A., Yao T.-P., McLaurin A., Vance J.M., Ito A., Yao T.-P., Vance J.M., Ito A., Yao T.-P., Ito A., Yao T.-P., Yao T.-P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Kennedy B.K., Austriaco N.R., Guarente L., Austriaco N.R., Guarente L., Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-Y., Jaruga E., Borghouts C., Jazwinski S.M., Jaruga E., Borghouts C., Jazwinski S.M., Borghouts C., Jazwinski S.M., Jazwinski S.M. A mutation in the ATP2 gene abrogates the age asymmetry between mother and daughter cells of the yeast Saccharomyces cerevisiae. Genetics. 2002;162:73–87. doi: 10.1093/genetics/162.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R.L., Williams J.A., Stadtman E.R., Shacter E., Williams J.A., Stadtman E.R., Shacter E., Stadtman E.R., Shacter E., Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- Lum R., Tkach J.M., Vierling E., Glover J.R., Tkach J.M., Vierling E., Glover J.R., Vierling E., Glover J.R., Glover J.R. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- McMurray M.A., Gottschling D.E., Gottschling D.E. An age-induced switch to a hyper-recombinatorial state. Science. 2003;301:1908–1911. doi: 10.1126/science.1087706. [DOI] [PubMed] [Google Scholar]
- O’Farrell P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Pappenberger G., McCormack E.A., Willison K.R., McCormack E.A., Willison K.R., Willison K.R. Quantitative actin folding reactions using yeast CCT purified via an internal tag in the CCT3/γ subunit. J. Mol. Biol. 2006;360:484–496. doi: 10.1016/j.jmb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Perrod S., Cockell M.C., Laroche T., Renauld H., Ducrest A.-L., Bonnard C., Gasser S.M., Cockell M.C., Laroche T., Renauld H., Ducrest A.-L., Bonnard C., Gasser S.M., Laroche T., Renauld H., Ducrest A.-L., Bonnard C., Gasser S.M., Renauld H., Ducrest A.-L., Bonnard C., Gasser S.M., Ducrest A.-L., Bonnard C., Gasser S.M., Bonnard C., Gasser S.M., Gasser S.M. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J. 2001;20:197–209. doi: 10.1093/emboj/20.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reits E.A.J., Neefjes J.J., Neefjes J.J. From fixed to FRAP: Measuring protein mobility and activity in living cells. Nat. Cell Biol. 2001;3:E145–E147. doi: 10.1038/35078615. [DOI] [PubMed] [Google Scholar]
- Requena J.R., Levine R.L., Stadtman E.R., Levine R.L., Stadtman E.R., Stadtman E.R. Recent advances in the analysis of oxidized proteins. Amino Acids. 2003;25:221–226. doi: 10.1007/s00726-003-0012-1. [DOI] [PubMed] [Google Scholar]
- Reverter-Branchat G., Cabiscol E., Tamarit J., Ros J., Cabiscol E., Tamarit J., Ros J., Tamarit J., Ros J., Ros J. Oxidative damage to specific proteins in replicative and chronological-aged Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:31983–31989. doi: 10.1074/jbc.M404849200. [DOI] [PubMed] [Google Scholar]
- Rogina B., Helfand S.L., Helfand S.L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Lindquist S., Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A. Paradigms and pitfalls of yeast longevity research. Mech. Ageing Dev. 2002;123:857–867. doi: 10.1016/s0047-6374(02)00023-4. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A., Guarente L., Guarente L. Extrachromosomal rDNA Circles—A cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smeal T., Claus J., Kennedy B., Cole F., Guarente L., Claus J., Kennedy B., Cole F., Guarente L., Kennedy B., Cole F., Guarente L., Cole F., Guarente L., Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- Stadtman E.R. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- Tissenbaum H.A., Guarente L., Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Weibezahn J., Schlieker C., Tessarz P., Mogk A., Bukau B., Schlieker C., Tessarz P., Mogk A., Bukau B., Tessarz P., Mogk A., Bukau B., Mogk A., Bukau B., Bukau B. Novel insights into the mechanism of chaperone-assisted protein disaggregation. Biol. Chem. 2005;386:739–744. doi: 10.1515/BC.2005.086. [DOI] [PubMed] [Google Scholar]
- Woldringh C.L., Fluiter K., Huls P.G., Fluiter K., Huls P.G., Huls P.G. Production of senescent cells of Saccharomyces cerevisiae by centrifugal elutriation. Yeast. 1995;11:361–369. doi: 10.1002/yea.320110409. [DOI] [PubMed] [Google Scholar]
- Yan L.J., Sohal R.S., Sohal R.S. Prevention of flight activity prolongs the life span of the housefly, Musca domestica, and attenuates the age-associated oxidative damage to specific mitochondrial proteins. Free Radic. Biol. Med. 2000;29:1143–1150. doi: 10.1016/s0891-5849(00)00423-8. [DOI] [PubMed] [Google Scholar]