Abstract
Integral β-barrel proteins (OMPs) are a major class of outer membrane proteins in Gram-negative bacteria. In Escherichia coli, these proteins are synthesized in the cytoplasm, translocated across the inner membrane via the Sec machinery, and assembled in the outer membrane through an unknown mechanism that requires the outer membrane YaeT complex and the periplasmic chaperones SurA, DegP, and Skp. Here, we have established the relationship between these three chaperones providing insight into the mechanism of OMP biogenesis using depletion analysis. Depletion of SurA alone results in a marked decrease in outer membrane density, while the loss of DegP and Skp has no effect on outer membrane composition. Furthermore, we demonstrate that SurA and YaeT interact directly in vivo. Based on these results, we suggest that SurA is the primary chaperone responsible for the periplasmic transit of the bulk mass of OMPs to the YaeT complex. The role of Skp and DegP is amplified in the absence of SurA. Evidence presented suggests that DegP/Skp function to rescue OMPs that fall off the SurA pathway. The seemingly redundant periplasmic chaperones do function in parallel, but the relative importance of the primary function of each pathway depends on whether or not cells are under stress.
Keywords: Periplasmic chaperones, outer membrane proteins, membrane biogenesis
In the Gram-negative bacterium Escherichia coli, proteins can be targeted to three different cellular compartments: the inner membrane, the periplasm, or the outer membrane. Proteins destined for the periplasm or the outer membrane contain a cleavable N-terminal signal sequence that directs them to the secretion machinery for translocation across the inner membrane (Dalbey and Chen 2004).
Three major classes of proteins are associated with the outer membrane: surface organelles such as pili, peripherally associated lipoproteins, and integral β-barrel proteins (OMPs). Protein subunits of pili, which assemble in the outer membrane, are targeted via the periplasmic chaperone/outer membrane usher pathway (Sauer et al. 2004). Lipoproteins are targeted to the outer membrane via the Lol pathway, which also includes both a dedicated periplasmic carrier protein and an outer membrane assembly site (Yokota et al. 1999). However, the role(s) that factors responsible for targeting OMPs to the bacterial outer membrane and to the outer membranes of bacteria-derived eukaryotic organelles, such as mitochondria and chloroplasts, are not as clearly defined (Paschen et al. 2003).
Two types of models have been proposed to explain how proteins are inserted into the outer membrane (Danese and Silhavy 1998). Bayer proposed that proteins traveling to the outer membrane do so through zones of adhesion between the inner and outer membranes, effectively bypassing the periplasm (Bayer 1968; Smit and Nikaido 1978). However, this model has been challenged; the appearance of these zones of adhesion may be an artifact of the fixation technique used by Bayer (Kellenberger 1990). A favored alternative model predicts that proteins pass through the periplasm prior to insertion in the outer membrane (Nikaido 1996; Danese and Silhavy 1998), interacting with periplasmic factors that assist in their folding and delivery to an outer membrane assembly site (de Cock et al. 1996; Eppens et al. 1997).
Consistent with the periplasmic intermediate model, numerous periplasmic chaperones have been identified (Duguay and Silhavy 2004). In addition, an outer membrane assembly site for OMPs has been discovered and partially characterized (Doerrler and Raetz 2005; Werner and Misra 2005; Wu et al. 2005). This assembly site contains the essential OMP YaeT (also known as Omp85 in Neisseria meningitides (Voulhoux and Tommassen 2004)), the essential lipoprotein YfiO, and three accessory lipoproteins YfgL, NlpB, and SmpA (Ruiz et al. 2005; Wu et al. 2005; Sklar et al. 2007).
The majority of periplasmic chaperones that have been identified can be classified into three distinct groups: factors that catalyze the formation and isomerization of disulfide bonds (Nakamoto and Bardwell 2004), peptidyl-prolyl cis/trans isomerases (PPIases), and those that have general chaperone activity such as Skp, DegP, and SurA (Duguay and Silhavy 2004). These latter periplasmic proteins are thought to function as chaperones in the OMP targeting pathway, delivering OMPs to the YaeT complex (Ruiz et al. 2006). The importance of these factors in the biogenesis of OMPs is illustrated by the fact that all of the genes for these proteins are regulated by the σE envelope stress response (Raivio and Silhavy 2001; Rhodius et al. 2006), which is activated in response to unfolded OMPs (Mecsas et al. 1993; Walsh et al. 2003).
DegP displays both protease and general chaperone activity and these activities are regulated in a temperature-dependent fashion (Lipinska et al. 1990; Spiess et al. 1999). The protease activity of DegP is well documented (Strauch et al. 1989). The chaperone activity was first demonstrated by Spiess et al. (1999), who discovered that DegP catalyzed the folding of the periplasmic protein MalS both in vitro and in vivo. Spiess et al. (1999) also showed that protease-deficient DegP was able to refold nonnative substrates such as citrate synthase, further demonstrating a general chaperone activity for DegP.
SurA is a member of the peptidyl-prolyl isomerase family but it also has general chaperone activity (Behrens et al. 2001). SurA was initially identified as a protein that is necessary for cell survival during stationary phase, but survival impairments are only manifested under certain conditions (Tormo et al. 1990). The physiological defects of surA mutants (mucoid colony formation, sensitivity to hydrophobic antibiotics, bile salts, and SDS) (Lazar and Kolter 1996; Rouviere and Gross 1996) suggest that the outer membrane of such mutants has been compromised. Indeed, cells that lack SurA contain reduced levels of OMPs (Rouviere and Gross 1996), and SurA was shown to participate in the folding and assembly of the outer membrane maltose transporter, LamB (Lazar and Kolter 1996; Rouviere and Gross 1996).
The general chaperone Skp has also been implicated in the folding of OMPs. Using affinity chromatography, it was demonstrated that Skp binds to denatured OMPs but not to denatured periplasmic or cytosolic proteins (Chen and Henning 1996). Additionally, it has been reported that skp-null mutants have decreased levels of LamB, OmpA, OmpC, and OmpF in their outer membrane (Chen and Henning 1996). The skp gene is located immediately downstream from yaeT (Voulhoux and Tommassen 2004), and both are regulated by the σE stress response (Rhodius et al. 2006).
Previous studies have revealed functional redundancy among periplasmic chaperones (Rizzitello et al. 2001). Synthetic lethal phenotypes were observed for null mutations in skp and surA and for null mutations in degP and surA, but not for skp and degP. Based on these observations, a model for parallel pathways for chaperone activity was proposed: Skp and DegP function in one pathway, while SurA acts in a separate, parallel pathway. The existence of parallel pathways raises numerous questions. For example, are the parallel pathways functionally equivalent? Do certain proteins prefer one pathway to the other? Do these pathways have different but partially overlapping roles in envelope biogenesis? Here we demonstrate a role for these periplasmic proteins in OMP biogenesis and clarify the relationship between Skp, SurA, and DegP.
Results
Construction of periplasmic chaperone depletion strains
The synthetic lethal phenotypes conferred by null mutations in genes for specific periplasmic chaperones suggest two separate pathways of chaperone activity (Rizzitello et al. 2001). To gain insight into the physiological significance of this genetic result, double-mutant depletion strains, skp and surA or degP and surA, were constructed with a wild-type, arabinose-inducible copy of surA on a low-copy-number plasmid vector. Unfortunately, it was difficult to determine if envelope proteins were being folded or assembled correctly upon depletion of SurA because the levels of envelope proteins were dramatically reduced (Rizzitello et al. 2001). The σE envelope stress response was strongly induced during the lengthy time period required for SurA depletion. This results in the production of sRNAs that inhibit OMP synthesis (Vogel and Papenfort 2006; Guisbert et al. 2007). Thus, it was impossible to distinguish between defects in the assembly of OMPs from an inhibition of their synthesis.
In order to be able to separate synthesis defects from targeting defects, we constructed depletion strains in which the copy number of the arabinose-inducible surA gene is reduced by inserting it into the chromosome at the λ-attachment site (Fig. 1A). Using these chromosomal depletion strains, we observed that depletion, as evidenced by decreased growth, occurs 6.5 cell generations (Fig. 1B) after subculturing into nonpermissive media, much faster than the required 10 cell generations with plasmid depletion strains. By depleting SurA much faster, we largely prevented the aforementioned OMP synthesis defects (Rizzitello et al. 2001). Using Western blot analysis, we detected substantial amounts of OMPs, such as OmpA and LamB, even after 7.5 h of growth in the absence of arabinose (Fig. 1C). Thus, we conclude that the σE stress response is not strongly induced during the course of our depletion studies.
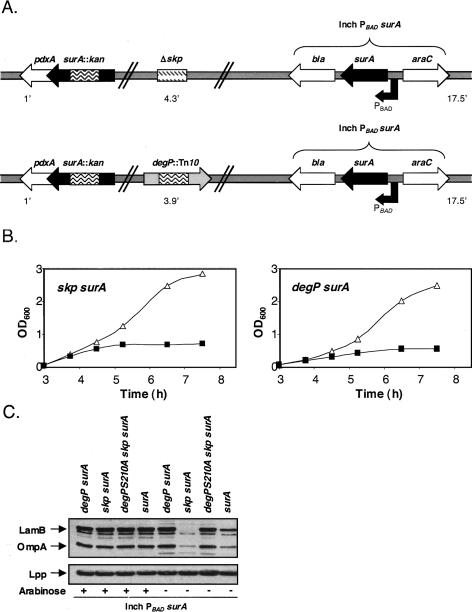
Figure 1.
(A) Genotype of chaperone depletion strains. An arabinose-inducible copy of the surA gene was introduced into the λ-att site while either the native copies of surA and skp were disrupted or the native copies of surA and degP were disrupted. The minutes in the chromosomal map where each locus is located are shown. (B) Growth of chaperone depletion strains requires arabinose. In the presence of arabinose (▵), chaperone depletion strains exhibit normal growth. In the absence of arabinose (◼), growth ceased after ∼6.5 generations. The results of a representative experiment are shown. (C) Steady-state levels of OmpA and LamB in various surA depletion strains. All depletion strains were grown in the presence (+) or absence (−) of arabinose for 6.5-h Braun’s lipoprotein (Lpp), which is assembled in the outer membrane by a process that does not require a general periplasmic chaperone or an OMP, remains unaffected and serves as a loading control.
Our ability to detect a larger amount of envelope proteins in a degP surA double-mutant depletion strain than a skp surA double-mutant depletion strain could be caused by the loss of DegP protease function in the former just as a degP surA depletion strain contains more OMPs than a surA depletion strain (Fig. 1C). In order to directly address this possibility, we introduced a protease deficient degP allele (degPS210A) into the skp surA double-mutant depletion strain. In the absence of DegP protease activity, the levels of OMPs in both depletion strains were equivalent when grown in the absence of arabinose (Fig. 1C), providing evidence that DegP is responsible for the degradation of envelope proteins in a skp surA double-mutant depletion strain.
Unfolded OmpA and LamB accumulate in skp surA and degP surA double-mutant depletion strains
Depletion of the essential members of the YaeT complex, YaeT and YfiO, in vivo results in the accumulation of misfolded OMPs (Wu et al. 2005; Malinverni et al. 2006). It follows therefore that if we disrupt the essential process of OMP targeting to the YaeT complex by disrupting redundant chaperone pathways, we may be able to detect the accumulation of misfolded OMPs. To determine if OmpA was assembled correctly after chaperone depletion, we took advantage of the fact that the folded β-barrel domain of this protein is stable in 2% SDS. As a result, folded OmpA migrates faster than predicted when run in a SDS polyacrylamide gel. When OmpA protein is heat denatured, it migrates according to its true molecular weight (Reithmeier and Bragg 1974; Freudl et al. 1986). This heat modifiable property of OmpA allowed us to directly assess its folding state during periplasmic chaperone depletion.
To preserve the native conformation of outer membrane proteins upon cell disruption, we used a previously described gentle-lysis protocol (Misra et al. 1991). Western blot analysis revealed that a significant amount of unfolded OmpA accumulated upon chaperone depletion in both the skp surA and degP surA double-mutant depletion strains (Fig. 2A,B), respectively. The larger amount of unfolded OmpA detected in the degP surA double-mutant depletion strain is due to the loss of DegP’s protease activity. Unfolded OmpA is more rapidly degraded if this protease is present.
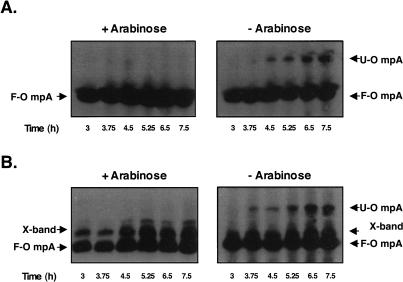
Figure 2.
Unfolded OmpA accumulates in chaperone depletion strains when grown in the absence of arabinose. Gentle lysis Western blots revealed that unfolded OmpA (U-OmpA) accumulates in a surA skp (A) and a surA degP double mutant (B) after growth in nonpermissive media. These samples were obtained from the growth curves shown in Figure 1B. Unfolded OmpA accumulation is observed ∼4 h after growth in the absence of arabinose. Folded OmpA (F-OmpA) and a nonspecific cross-reacting band (X-band) are also shown.
To assess whether accumulation of unfolded OmpA was specific to this OMP, we investigated the folding status of LamB upon chaperone depletion. LamB exists in the outer membrane as a homotrimer of β-barrels, where it serves as the transporter of maltodextrins and as the receptor for bacteriophage λ (Schirmer et al. 1995). Trimeric LamB is stable in 2% SDS and, here again, heat modifiability provides a means to monitor LamB folding upon chaperone depletion.
The expression of LamB is regulated by the presence of the inducer maltose (Boos and Shuman 1998). However, the use of maltose as an inducer interferes with the expression of the ectopic surA gene, which is under control of the arabinose promoter. To avoid this complication we used uninduced cells, as the basal levels of lamB expression were sufficient to detect and monitor this protein in these chaperone depletion strains. Following gentle lysis, we performed Western blot analysis of SDS gels using antibodies raised against LamB trimer and unfolded LamB monomer. In the presence of arabinose, LamB was targeted correctly, efficiently forming trimers in the outer membrane (Fig. 3A). In contrast, when surA was depleted in a degP mutant, unfolded LamB monomer accumulated (Fig. 3B) and failed to form trimers in the outer membrane. LamB trimerization was also prevented in the skp surA double-mutant depletion strain; however, LamB monomers did not accumulate (data not shown) probably because DegP degrades unfolded LamB.
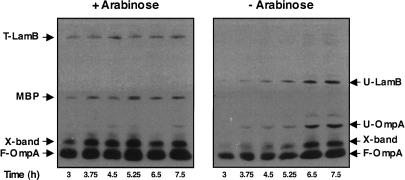
Figure 3.
LamB is unable to fold and form trimers in a degP surA double-mutant depletion strain. Gentle Lysis Western blots revealed that LamB is unable to assemble correctly into trimers (T-LamB) when a degP surA double-mutant depletion strain is grown under nonpermissive conditions. In addition, chaperone depletion leads to the accumulation of unfolded LamB (U-LamB) and unfolded OmpA (U-OmpA). MBP, folded LamB (F-LamB), folded OmpA (F-OmpA), and a nonspecific cross-reacting band (X-band) are also shown.
Results presented in this section demonstrate that periplasmic chaperones are required for the proper folding and targeting of the outer membrane β-barrel proteins OmpA and LamB. In addition, they confirm that SurA and DegP/Skp can perform redundant chaperone function.
Depletion of redundant chaperones reduces the density of the outer membrane
In the previous section we examined the assembly of two OMPs, OmpA and LamB, during redundant chaperone depletion. However, the synthetic phenotypes exhibited by skp surA and degP surA double-mutant depletion strains suggests that the two parallel chaperone pathways are responsible for the delivery of the majority of OMPs to the outer membrane (Rizzitello et al. 2001). Following this logic, rather than laboriously looking at the assembly of all of the individual OMPs, we decided to monitor the delivery of the bulk mass of OMPs during redundant chaperone depletion. Specifically, the amount of OMPs delivered to the outer membrane should decrease. As a consequence of this, the protein to lipid ratio in the outer membrane should decrease resulting in a decrease in outer membrane density.
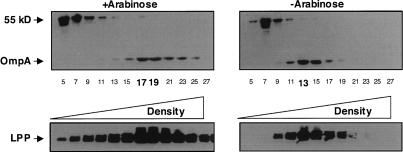
When the degP surA double-mutant depletion strain was grown in the presence of arabinose, the densities of the inner membrane and outer membrane were 1.131 g mL−1 and 1.225 g mL−1 respectively, which is comparable with the density of a wild-type strain. In contrast, when the degP surA double-mutant depletion strain was grown in the absence of arabinose, the densities of the inner membrane and outer membrane were 1.135 g mL−1 and 1.186 g mL−1, respectively (Fig. 4). The decrease in outer membrane density is a consequence of the inability of OMPs to be properly targeted and inserted into the outer membrane during periplasmic chaperone depletion. It cannot be due to decreased synthesis because the steady-state levels of OMPs are the same when this strain is grown for an equivalent amount of time in the presence or absence of arabinose (Fig. 1C).
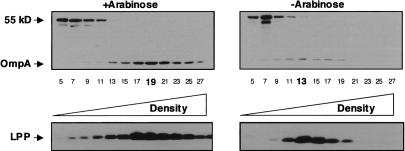
Figure 4.
Depletion of redundant periplasmic chaperones results in a decrease in outer membrane density. Inner and outer membranes were isolated from a degP surA double-mutant depletion strain. Cells were grown in the presence or absence of arabinose 6.5 h after the initial subculture and membranes were separated using sucrose density centrifugation. Odd-numbered fractions were analyzed by Western blot using an antibody that recognizes OmpA and an unknown inner membrane protein (55 kDa). Fraction density increases from left to right. The fractions corresponding to peak outer membrane densities for each condition are highlighted in bold. Lpp serves as an additional marker for the outer membrane.
LamB prefers the SurA assembly pathway to the Skp/DegP assembly pathway
In surA mutants unfolded LamB monomers accumulate (Rouviere and Gross 1996). This could indicate that LamB prefers the SurA targeting pathway to the Skp/DegP targeting pathway for its assembly. Alternatively, since these experiments were done under conditions where LamB synthesis was strongly induced, it may be that high level expression of OMPs requires the full participation of both chaperone pathways. To distinguish these possibilities we needed to establish if both chaperone pathways were equally proficient for LamB assembly. In particular, we wished to determine if LamB could be assembled efficiently in strains lacking Skp and DegP, but not SurA under conditions where LamB synthesis was strongly induced.
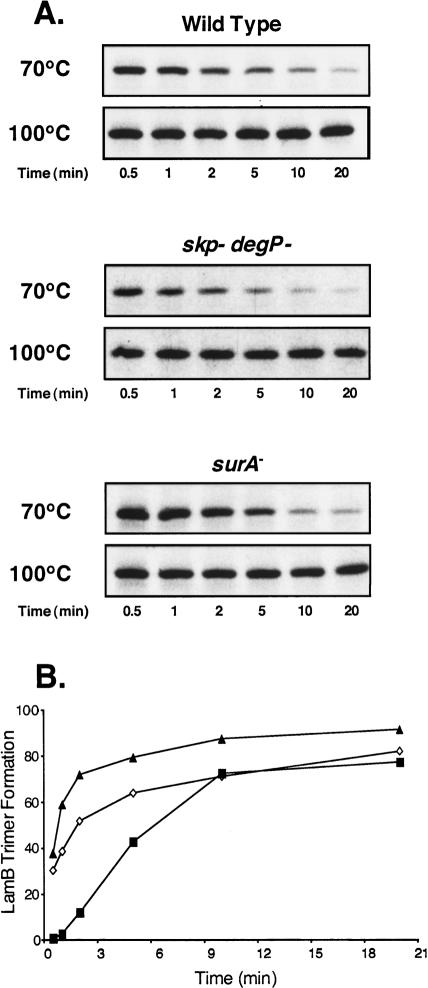
We monitored the kinetics of LamB assembly using pulse chase analysis, followed by immunoprecipitation, selective sample heating, and then SDS gel electrophoresis. Sample boiling, which denatures all proteins, shows that the amount of labeled LamB present in the wild-type, skp degP double mutant, and surA mutant strains was equivalent (Fig. 5A). However, by heating samples to 70°C, we could follow the disappearance of LamB monomers as they are assembled into stable trimers. Figure 5A shows the expected LamB assembly defect in SurA mutants. This assembly defect is not observed in a degP skp double mutant (Fig. 5A,B). In fact, LamB forms trimers faster in this strain, possibly as a result of the presence of increased periplasmic folding factors regulated by the σE envelope stress response (see below). We conclude that while LamB assembly can proceed by both the SurA and the DegP/Skp chaperone pathways, the SurA pathway is more efficient.
Figure 5.
Assembly kinetics of LamB in periplasmic chaperone mutants. Wild-type cells, skp degP mutants, and surA mutants were pulse-labeled with [35S]methionine for 30 sec and then chased with cold methionine for the times indicated (minutes). Gentle lysis was performed and samples from each time point were heated to 70°C or 100°C. Following this incubation, LamB was immunoprecipitated using antibody that recognizes the unfolded monomer form of LamB. (B) LamB trimer formation over time was calculated by plotting the inverse of the ratio of unfolded LamB/total amounts of LamB over time for wild-type (◇), surA (◼), and skp degP (▴). Levels of LamB shown in A were quantified using NIH ImageJ software.
Depletion of SurA reduces the density of the outer membrane
Our inability to detect defects in the kinetics of LamB assembly in degP skp double mutants led us to conclude that SurA is responsible for the assembly of LamB in a wild-type cell. Perhaps SurA is responsible for the assembly and targeting of most OMPs. In order to test this possibility, we looked at the density of the outer membrane when SurA is depleted in an otherwise wild-type cell.
The densities of the inner membrane and outer membrane were 1.134 g mL−1 and 1.219 g mL−1, respectively, when the surA depletion strain was grown in the presence of arabinose. However, when SurA was depleted from cells, the densities of the inner membrane and outer membrane were 1.131 g mL−1 and 1.186 g mL−1, respectively (Fig. 6). In contrast, the densities of the inner membrane and outer membrane were 1.134 g mL−1 and 1.219 g mL−1, respectively, in degP skp double mutants, implying that most OMPs are properly localized to the outer membrane when SurA is present.
Figure 6.
Depletion of SurA results in a decrease in outer membrane density. Inner and outer membranes were isolated from a surA depletion strain. Cells were grown in the presence or absence of arabinose 6.5 h after the initial subculture and separated using sucrose density centrifugation. Odd-numbered fractions were analyzed by Western blot using an antibody that recognizes OmpA and an unknown inner membrane protein (55 kDa). Fraction density increases from left to right. The fractions corresponding to peak outer membrane densities for each condition are in bold.
These experiments demonstrate that the use of a surA depletion strain instead of a surA mutant is more informative. We know that there is an OMP synthesis defect in surA mutants (Rouviere and Gross 1996). In contrast, the lower steady-state levels of OMPs in a surA depletion strain result from the degradation of misfolded OMPs by DegP and not from a defect in OMP synthesis (Fig. 1C). Therefore, the observed decrease in outer membrane density during SurA depletion reflects the inability to properly target OMPs to the outer membrane.
Decreases in outer membrane density during SurA depletion are similar to those observed during depletion of YaeT (Doerrler and Raetz 2005; Wu et al. 2005) or the essential lipoprotein member of the YaeT complex, YfiO (Malinverni et al. 2006). We considered the possibility that the shift in outer membrane density that occurs upon SurA depletion was actually a secondary effect caused by lower levels of YaeT or YfiO. In order to test this possibility, we monitored the steady-state levels of YaeT and YfiO in a surA depletion strain in the presence or absence of arabinose. The levels of YfiO and YaeT were equivalent in our surA depletion strain after 6.5 h of growth in the presence or absence of arabinose (data not shown). Therefore, the decrease in outer membrane density was likely a direct consequence of the loss of SurA.
Because we are able to observe decreases in OM density when we deplete only surA, we suggest that SurA is the principal chaperone required for the delivery of most OMPs to the outer membrane.
SurA can be cross-linked to the YaeT complex in vivo
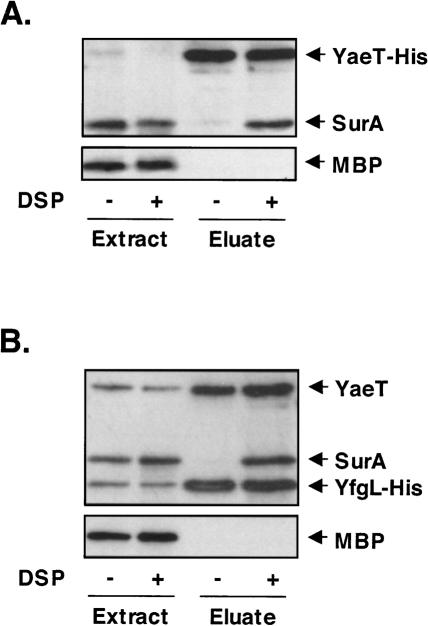
Despite repeated attempts, we have been unable to copurify any of the periplasmic chaperones with the YaeT complex. Apparently if there is an interaction between these periplasmic chaperones and the YaeT complex it is weak or transient, and does not withstand the various purification protocols used. Thus, we employed an in vivo cross-linking strategy, hoping, to covalently attach the chaperone proteins to a component of the YaeT complex, thereby allowing the identification of interacting chaperones following copurification using affinity chromatography. As shown in Figure 7A, when the cross-linker Di-thiobis(succinimidyl propionate) (DSP) is added to cells that produce His-tagged YaeT, SurA copurified with YaeT-His. The absence of another periplasmic protein, maltose-binding protein (MBP), in the same sample suggests that SurA is specifically cross-linked to YaeT. As expected, when DSP is omitted, neither MBP nor SurA was present, even though YaeT is still quantitatively isolated using a Ni column.
Figure 7.
SurA cross-links to the YaeT complex in vivo. Immunoprecipitation experiments were performed in the presence or absence of the cross-linker DSP. Whole-cell extracts and Ni-NTA column-enriched eluate fractions of wt/pYaeT-His (A) and yfgL−/pYfgL-His (B) were subjected to Western blot analysis using antibodies that recognize MBP, SurA, YaeT, and monoclonal anti-His antibody.
As YaeT itself is an OMP, it is possible that we cross-linked SurA to YaeT while it was in transit through the periplasm. In order to demonstrate that SurA can be cross-linked to the YaeT complex in the outer membrane, we repeated the experiment using a His-tagged lipoprotein member of the complex, YfgL. As shown in Figure 7B, His-tagged YfgL was able to copurify with SurA and YaeT in the presence of DSP, providing evidence that SurA is cross-linked to a functional YaeT complex in the outer membrane.
We were unable to detect Skp or DegP in any of our cross-linking experiments, even when these experiments were performed in a surA mutant (data not shown). Although we were unable to detect an interaction between these chaperones and the YaeT complex, an interaction between these proteins may still exist. The interactions between these chaperones and the YaeT complex may be quite weak or indirect, consistent with their role as a secondary OMP folding pathway. It is also possible that none of the lysine residues in Skp or DegP are positioned in a way to allow cross-linking to the YaeT complex. Regardless, a stronger interaction between SurA and the YaeT complex than with other periplasmic chaperones is consistent with our proposal that SurA is the primary chaperone responsible for the delivery of the bulk mass of OMPs to the outer membrane.
A role for Skp/DegP in OMP assembly
The loss of Skp/DegP does not result in a decrease in outer membrane density, nor does it affect the kinetics of assembly of LamB. In order to assess the possible function of Skp/DegP in OMP assembly, we monitored the activation of the σE stress response in skp degP mutants. Even subtle defects in OMP assembly result in σE activation (Sklar et al. 2007). The rpoHP3 promoter is under direct control of σE (Erickson and Gross 1989; Danese and Silhavy 1997). The σE stress response is induced in this double mutant, as we observed a threefold increase in expression from an rpoHP3′-lacZ+ fusion. Indeed, we think it is likely that this elevated σE response explains the more rapid assembly of LamB trimer in skp/degP mutants than in wild-type cells (Fig. 5). We conclude that in the absence of Skp/DegP, misfolded OMPs accumulate to sufficient levels to activate this stress response. Thus, Skp/DegP must participate in some aspect of OMP assembly.
Localization of unfolded OMPs during periplasmic chaperone depletion
By definition, when OMPs are properly targeted to and assembled in the outer membrane, they are properly folded. This raises the question of whether folding, targeting, and assembly are coupled reactions. To address this question and determine which processes require periplasmic chaperones, we sought to determine the cellular location of the misfolded OmpA and LamB species that accumulate upon chaperone depletion.
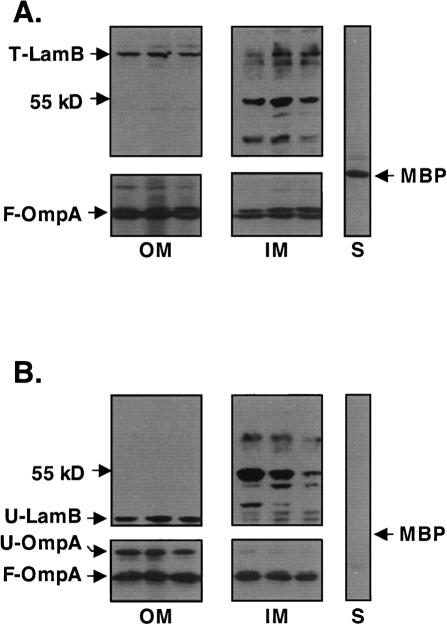
The degP surA double-mutant depletion strain was grown in the presence or absence of the inducer arabinose. In the presence of arabinose, when SurA is present, both trimeric LamB and folded OmpA properly fractionated with the outer membrane as expected (Fig. 8A). Under chaperone depletion conditions, unfolded OmpA and unfolded LamB could also be detected in outer membrane fractions (Fig. 8B). However, we had to overexpose the blots to detect these molecules. Even with overexposure, we could not detect any LamB or OmpA in the soluble fractions. We believe that most of the unfolded OMPs that accumulate during periplasmic chaperone depletion are aggregated and are lost during the fractionation procedure. Controls including the periplasmic MBP and an unknown 55-kDa inner membrane protein (Braun and Silhavy 2002) demonstrated the reliability of the fractionation protocol with chaperone-depleted cells.
Figure 8.
Unfolded LamB and OmpA associate with the outer membrane in a degP surA double-mutant depletion strain. Bacterial cells grow in the presence (A) or absence (B) of arabinose, were fractionated into soluble cytoplasm and periplasm (S) and insoluble inner membranes (IM) and outer membranes (OM). Western blot analysis revealed that the unfolded OmpA that accumulates during nonpermissive growth fractionates with the outer membrane. Western blots were overexposed to visualize unfolded OmpA during fractionation. An unknown 55-kDa inner membrane protein is shown as a control.
Folding of periplasmic proteins is unaffected during chaperone depletion
In addition to the OMPs OmpA and LamB, we monitored the steady-state levels of the periplasmic MBP in the degP surA and skp surA mutant strains. Although substantial levels of MBP were present when the cells were grown in the presence of arabinose, we were unable to detect MBP in cells grown under the nonpermissive condition (Fig. 3). We ruled out the possibility that under the chaperone depletion conditions the damaged cells were leaking MBP into the extracellular environment, as we could not detect any MBP in the culture media. This lack of MBP could be a consequence of the synthesis defect for envelope proteins during depletion of periplasmic chaperones that we previously observed (Rizzitello et al. 2001), or this could result from the degradation of misfolded MBP. In order to test this hypothesis, we expressed MBP using the Ptac promoter and the native malE Shine-Dalgarno sequence. Expression of MBP from a nonnative promoter during periplasmic chaperone depletion restored steady-state levels of MBP (data not shown). Based on these results, we conclude that lack of MBP during periplasmic chaperone depletion results from a down-regulation of transcription form the native malE promoter and not from an inability of MBP to fold properly. Apparently, this promoter is more sensitive to this stress-induced down-regulation than the malK-lamB promoter. Unidentified periplasmic chaperones likely assist MBP folding.
Discussion
Numerous periplasmic proteins have been implicated in OMP maturation (Duguay and Silhavy 2004; Mogensen and Otzen 2005). Previous work demonstrating functionally redundant periplasmic chaperones proposed a model suggesting parallel pathways for chaperone activity: Skp and DegP function in one pathway, while SurA acts in a separate, parallel pathway (Rizzitello et al. 2001). What is the physiological basis for redundant pathways of chaperone activity in the periplasm? Based on the data presented above, we suggest that SurA is the primary periplasmic chaperone responsible for the assembly of most OMPs in the outer membrane. In contrast, we suggest that the role of Skp/DegP is to rescue OMPs that have fallen off the normal assembly pathway. Therefore, under normal growth conditions Skp/DegP play a minor role since the fraction of mistargeted OMPs is small. Under stress conditions, or in the absence of SurA, the role of Skp/DegP is greatly amplified.
SurA is the primary periplasmic chaperone
When SurA is depleted from cells the levels of both LamB and OmpA are reduced. Furthermore, the density of the OM decreases dramatically when SurA is depleted from cells, which is a direct consequence of the inability of the bulk mass of OMPs to be properly targeted to the outer membrane. Indeed, the changes in OM density we observe upon depletion of SurA are very similar to those observed upon depletion of the essential components of the YaeT complex (Doerrler and Raetz 2005; Werner and Misra 2005; Wu et al. 2005). In contrast, loss of both DegP and Skp neither result in a decrease in outer membrane density nor slower assembly of LamB.
If SurA serves as the primary periplasmic chaperone for OMPs, it must be involved in both the early and late steps of OMP biogenesis. Based on a detailed kinetic analysis of LamB assembly in various mutant backgrounds, Ureta et al. (2007) have suggested that SurA interacts with LamB during, or very shortly after, translocation, but prior to signal sequence cleavage. Here, we have shown that SurA interacts directly with the YaeT complex in vivo (Fig. 7). Taken together, these data are consistent with the idea that SurA is escorting OMP assembly intermediates from one side of the periplasm to the other.
Previous work performed with surA-null mutants had indicated that SurA plays an important role in the folding and assembly of OMPs (Lazar and Kolter 1996; Rouviere and Gross 1996). However, delineating the role of SurA and distinguishing its function from other periplasmic chaperones using surA-null mutations is complicated by the strong induction of the σE stress response that occurs in these mutants (Rouviere and Gross 1996). This stress response not only increases the levels of DegP/Skp, it also decreases the synthesis of OMPs (Erickson and Gross 1989; Vogel and Papenfort 2006; Guisbert et al. 2007), and together these effects minimize assembly defects. Using a SurA depletion strain allows us to more clearly observe the folding and assembly of OMPs in the absence of this periplasmic chaperone before full induction of the σE stress response occurs.
We know that simultaneous disruption of the SurA and DegP/Skp pathways leads to dramatic changes in outer membrane composition and the accumulation of misfolded OMPs. What then is the true function of the DegP/Skp pathway?
DegP/Skp function to rescue off-pathway OMP assembly intermediates
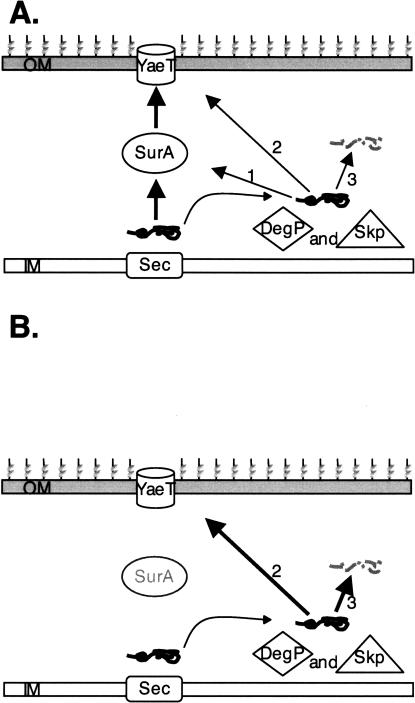
We suggest that the primary role of the DegP/Skp is to rescue OMPs that have fallen off the assembly pathway (Fig. 9). As long as SurA is active, the fraction of off-pathway intermediates remains small. However, in the absence of SurA off-pathway intermediates would accumulate (Ureta et al. 2007), amplifying the role of the DegP/Skp pathway. The σE stress response monitors the assembly of OMPs, and even in wild-type unstressed cells, this stress response is partially activated. We know this because ompR mutations, which prevent the expression of the genes for two major OMPs, OmpF, and OmpC, decrease σE activity (Mecsas et al. 1993). The most likely explanation for this partial activation is that under normal conditions some fraction of OMPs falls off the assembly pathway. The ompR mutation reduces the overall load on the assembly machinery and this, in turn, decreases the levels of misassembled OMPs. We propose that DegP/Skp can rescue OMPs that fall off the assembly pathway during biogenesis. DegP/Skp can perform three functions when they encounter these off-pathway intermediates; redirect these substrates to the SurA pathway, deliver these intermediates to the YaeT complex directly, or degrade them. Although we have no evidence that Skp/DegP can deliver substrates to SurA, we know that DegP/Skp can interact directly or indirectly with the YaeT complex, because they are required for OMP assembly in the absence of SurA. We also know that DegP largely degrades mistargeted OMPs.
Figure 9.
A model for the periplasmic chaperone-mediated biogenesis of OMPs in E. coli. OMPs are translocated across the inner membrane (IM) via the Sec YEG complex and are released into the periplasm. (A) In wild-type cells, most OMPs are escorted across the periplasm by SurA, while off-pathway OMPs are rescued by DegP/Skp. The DegP/Skp pathway can redeliver OMPs back to the SurA pathway (1) or deliver OMPs directly to the YaeT complex in the outer membrane (OM) independent of SurA (2), or DegP’s protease activity can degrade these off-pathway OMPs. (B) In the absence of SurA, the role of DegP/Skp is enhanced due to the increased accumulation of off-pathway OMP intermediates.
We have considered the alternative possibility that certain OMPs prefer the SurA pathway for their assembly, while others prefer the DegP/Skp pathway. Although we do not favor this model, we cannot dismiss it. The bulk mass of OMPs constitutes only a few different proteins (Nikaido 2003) and all of them are affected in surA mutants (Lazar and Kolter 1996; Rouviere and Gross 1996). Blocking major OMP assembly would have dramatic effects on OM density even if minor OMP assembly was unaffected. Therefore, it remains possible that one or more minor OMPs prefer the DegP/Skp pathway.
A third possibility is that the SurA pathway is responsible for the assembly of OMPs, while the Skp/DegP is primarily involved in the folding of periplasmic proteins. In the absence of SurA, Skp/DegP can assemble OMPs, albeit not as efficiently as SurA. SurA has not been implicated in the folding of periplasmic proteins. Indeed, SurA has been shown to preferentially recognize in vitro synthesized OMPs over other equally sized proteins (Behrens et al. 2001). In contrast, Skp has been reported to facilitate the folding of certain heterologous proteins produced by recombinant methods (Bothmann and Pluckthun 2000), and DegP has been shown to catalyze the folding of the periplasmic protein MalS (Spiess et al. 1999). However, it seems unlikely that the Skp and DegP are the primary chaperones responsible for the folding of periplasmic proteins because degP skp double mutants exhibit no overt protein folding defects. Indeed, MBP folding appears to occur normally in cells lacking both SurA and Skp or SurA and DegP. The identity of the periplasmic chaperones that facilitate the folding of periplasmic proteins, especially those that lack disulfide bonds (Nakamoto and Bardwell 2004), remains an important open question.
What happens when off-pathway OMP assembly intermediates accumulate?
In surA mutants a significant fraction of LamB falls off the assembly pathway, and the mistargeted molecules likely remain associated with the inner membrane because the signal sequence remains attached (Ureta et al. 2007). Similar LamB off-pathway intermediates likely occur in spheroplasts, which are effectively depleted of most periplasmic proteins including SurA. Previously Skp was proposed to contribute to the early steps of OMP biogenesis based on its ability to associate with the inner membrane (de Cock et al. 1996) and its ability to be cross-linked to various OMPs as they emerge from the secretion machinery in spheroplasts (Schafer et al. 1999; Harms et al. 2001). We suggest instead that these experiments reveal the true function of Skp, namely to rescue OMP assembly intermediates that have fallen off pathway. The seemingly redundant periplasmic chaperones do function in parallel but the relative importance of the primary function of each pathway depends on whether or not cells are under stress.
Materials and methods
Strains and microbiological techniques
All strains were constructed using standard microbiological techniques. The bacterial strains used in this study are derivatives of E. coli K-12 strain MC4100. The chaperone depletion strains were constructed using the following methodology: pAER1, a plasmid that contains a copy of the surA gene under control of the arabinose promoter (Rizzitello et al. 2001), was integrated to the λ-att site with λ-InCh1 as described previously (Boyd et al. 2000), creating strain MB292. Strains JGS189 (MC4100 arar degP∷Tn10) and JGS190 (MC4100 arar Δskp) were transduced with P1 lysate raised from strain MB292 creating strains JGS195 [JGS189 Δ(λatt-lom)∷bla PBAD surA araC] and JGS196 [JGS190Δ(λatt-lom)∷bla PBAD surA araC], respectively. In the final step of depletion strain construction, surA∷kan was introduced via P1 transduction into JGS195 and JGS196 in the presence of arabinose, creating strains JGS199 and JGS200. Both depletion strains (JGS199 and JGS200) required arabinose for normal growth.
λ-red-mediated recombination
Homologous recombination using single-stranded oligionucleotides (Costantino and Court 2003) was used to create the DegPS210A allele.
The recdegPS210A oligonucleotide (GTTCGCCGTTCAGGTT AACCAGCGCACCGCCGGCGTTACCACGGTTGATCGCTG CATCGGTCTGGATGAA) was used to change Ser210 to Ala and a silent mutation at Gly211, creating a unique NgoMIV restriction site. After the expression of the Red genes of HME6mutS was induced, 5 pmol of the recdegPS210A oligonucleotide was used for electroporation. Electroporated cells were diluted and grown on Luria-Bertani (LB) agar at 30°C. Individual colonies were screened for mixed recombinants by partial gain of an NgoMIV restriction site as determined by NgoMIV (New England Biolabs) digestion of PCR products that amplify degP. Mixed recombinant colonies were purified to allow segregation, and then individual colonies were screened for gain of the NgoMIV site and partial temperature sensitivity at 42°C.
Media and growth conditions
LB and M63 media were prepared as described previously (Silhavy et al. 1984). When necessary, media was supplemented with 20 μg/mL chloramphenicol, 25 μg/mL kanamycin, or 25 μg/mL tetracycline. All bacterial cultures were grown under aerobic conditions at 37°C unless otherwise noted.
Western blot analysis
Overnight cultures of strain JGS199 and JGS200 were grown in the presence 0.2% L-arabinose (wt/vol). Overnight cultures were washed twice with fresh media before subculturing into media lacking or containing L-arabinose. JGS199 was subcultured 1:2000 into fresh media while JGS200 was subcultured 1:10,000 into fresh media. Growth was monitored by measuring the OD600 of the cultures every 45 min after subculturing. An equal number of cells from both cultures was harvested by centrifugation and immediately frozen in a dry ice/ethanol bath. In order to preserve the native conformation of OMPs, these frozen cell pellets were gently lysed as described previously (Misra et al. 1991). Samples were resuspended in sample buffer containing 3% (wt/vol) SDS, 10% (vol/vol) glycerol, and 5% (vol/vol) β-mercaptoethanol in 70 mM Tris-HCl (pH 6.8). Samples were heated to 37°C for 10 min. SDS-PAGE analysis was performed using 10% acrylamide gels, and gels were electrophoresed at 50 V in order to ensure that any folded OMPs collected using gentle lysis would not be denatured during electrophoresis. After electrophoresis, gels were transferred to nitrocellulose membranes and probed with antibodies used to detect LamB (1/15,000), LamB trimer (1/15,000), OmpA (1/15,000), Lpp (1/15,000), and MBP (1/100,000) (Misra et al. 1991). Donkey anti-rabbit IgG horseradish peroxidase (Amersham Pharmacia Biotech) conjugate was used as the secondary antibody at a concentration of 1/8000. ECL (Amersham Pharmacia Biotech) and XAR film (Kodak) were used to visualize the protein bands. Protein bands were analyzed with gel image analysis software (ImageJ software).
Cell fractionation
The following cell fraction is based on the techniques described by Braun and Silhavy (2002) with some modifications. Depletion Strains JGS199 and JGS200 were grown as described above in the presence or absence of L-arabinose. After 6.5 h of growth in the presence or absence of arabinose, 100 mL of cell cultures were harvested by centrifugation. Cell pellets were resuspended in 3 mL of a buffer containing 10 mM Tris (pH 7.5), 1 mM EDTA, and 0.1 mM PMSF. Cell suspensions were passed through a French Pressure cell twice at 14,000 psi in order to ensure good lysis of the cells. Unbroken cells and debris were collected by centrifuging the lysate at 2000g for 10 min at 4°C. The supernatents (2.5 mL) were placed on top of a two-step sucrose gradient of 0.3 mL of 65% (wt/wt) and 1 mL of 25% (wt/wt) sucrose in 10 mM Tris (pH 7.5), and 1 mM EDTA, and were centrifuged for 2 h in a Beckman SW55 Ti rotor (4°C, 368,000g). The top 3 mL were collected and designated as the soluble contents (periplasm and cytoplasm). The bottom 0.7 mL containing the membranes was diluted with 1.4 mL of a buffer containing 10 mM Tris (pH 7.5) and 1 mM EDTA; was layered on top of a layered sucrose gradient composed of 0.5 mL of 65%, 0.5 mL of 55%, 1 mL of 50%, 2 mL of 45%, 2 mL of 40%, 2 mL of 35%, and 1.5 mL of 30% sucrose in 10 mM Tris (pH 7.5) and 1 mM EDTA (wt/wt); and was centrifuged for 17 h in a Beckman SW41 Ti rotor (4°C, 222,000g). Four-hundred-microliter samples were collected and stored at −20°C for future experiments. Odd-numbered fractions were used for SDS-PAGE electrophoresis using 12% acrylamide gels. Western Blot analysis was performed as described above using antibodies directed against the OMP Imp at a concentration of 1/7000.
Pulse labeling and immuoprecipitation
Pulse labeling was performed as described previously (Ureta et al. 2007). In order to preserve the native conformation of OMPs, the labeled cells were gently lysed as described above and immunoprecipated using antisera to LamB monomer, LamB trimer, and MBP followed by Protein A-Sepharose beads CL4B (Amersham Pharmacia). The samples were resuspended in buffer containing 40 mM Tris (pH 7.5), 20 mM EDTA, 20% glycerol (vol/vol), 6% SDS (wt/vol), and 10% β-mercaptoethanol. Resuspended samples were incubated at room temperature for 1 h, at which point the Protein A-Sepharose beads were removed by centrifugation. Samples were aliquoted into separate tubes prior to heating.
In vivo cross-linking and affinity purification
Cross-linking experiments are based on techniques described by Thanabalu et al. (1998) with modifications. Cells were grown in 250 mL of LB media to OD600nm of ∼0.6. The cells were harvested by centrifugation at 5000g for 10 min. The cell pellet was washed with 25 mL of 20 mM potassium phosphate (pH 7.2) and 150 mM NaCl, resuspended in 25 mL of the same buffer, and subsequently incubated for 15 min at 37oC. DSP dissolved in DMSO was added to the cell suspension at a final concentration of 80 μg/mL, and the cells were incubated for 30 min at 37oC. The cross-linking reaction was quenched by addition 1 M Tris-HCl (pH 7.4) to a final concentration of 20 mM, and cells were harvested by centrifugation at 5000g for 10 min. Cell pellets were resuspended in 5 mL of 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 5 mM MgCl2 1% ZW3-14 containing lysozyme (50 μg/mL), DNase I (50 μg/mL), and RNase I (50 μg/mL), and were lysed by shaking for 20 min at room temperature. In order to remove cell debris after lysis, the mixture was then centrifuged at 10,000g for 10 min. The cleared lysate (whole-cell extract) was supplemented with 20 mM Imidazole (pH 8.0), and loaded onto a 0.5-mL Ni-NTA column. The column was then washed with 10 mL of 50 mM potassium phosphate buffer (pH 8.0), with 300 mM NaCl, 20 mM Imidazole, 0.1% Triton X-100, and 0.1% SDS. The column was eluted with 5 mL of 50 mM potassium phosphate buffer (pH 8.0), 300 mM NaCl, and 200 mM Imidazole. The eluate was concentrated via ultrafiltration at 5000g for 30 min. The concentrated sample was used for SDS-PAGE and Western blot analysis as described above.
β-Galactosidase assays
Cultures of strains grown in LB overnight were subcultured 1:1000 into fresh LB broth and grown with aeration at 37°C until the OD600 was 0.4. β-Galactosidase assays were performed as described previously (Slauch and Silhavy 1991). The data presented were from a single representative experiment of three independent experiments.
Acknowledgments
We thank Professor Masayori Inouye for the generous gift of Lpp antibody. We also thank members of the Silhavy and Kahne laboratories for their critical reading of this manuscript. We thank Susan DiRenzo for her assistance in the preparation of this manuscript. This work was supported by an NIH Genetics and Molecular Biology training grant (T32JM07388). T.J.S. was supported by an NIGMS award (GM34821). D.K. was supported by an NIGMS award (GM66174).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1581007
References
- Bayer M.E. Areas of adhesion between wall and membrane of Escherichia coli. J. Gen. Microbiol. 1968;53:395–404. doi: 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- Behrens S., Maier R., de Cock H., Schmid F.X., Gross C.A., Maier R., de Cock H., Schmid F.X., Gross C.A., de Cock H., Schmid F.X., Gross C.A., Schmid F.X., Gross C.A., Gross C.A. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 2001;20:285–294. doi: 10.1093/emboj/20.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos W., Shuman H., Shuman H. Maltose/maltodextrin system of Escherichia coli: Transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 1998;62::204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmann H., Pluckthun A., Pluckthun A. The periplasmic Escherichia coli peptidylprolyl cis,trans-isomerase FkpA. I. Increased functional expression of antibody fragments with and without cis-prolines. J. Biol. Chem. 2000;275:17100–17105. doi: 10.1074/jbc.M910233199. [DOI] [PubMed] [Google Scholar]
- Boyd D., Weiss D.S., Chen J.C., Beckwith J., Weiss D.S., Chen J.C., Beckwith J., Chen J.C., Beckwith J., Beckwith J. Towards single-copy gene expression systems making gene cloning physiologically relevant: λ InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 2000;182::842–847. doi: 10.1128/jb.182.3.842-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M., Silhavy T.J., Silhavy T.J. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 2002;45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- Chen R., Henning U., Henning U. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol. Microbiol. 1996;19:1287–1294. doi: 10.1111/j.1365-2958.1996.tb02473.x. [DOI] [PubMed] [Google Scholar]
- Costantino N., Court D.L., Court D.L. Enhanced levels of λ Red-mediated recombinants in mismatch repair mutants. Proc. Natl. Acad. Sci. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey R.E., Chen M., Chen M. Sec-translocase mediated membrane protein biogenesis. Biochim. Biophys. Acta. 2004;1694:37–53. doi: 10.1016/j.bbamcr.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Danese P.N., Silhavy T.J., Silhavy T.J. The σE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes & Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- Danese P.N., Silhavy T.J., Silhavy T.J. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet. 1998;32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- de Cock H., van Blokland S., Tommassen J., van Blokland S., Tommassen J., Tommassen J. In vitro insertion and assembly of outer membrane protein PhoE of Escherichia coli K-12 into the outer membrane. Role of Triton X-100. J. Biol. Chem. 1996;271:12885–12890. doi: 10.1074/jbc.271.22.12885. [DOI] [PubMed] [Google Scholar]
- Doerrler W.T., Raetz C.R., Raetz C.R. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J. Biol. Chem. 2005;280:27679–27687. doi: 10.1074/jbc.M504796200. [DOI] [PubMed] [Google Scholar]
- Duguay A.R., Silhavy T.J., Silhavy T.J. Quality control in the bacterial periplasm. Biochim. Biophys. Acta. 2004;1694:121–134. doi: 10.1016/j.bbamcr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Eppens E.F., Nouwen N., Tommassen J., Nouwen N., Tommassen J., Tommassen J. Folding of a bacterial outer membrane protein during passage through the periplasm. EMBO J. 1997;16:4295–4301. doi: 10.1093/emboj/16.14.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J.W., Gross C.A., Gross C.A. Identification of the σE subunit of Escherichia coli RNA polymerase: A second alternate σ factor involved in high-temperature gene expression. Genes & Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- Freudl R., Schwarz H., Stierhof Y.D., Gamon K., Hindennach I., Henning U., Schwarz H., Stierhof Y.D., Gamon K., Hindennach I., Henning U., Stierhof Y.D., Gamon K., Hindennach I., Henning U., Gamon K., Hindennach I., Henning U., Hindennach I., Henning U., Henning U. An outer membrane protein (OmpA) of Escherichia coli K-12 undergoes a conformational change during export. J. Biol. Chem. 1986;261:11355–11361. [PubMed] [Google Scholar]
- Guisbert E., Rhodius V.A., Ahuja N., Witkin E., Gross C.A., Rhodius V.A., Ahuja N., Witkin E., Gross C.A., Ahuja N., Witkin E., Gross C.A., Witkin E., Gross C.A., Gross C.A. Hfq modulates the σE-mediated envelope stress response and the σ32-mediated cytoplasmic stress response in Escherichia coli. J. Bacteriol. 2007;189:1963–1973. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms N., Koningstein G., Dontje W., Muller M., Oudega B., Luirink J., de Cock H., Koningstein G., Dontje W., Muller M., Oudega B., Luirink J., de Cock H., Dontje W., Muller M., Oudega B., Luirink J., de Cock H., Muller M., Oudega B., Luirink J., de Cock H., Oudega B., Luirink J., de Cock H., Luirink J., de Cock H., de Cock H. The early interaction of the outer membrane protein phoe with the periplasmic chaperone Skp occurs at the cytoplasmic membrane. J. Biol. Chem. 2001;276:18804–18811. doi: 10.1074/jbc.M011194200. [DOI] [PubMed] [Google Scholar]
- Kellenberger E. The ‘Bayer bridges’ confronted with results from improved electron microscopy methods. Mol. Microbiol. 1990;4:697–705. doi: 10.1111/j.1365-2958.1990.tb00640.x. [DOI] [PubMed] [Google Scholar]
- Lazar S.W., Kolter R., Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska B., Zylicz M., Georgopoulos C., Zylicz M., Georgopoulos C., Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J. Bacteriol. 1990;172:1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinverni J.C., Werner J., Kim S., Sklar J.G., Kahne D., Misra R., Silhavy T.J., Werner J., Kim S., Sklar J.G., Kahne D., Misra R., Silhavy T.J., Kim S., Sklar J.G., Kahne D., Misra R., Silhavy T.J., Sklar J.G., Kahne D., Misra R., Silhavy T.J., Kahne D., Misra R., Silhavy T.J., Misra R., Silhavy T.J., Silhavy T.J. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- Mecsas J., Rouviere P.E., Erickson J.W., Donohue T.J., Gross C.A., Rouviere P.E., Erickson J.W., Donohue T.J., Gross C.A., Erickson J.W., Donohue T.J., Gross C.A., Donohue T.J., Gross C.A., Gross C.A. The activity of σE, an Escherichia coli heat-inducible σ-factor, is modulated by expression of outer membrane proteins. Genes & Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- Misra R., Peterson A., Ferenci T., Silhavy T.J., Peterson A., Ferenci T., Silhavy T.J., Ferenci T., Silhavy T.J., Silhavy T.J. A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J. Biol. Chem. 1991;266:13592–13597. [PubMed] [Google Scholar]
- Mogensen J.E., Otzen D.E., Otzen D.E. Interactions between folding factors and bacterial outer membrane proteins. Mol. Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- Nakamoto H., Bardwell J.C., Bardwell J.C. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim. Biophys. Acta. 2004;1694:111–119. doi: 10.1016/j.bbamcr.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Outer membrane. In: Neidhardt F.C., et al., editors. Escherichia coli and Salmonella: Cellular and molecular biology. ASM Press; Washington, DC: 1996. pp. 29–47. [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen S.A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W., Cyrklaff M., Hell K., Rapaport D., Neupert W., Hell K., Rapaport D., Neupert W., Rapaport D., Neupert W., Neupert W. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- Raivio T.L., Silhavy T.J., Silhavy T.J. Periplasmic stress and ECF σ factors. Annu. Rev. Microbiol. 2001;55:591–624. doi: 10.1146/annurev.micro.55.1.591. [DOI] [PubMed] [Google Scholar]
- Reithmeier R.A., Bragg P.D., Bragg P.D. Purification and characterization of heat-modifiable protein from the outer membrane of Escherichia coli. FEBS Lett. 1974;41:195–198. doi: 10.1016/0014-5793(74)81210-x. [DOI] [PubMed] [Google Scholar]
- Rhodius V.A., Suh W.C., Nonaka G., West J., Gross C.A., Suh W.C., Nonaka G., West J., Gross C.A., Nonaka G., West J., Gross C.A., West J., Gross C.A., Gross C.A. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzitello A.E., Harper J.R., Silhavy T.J., Harper J.R., Silhavy T.J., Silhavy T.J. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouviere P.E., Gross C.A., Gross C.A. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes & Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- Ruiz N., Falcone B., Kahne D., Silhavy T.J., Falcone B., Kahne D., Silhavy T.J., Kahne D., Silhavy T.J., Silhavy T.J. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Ruiz N., Kahne D., Silhavy T.J., Kahne D., Silhavy T.J., Silhavy T.J. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- Sauer F.G., Remaut H., Hultgren S.J., Waksman G., Remaut H., Hultgren S.J., Waksman G., Hultgren S.J., Waksman G., Waksman G. Fiber assembly by the chaperone–usher pathway. Biochim. Biophys. Acta. 2004;1694:259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Schafer U., Beck K., Muller M., Beck K., Muller M., Muller M. Skp, a molecular chaperone of Gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 1999;274:24567–24574. doi: 10.1074/jbc.274.35.24567. [DOI] [PubMed] [Google Scholar]
- Schirmer T., Keller T.A., Wang Y.F., Rosenbusch J.P., Keller T.A., Wang Y.F., Rosenbusch J.P., Wang Y.F., Rosenbusch J.P., Rosenbusch J.P. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- Silhavy T.J., Berman M.L., Enquist L.W., Berman M.L., Enquist L.W., Enquist L.W. Experiments with gene fusions. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1984. [Google Scholar]
- Sklar J.G., Wu T., Gronenberg L.S., Malinverni J.C., Kahne D., Silhavy T.J., Wu T., Gronenberg L.S., Malinverni J.C., Kahne D., Silhavy T.J., Gronenberg L.S., Malinverni J.C., Kahne D., Silhavy T.J., Malinverni J.C., Kahne D., Silhavy T.J., Kahne D., Silhavy T.J., Silhavy T.J. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch J.M., Silhavy T.J., Silhavy T.J. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 1991;173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J., Nikaido H., Nikaido H. Outer membrane of Gram-negative bacteria. XVIII. Electron microscopic studies on porin insertion sites and growth of cell surface of Salmonella typhimurium. J. Bacteriol. 1978;135:687–702. doi: 10.1128/jb.135.2.687-702.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C., Beil A., Ehrmann M., Beil A., Ehrmann M., Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- Strauch K.L., Johnson K., Beckwith J., Johnson K., Beckwith J., Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanabalu T., Koronakis E., Hughes C., Koronakis V., Koronakis E., Hughes C., Koronakis V., Hughes C., Koronakis V., Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from E. coli: Reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 1998;17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo A., Almiron M., Kolter R., Almiron M., Kolter R., Kolter R. surA, an Escherichia coli gene essential for survival in stationary phase. J. Bacteriol. 1990;172:4339–4347. doi: 10.1128/jb.172.8.4339-4347.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ureta A.R., Endres R.G., Wingreen N.S., Silhavy T.J., Endres R.G., Wingreen N.S., Silhavy T.J., Wingreen N.S., Silhavy T.J., Silhavy T.J. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J. Bacteriol. 2007;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Papenfort K., Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Voulhoux R., Tommassen J., Tommassen J. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 2004;155:129–135. doi: 10.1016/j.resmic.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Walsh N.P., Alba B.M., Bose B., Gross C.A., Sauer R.T., Alba B.M., Bose B., Gross C.A., Sauer R.T., Bose B., Gross C.A., Sauer R.T., Gross C.A., Sauer R.T., Sauer R.T. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- Werner J., Misra R., Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T.J., Kahne D., Malinverni J., Ruiz N., Kim S., Silhavy T.J., Kahne D., Ruiz N., Kim S., Silhavy T.J., Kahne D., Kim S., Silhavy T.J., Kahne D., Silhavy T.J., Kahne D., Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Yokota N., Kuroda T., Matsuyama S., Tokuda H., Kuroda T., Matsuyama S., Tokuda H., Matsuyama S., Tokuda H., Tokuda H. Characterization of the LolA–LolB system as the general lipoprotein localization mechanism of Escherichia coli. J. Biol. Chem. 1999;274:30995–30999. doi: 10.1074/jbc.274.43.30995. [DOI] [PubMed] [Google Scholar]