Abstract
The loss of telomere function can result in telomeric fusion events that lead to the types of genomic rearrangements, such as nonreciprocal translocations, that typify early-stage carcinogenesis. By using single-molecule approaches to characterize fusion events, we provide a functional definition of fusogenic telomeres in human cells. We show that approximately half of the fusion events contained no canonical telomere repeats at the fusion point; of those that did, the longest was 12.8 repeats. Furthermore, in addition to end-replication losses, human telomeres are subjected to large-scale deletion events that occur in the presence or absence of telomerase. Here we show that these telomeres are fusogenic, and thus despite the majority of telomeres being maintained at a stable length in normal human cells, a subset of stochastically shortened telomeres can potentially cause chromosomal instability. Telomere fusion was accompanied by the deletion of one or both telomeres extending several kilobases into the telomere-adjacent DNA, and microhomology was observed at the fusion points. This contrasted with telomere fusion that was observed following the experimental disruption of TRF2. The distinct error-prone mutational profile of fusion between critically shortened telomeres in human cells was reminiscent of Ku-independent microhomology-mediated end-joining.
Keywords: Telomere, telomerase, genomic instability, mutation, DNA repair, neoplasia
Telomeres are the structures that cap the ends of linear chromosomes. In humans, they are composed of the hexameric DNA sequence TTAGGG tandemly repeated into arrays of up to 25 kb. In human cells, telomeric function is conferred by a complex of proteins that associate with telomeres via interactions with the key TTAGGG repeat-binding proteins TRF1, TRF2, and Pot1 (Chong et al. 1995; Broccoli et al. 1997; Baumann and Cech 2001). Telomeres prevent the natural end of the chromosome from being recognized as a double-stranded DNA break (d’Adda di Fagagna et al. 2003) and counteract the end-replication losses that occur as a consequence of semiconservative replication of linear DNA molecules (Olovnikov 1971; Harley et al. 1990). This function is mediated by the reverse transcriptase telomerase, which catalyzes the RNA-templated addition of telomere repeats de novo at the chromosomal terminus (Greider and Blackburn 1985). The majority of human somatic cells do not express sufficient levels of telomerase to counteract end-replication losses, and thus telomeric sequences erode at a rate of ∼60–120 base pairs (bp) per cell division (Harley et al. 1990; Baird et al. 2003). Telomerase is expressed at high levels in immortal cell lines, the stem cell compartments of actively proliferating tissues, and >90% of human malignancies (Kim et al. 1994; Kolquist et al. 1998).
Gross chromosomal rearrangements are common during epithelial carcinogenesis (Mitelman et al. 1997; Shih et al. 2001). They occur early in tumor development, after which the genome appears to stabilize with advancing malignancy, roughly coincident with the activation of telomerase (Kim et al. 1994; Chadeneau et al. 1995; Meyerson et al. 1997). Late-generation telomerase knockout mice (terc−/−) show telomere loss, genomic instability, a higher rate of tumor formation, and a lower age of onset (Rudolph et al. 1999; O’Hagan et al. 2002). Crossing telomerase knockout mice with long telomeres with those with short telomeres revealed that fusion occurred preferentially between chromosomes containing the short telomeres (Hemann et al. 2001). Tumors derived from terc−/− p53+/− mice contain high frequencies of chromosomes lacking telomeric signals, anaphase bridges, nonreciprocal translocations, and end-to-end fusions (Artandi et al. 2000). Thus, short dysfunctional telomeres can drive the earliest stages of cancer; indeed, telomere dysfunction has been proposed to contribute to the nonreciprocal translocations that are common in adult carcinomas (Atkin 1986; Gisselsson et al. 2001, 2004; Rudolph et al. 2001).
The end-capping function of human telomeres is mediated by TRF2 and associated proteins (van Steensel et al. 1998). Inhibition of TRF2 function results in telomere fusion events that are dependent on factors involved in nonhomologous end-joining (NHEJ) (Smogorzewska et al. 2002; Celli and de Lange 2005). These types of fusion usually contain many kilobases of telomeric repeat DNA (van Steensel et al. 1998) and result from the covalent linkage of the G-strand of one chromosome end to the C-strand of the other (Smogorzewska et al. 2002). These data are consistent with a role of TRF2 in preventing aberrant fusion of functional telomeres by NHEJ. However, in contrast to TRF2-deficient cells, telomere fusion has been observed between short telomeres in the absence of NHEJ components (Baumann and Cech 2000; Heacock et al. 2004; Maser et al. 2007). Thus, the mechanistic basis of fusion between critically shortened telomeres in human cells is still unclear.
We described previously that, in addition to the predicted end-replication losses that result in a gradual loss of telomeric sequences as a function of cell division (Baird et al. 2006; Britt-Compton et al. 2006), telomeres can be subject to large-scale, apparently stochastic, deletion events, creating telomeres that can be shorter than those observed in senescent cell cultures (Baird et al. 2003). These telomeric deletion events occur in both the presence and absence of telomerase in normal human cells and tissues (Baird et al. 2006). If telomeric deletion results in dysfunctional and fusogenic telomeres, it may contribute significantly to genomic instability that can lead to oncogenic mutation in human tissues. However, thus far, there has been no definition of the length at which human telomeres become dysfunctional.
Here, by the sequence analysis of telomere fusion events directly from human cells, we provide a functional definition of the length at which telomeres become fusogenic. Furthermore, we show that fusion events display a distinct mutational profile that appears consistent with Ku-independent end-joining being the predominant mechanism for the fusion of critically shortened telomeres in human cells.
Results
Telomere dynamics in cells that escaped senescence
In order to investigate the fate of dysfunctional telomeres, we took advantage of the allelic variation and homogeneity of the telomere length profiles in clonal derivatives of the normal human fetal lung diploid fibroblast strain MRC5 (Baird et al. 2003). We chose two clones that displayed widely divergent allelic length distributions at the XpYp telomere and had relatively short telomeres at 17p. At three to four population doublings (PD) prior to the point of senescence, we forced the expression of the human papilloma virus (HPV)16 E6E7 oncoproteins, which abrogate the function of p53 and pRb, respectively, thus allowing the cells to bypass senescence and continue to divide to crisis (Shay et al. 1993). The expression of E6E7 extended the replicative life span of both clones by 26–28 PDs, whereas the untransfected or empty vector controls underwent replicative senescence (Supplementary Fig. 1).
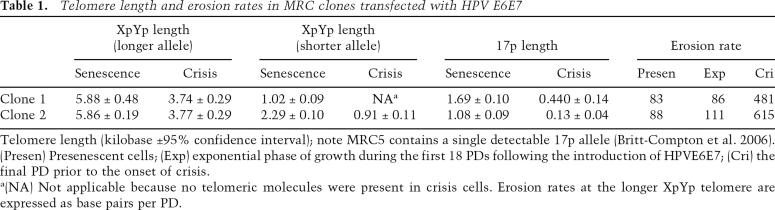
We then used allele-specific single-telomere length analysis (STELA), to track the erosion of the specific XpYp telomeric alleles and the 17p telomere as the cells continued to divide through to crisis. STELA is a long-range single-molecule PCR technique that determines the length of the double-stranded telomere repeat region from specific chromosome ends. Each DNA is analyzed with several PCR reactions containing typically four to 10 amplifiable molecules; single bands on the gel represent single telomeric molecules. Some cells, such as MRC5, contain XpYp telomere-adjacent single-nucleotide heterozygosities that can be utilized for allele-specific analysis of the two alleles at the XpYp telomere. In the early stages of culture of both MRC5 clones following the expression of HPV E6E7, the longer XpYp allele continued to shorten at a rate that was indistinguishable from that seen in the parental clone prior to replicative senescence (Table 1); thus, the expression of HPV E6E7 did not appear to directly influence telomere dynamics in these cells. As the rate of population expansion decreased, the rate of telomere erosion increased (Table 1). The final stages of culture were characterized by long periods of time with small increases in cell number (Supplementary Fig. 1). However, during these periods a significant amount of telomere erosion was observed at the larger XpYp allele—such that in the final PD of culture, these clones exhibited >480 bp of telomere loss (Table 1). These data indicate that despite limited increases in cell number of the population as a whole, there was still significant cell turnover. This accounts for the apparently sudden disappearance of the shorter XpYp allele in clone 1 and the 17p telomere in clone 2 in the final stages of culture (Fig. 1; Supplementary Fig. 2).
Table 1.
Telomere length and erosion rates in MRC clones transfected with HPV E6E7
Telomere length (kilobase ±95% confidence interval); note MRC5 contains a single detectable 17p allele (Britt-Compton et al. 2006). (Presen) Presenescent cells; (Exp) exponential phase of growth during the first 18 PDs following the introduction of HPVE6E7; (Cri) the final PD prior to the onset of crisis.
a(NA) Not applicable because no telomeric molecules were present in crisis cells. Erosion rates at the longer XpYp telomere are expressed as base pairs per PD.
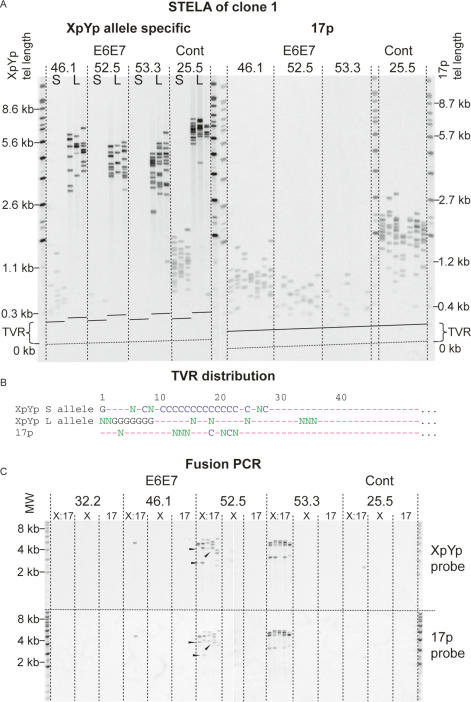
Figure 1.
Telomere loss, with a commensurate increase in the frequency of fusion, in MRC5 clone 1 expressing HPV E6E7 oncoproteins. (A) Allele-specific STELA at the XpYp telomere and 17p telomere. (S) Shorter allele; (L) longer allele. The dashed lines marked “TVR” represent the distal limit of telomere repeat variants and the beginning of the region of the telomere repeats containing pure TTAGGG repeats. The numbers above represent the number of PDs from the point of single-cell cloning; crisis occurred at PD 53.3 and the control cells underwent replicative senescence at PD 25.5. Telomere length at the XpYp and 17p telomeres is indicated on the left and right sides; this is calculated by subtracting the fragment size from the known amount of DNA between the PCR priming site and the start of the telomere repeat array. (B) TVR distributions from XpYp and 17p. (G) TGAGGG; (C) TCAGGG; (N) other repeat variants; (—) TTAGGG obtained by TVR-PCR at the XpYp telomere and sequence analysis of fusions containing the 17p telomere. (C) Fusion PCR. (X:17) Primers for XpYp and 17p; (X) single XpYp primer; (17) single 17p primer. Products were detected with the XpYp- and 17p-specific probes as indicated on the right. Arrowheads indicate fusion products that display differential hybridization.
In clone 1, the shorter telomeric allele at XpYp continued to erode below the length of the senescent control cultures (Fig. 1A; Table 1). Erosion continued to an apparent telomere length threshold, at which the number of telomeres reduced such that in crisis cells, no telomeres derived from the shorter allele were present, whereas the longer allele was still intact (Fig. 1A; Table 1). To refine the minimum threshold telomere length below which telomeres were no longer present, we determined the XpYp telomere variant repeat (TVR) distributions using TVR-PCR (Baird et al. 1995). The shorter XpYp allele contains an interspersion pattern of TTAGGG and telomere repeat variants that include TCAGGG and TGAGGG, extending 28 repeats (168 bp) into the telomere repeat array (Fig. 1B). Thus, we were able to estimate that the lower telomere length threshold was nine TTAGGG repeat units distal to the last variant repeat. The 17p telomere also continued to erode beyond the point of senescence to a lower length threshold (Fig. 1A; Table 1) that, by taking into account the variant repeat composition, we calculated to be three TTAGGG repeat units. The number of 17p telomeres reduced with ongoing cell division such that in the crisis cells we estimated that 85% of cells had lost the 17p telomere (Fig. 1A; Table 1). Similar results were observed in clone 2. However, due to the shorter XpYp telomere being >1 kb longer than clone 1, this allele was still detectable in crisis cells, whereas the 17p telomere, which was 0.6 kb shorter than clone 1, was no longer detectable in >94% of cells (Table 1; Supplementary Fig. 2A).
The fate of dysfunctional telomeres
We next examined the fate of the XpYp and 17p telomeres that had become undetectable by STELA. We considered that the telomere may continue to erode by end-replication losses into the telomere-adjacent sequences. To test this, we reasoned that in clone 1, where the shorter allele became undetectable in crisis cells, we would observe a loss of heterozygosity (LOH) at single-nucleotide polymorphism (SNP) alleles linked to this allele but not with SNP alleles linked to the longer allele. We typed by sequence analysis the 855 bp of XpYp telomere-adjacent DNA that contain 13 SNPs (Baird et al. 1995) that are heterozygous in MRC5 and observed no evidence of LOH (Supplementary Fig. 3). These data indicated that the shorter allele did not continue to erode into the telomere-adjacent DNA. We therefore reasoned that, at a specific threshold length, telomeres may become fusogenic and fused telomeres would no longer be detected by STELA. To test this hypothesis, we developed a PCR-based assay that would allow the detection of single telomere–telomere fusion molecules between specific chromosome ends. This utilized oligonucleotide primers targeted to the telomere-adjacent sequences of specific telomeres, up to 3 kb proximal to the first telomere repeat and orientated 5′–3′ toward the chromosomal terminus. By using long-range single-molecule amplification and the detection of specific products by Southern hybridization with telomere-adjacent probes, we were able to detect and quantify the presence of telomere–telomere fusions at the single-molecule level. Focusing on fusions between the XpYp and 17p telomeres in both clones, we observed an increase in the frequency and diversity of fusions that appeared to correlate with a decrease in the number of detectable telomeres using STELA (Fig. 1C; Supplementary Fig. 2B). We also observed that some molecules could only be detected with one of the two telomere-adjacent probes (Fig. 1C, arrowheads). This pattern of hybridization was consistent with either XpYp:17p fusion accompanied by a deletion of the telomere-adjacent DNA at one of the telomeres, or intra-allelic fusion, such as fusion between sister chromatids. We were unable to detect fusion between 17p and XpYp with the chromosome ends of 2p, 11q, and 12q (data not shown). However, STELA at these ends revealed that the telomeres were comparatively long (2p, 5.9 kb; 11q, 3.6 kb; and 12q, 6.3 kb) with no telomeres within the range that yielded telomere fusions at 17p and XpYp. We concluded that these ends, not having eroded beyond the critical telomere length, were still fully capped and not fusogenic. Fusions were also detected in a sample of HCA2 (MJ) normal human dermal fibroblast cells expressing HPV E6E7 at crisis that had lost one telomeric allele at XpYp (data not shown).
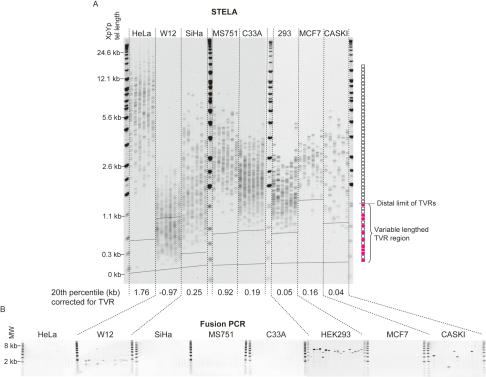
In order to understand if our findings regarding telomeric fusion in fibroblast cells transfected with HPV E6E7 could be more generally applied and were not specific to HPV E6E7-expressing cells, we analyzed telomere length and fusion at XpYp and 17p in a panel of eight telomerase-expressing epithelial cell lines. This included six cervical cell lines isolated from either carcinoma tissue (HeLa, SiHa, MS751, C33A, and CaSki) or a low-grade lesion (W12). With the exception of C33A, all of the cervical cell lines expressed HPV. We also included the breast cancer cell line MCF7 and the experimentally transformed line HEK 293 (Fig. 2; Graham et al. 1977; Kyo et al. 1996; Pett et al. 2004). Fusions were detected in W12 at a frequency of 1 × 10−4 ± 3 × 10−5 (±standard error), CaSki at a frequency of 2 × 10−5 ± 6 × 10−6, and HEK 293 at a frequency of 5 × 10−4 ± 4 × 10−5 (Fig. 2). Telomere fusion was independent of HPV status but appeared to be dependent on the telomere length distributions. The start of the pure TTAGGG repeat region of the XpYp telomere varies considerably due to the presence of variable quantities of TVRs (Baird et al. 1995). For example, MCF7 has least one XpYp telomere allele that contains extensive TVRs such that the beginning of the pure TTAGGG repeats is 1.3 kb into telomere repeat array, whereas the beginning of the pure TTAGGG repeats is at 0.14 kb in SiHa. By taking the TVR region into account and determining the 20th percentile of the distributions, it was clear that the cell lines that displayed the shortest telomeres in the lower range of the distribution also displayed fusions (Fig. 2). Thus, the distinction between the presence or absence of fusions was dependent on the existence of telomeres within the telomere length distribution that contained small amounts of TTAGGG repeats.
Figure 2.
XpYp telomere length distributions and fusion frequencies in eight telomerase-expressing epithelial cell lines. (A) STELA at the XpYp telomere. The 20th percentile length, taking into account the position of the distal extent of the TVR, is indicated below. The distal extent of the highly variable TVR region from the longest TVR allele of each cell line is indicated by a dashed line across the gel image; the TVR region of the second allele may be less extensive. TVR and TTAGGG repeat regions are illustrated on the right, blank boxes represent TTAGGG repeats, and red filled boxes represent TVRs. (B) Fusion PCR between the XpYp and 17p telomeres.
Our data indicated that if a cell population maintains a telomere length distribution that extends below a critical length, it can be subjected to telomeric fusion. Most normal human cells and tissues display telomere length profiles in which the vast majority of telomeres are longer than those that we identified as causing fusion. However, these distributions are superimposed with rare stochastically deleted telomeres that can be within the fusogenic range (Baird et al. 2003, 2006; Britt-Compton et al. 2006). Importantly, these events occur in both proliferating and senescent cell populations, which indicates that cells with an intact DNA damage checkpoint response can tolerate at least one dysfunctional telomere without the induction of growth arrest. Using our previous data on allele-specific telomere lengths in normal diploid fibroblast cells (Baird et al. 2003), we estimated the frequency of telomeres within the fusogenic range (<10× TTAGGG) to be 9 × 10−3 ± 3 × 10−3. Assuming a similar frequency at other chromosome ends, the probability of any two specific fusogenic chromosome ends occurring at the same time in the same cell will be extremely low; i.e., less than the combined frequency of 8 × 10−5. The rate of fusion between these ends is likely to be lower than this, as, for example, they may be physically and temporally separated or they may subjected to fusion/end-joining with another telomere or nontelomeric double-strand break. However, given that we developed a highly specific and sensitive single-molecule telomere fusion assay, we used this to test whether fusion could be detected between the XpYp and 17p telomeres in normal cells. We undertook a large-scale analysis of normal fibroblast populations and detected similar frequencies of fusion between the XpYp and 17p telomeres. We detected fusion at a frequency of 4 × 10−6 in proliferating HCA2 cells (±1 × 10−6, n = 4 × 106 cells analyzed) and in two separate senescent IMR90 clones at rates of 3 × 10−6 (±2 × 10−6, n = 9 × 105 cells) and 4 × 10−6 (±4 × 10−6, n = 3 × 105 cells), for a total combined frequency of 4 × 10−6 (±9 × 10−7, n = 5 × 106 cells). From these data we conclude that normal human cells can be subjected to telomeric fusion, and that this may occur despite the majority of cells displaying an apparently normal telomere length distribution. Furthermore, the presence of rare fusion events is consistent with the concept that sporadic telomeric deletion can generate fusogenic chromosome ends in normal cells. It will therefore be of interest to determine if sporadic fusion events can occur in normal and diseased human tissue.
Internal structure of telomeric fusions
In order to understand the nature of the telomeric fusions, we examined the internal structure by direct sequence analysis of the amplified fusion products. We sequenced 79 telomere fusions, of which 60 were unique and 12 were sequenced more than once within the same cell strain. The fusion assay resulted from the amplification of single DNA molecules, and therefore, if the same fusion is identified in separate reactions, it will have been derived from separate cells. Thus, we identified the same fusion events in separate cells within the same cell strain. Furthermore, as the MRC5 E6E7 cells approached crisis, the diversity of fusion events reduced such that at crisis the population was dominated by a limited number of identical fusion events (PD 53.3 in Fig. 1C; PD 49.6 in Supplementary Fig. 2B). These data indicate that, once formed, fusions can be stably maintained throughout the cell cycle, and that specific fusion events can dominate the culture as a consequence of clonal growth. These data are consistent with observations that nonreciprocal translocations in carcinomas are frequently clonal (Atkin 1986; Artandi et al. 2000).
Telomeric deletion and insertion
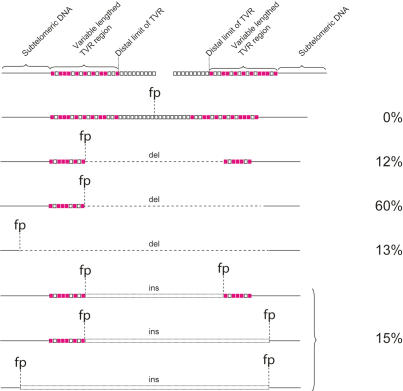
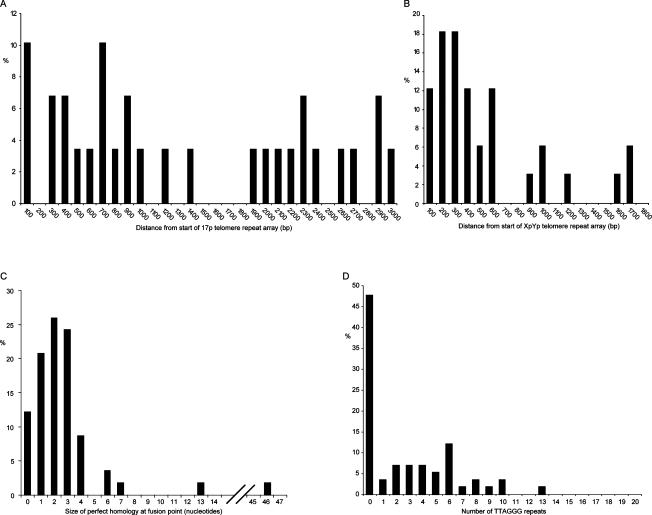
A particularly striking aspect of our data was that all of the fusion events sequenced involved the deletion of one or both telomeres (Figs. 3, 4). The deletion events included the proximal 1- to 2-kb TVR region of the telomere that contains telomere repeat variants (Baird et al. 1995) and extended into the telomere-adjacent DNA up to the limit of detection with our assays (mean subtelomeric deletion, 866 bp; maximum, 2965 bp) (Fig. 5). The fusion assay detects deletions in the telomere-adjacent DNA of 17p up to a limit of 3010 bp from the start of the telomere repeat array. The distribution of fusion events within this region appeared to be random, with no clear relationship between the distance from the telomere and the frequency of fusion (Fig. 5A). At the XpYp telomere, the fusion assay extends up to 1627 bp into the telomere-adjacent DNA. Here, 79% of deletion events at XpYp were clustered within 600 bp of the telomere repeat array, and beyond 600 bp the frequency appeared to be random (Fig. 5B). We conclude from these data that subtelomeric deletion can be extensive and is likely to extend beyond the limit of our fusion assay. Furthermore, with the exception of two separate fusion events that occurred at the same base in the XpYp telomere-adjacent DNA (−203 bp) but had different length 17p telomeres, there were no detectable hotspots for deletion points. The two MRC5 fibroblast clones expressing E6E7 showed subtly different mutational profiles. Clone 1, which showed loss of the shorter XpYp allele prior to loss of 17p, had a bias toward XpYp deletion, fused to short 17p telomeres, whereas the converse was true with clone 2, which showed the complete loss of 17p but no loss at XpYp. Thus, the telomere length dynamics were consistent with the type of telomere fusion observed by sequence analysis.
Figure 3.
Sequence data illustrating examples of the different classes of XpYp:17p fusion, isolated from MRC5 cells expressing E6E7 (A–F) and HEK 293 (G) cells. Arrows indicate informative subtelomeric SNP used to identify the XpYp telomeric alleles involved in the fusions identified from MRC5 cells. Telomere repeats (TTAGGG) in black text are identified below as “T” and variants are identified in red text as “V.” The fusion points, size of deletion (Δ) in base pairs, and microhomology (underlined in bold) are indicated below. (A) The XpYp telomere derived from the shorter allele fused to a 17p subtelomeric deletion. (B) The XpYp telomere derived from the longer allele fused to a17p subtelomeric deletion. (C) The converse of A: The 17p telomere is fused to an XpYp subtelomeric deletion of the shorter allele. (D) Fusion between subtelomeric deletions of both the XpYp shorter allele and 17p. (E) An example of XpYp:17p fusion that contains an insertion of XpYp telomere-adjacent DNA derived from the same allele in reverse orientation. Horizontal arrows below the sequence trace indicate the orientation of the sequences involved in the fusion. (F) An example of a fusion between sister chromatids; the 17p telomere is fused to a subtelomeric deletion of the 17p telomere-adjacent DNA. (G) An example of fusion between the telomere repeat variant-containing regions of XpYp and 17p from HEK 293 cells.
Figure 4.
Summarizing telomere fusion data, the various different classes of fusion molecules are illustrated. Open squares represent TTAGGG repeats, filled squares represent TVRs. (fp) Fusion point; (del) deletion; (ins) insertion.
Figure 5.
Histograms summarizing telomere fusion data. (A) The size of subtelomeric deletion events at 17p. (B) Deletion events at XpYp. (C) The extent of sequence homology at the fusion point. (D) The number of TTAGGG repeats immediately adjacent to the fusion point.
Nucleotide homology was observed at the fusion point in 89% of fusions (Figs. 3, 5C). The mean length of homology was 3.1 nucleotides (nt), and the largest region of homology (46 nt) was observed in a fusion that involved the 17p telomere with an interstitial telomeric repeat sequence. Additional nucleotide homology was observed at varying distances from the fusion points, with 42% of fusion events displaying a mean of 49% homology extending up to 28 bases from the fusion point (mean, 13 bases). Interestingly, this additional homology was highly polar, occurring on one side of the fusion point only (P < 0.001, z-test).
The majority of fusions involved the XpYp and 17p telomeres only. However, a subset of 15% was more complex (Fig. 4), containing insertions at the fusion point of additional sequences, some of which were derived from nontelomeric genomic loci and the length of which varied between 49 and 1650 bp. Thus telomeres can become fused to nontelomeric double-strand breaks, and several ligation steps between different sequences may be required before the two chromosome ends are fused. Two of these types of fusion events contained an insertion, in reverse orientation, of the telomere-adjacent sequence derived from the same telomeric allele involved in the fusion event (Fig. 3E). Such events are consistent with an intramolecular rearrangement prior to fusion with another chromosome end.
Fusion events were observed that resulted in the joining of homologous chromosomes (5%); i.e., XpYp:XpYp or 17p:17p (Fig. 3F). As with the other classes of fusion, all of these events involved deletion within the subtelomeric DNA of one of the fusion partners. Interestingly, these deletions were among the largest detected (mean, 2.1 kb). These types of events could be a consequence of both inter- or intra-allelic fusion; unfortunately, none of the events at XpYp occurred within the region of informative telomere-adjacent polymorphism, and thus it was not possible to determine which alleles were involved. However, one such event occurred at the 17p telomere in MRC5 cells; MRC5 contains a single 17p telomere bearing the sequence we were analyzing (Britt-Compton et al. 2006), and thus a 17p:17p fusion in these cells was consistent with intra-allelic fusion between sister chromatids. However, using artificial fusion constructs to test our assays, we were unable to detect sister chromatid-type fusion events containing telomere repeats on both sides of the fusion point, and thus these data may represent an underestimated frequency of sister chromatid fusion. Nevertheless, the existence of sister chromatid fusion events between short telomeres demonstrates that fusion can occur between the leading and lagging strands following replication (Smogorzewska et al. 2002), and not solely between the leading strands (Bailey et al. 2001). Furthermore, the fact that these events involve deletion and microhomology implies that they are processed in the same way as chromatid-type fusions.
Telomeric deletion events
We were able to utilize the extensive XpYp telomere-adjacent sequence polymorphism to identify the telomeric alleles involved in the fusion events. We observed that 83% of fusion events from MRC5 cells involved the shorter telomeric allele (Fig. 3A,C–E). The remaining 17% involved the longer allele (Fig. 3B). The majority of these events involved the deletion of the longer allele, which extended into the telomere-adjacent DNA. The mean length of the longer telomeric alleles prior to fusion in both clones was >3.7 kb (Fig. 1A), and the maximum length of TTAGGG repeats observed at a fusion point was 39 bp (Fig. 3B); thus, the long telomere had been subjected to a deletion event prior to fusion. These data imply that the telomeric deletion events that we observed previously with STELA in normal cells and tissues can be subjected to fusion (Baird et al. 2003, 2006; Britt-Compton et al. 2006).
TTAGGG repeat content
STELA, coupled with the fusion PCR, indicates the existence of a telomere length threshold below which telomeric fusions become detectable. We were able to refine this threshold by examining the telomere repeat composition of telomeric fusion events. Nearly half (47%) of the fusions contained no TTAGGG repeats at the fusion point (Fig. 5D); instead, the fusion event had occurred within the subtelomeric sequences or within the TVR-containing regions (Figs. 3, 4). These regions are highly variable in both length (0–2 kb) and repeat content (Baird et al. 1995). This extreme variability, coupled with the exquisite specificity of TRF1, TRF2, and POT1 to TTAGGG repeats (Bianchi et al. 1999; Baumann and Cech 2001; Lei et al. 2002), implies that telomere repeat variants do not confer telomeric function. This view is consistent with the data shown here, where it appears that a telomere that has shortened to within the TVR region becomes fusogenic. Of the those fusions that did contain (TTAGGG)n at the fusion point, the length ranged from 0.33 repeat to a maximum of 12.8 repeats (Fig. 3C), with a mean of 4.7 repeats (SD = 3.0) (Fig. 5D). Some telomeres contained extensive regions of telomere repeat variants, but still contained <12.8 repeats at the break point. The most extreme example was the fusogenic 17p telomere observed in HCA2 cells at crisis. This telomere contained a complex interspersion pattern of 114 TTAGGG and variant repeats, with a block of just five TTAGGG repeats immediately adjacent to the fusion point.
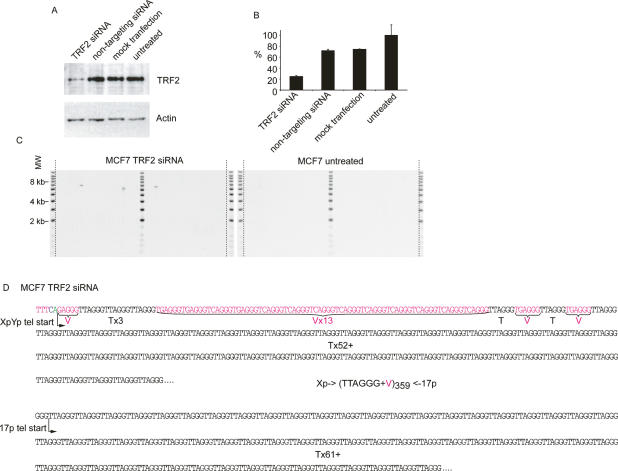
In the entire data set, we observed a single fusion event that contained TTAGGG repeats at both sides of the fusion point. Detected in telomerase-expressing HEK293 cells this fusion contained just 1.5 TTAGGG repeats from the XpYp telomere and 5.5 repeats from 17p (Fig. 3G), and occurred within the complex interspersion of telomere repeat variants and TTAGGG in the proximal regions of the two telomeres. Thus, a particularly striking feature of our data was that we did not detect telomere fusions containing >1.5 TTAGGG repeats on each side of the fusion point. We considered the possibility that, while our PCR-based telomere assays are capable of amplifying very large telomere repeat arrays from single DNA molecules (for example, >27 kb in HeLa cells) (Fig. 2), the inverted repeats that would result from the fusion of two TTAGGG repeat-containing telomeres might form secondary structures that could be refractory to PCR. As a consequence, these structures would not be represented in our assays. To test this, we generated telomere–telomere fusions that contain large amounts of TTAGGG repeats on both sides of the fusion point by the knockdown of TRF2 (van Steensel et al. 1998). We undertook short interfering RNA (siRNA)-mediated knockdown of TRF2 in MCF7 cells, a cell line in which, despite analyzing >5 × 106 cells, we were unable to detect a single telomeric fusion between XpYp and 17p (Fig. 2). Knockdown was confirmed by real-time RT–PCR and Western blotting (Fig. 6A,B), and was sufficient for us to detect a significant increase in the frequency of 17p:XpYp telomere fusion (8 × 10−6 ± 3 × 10−6, P < 0.02) (Fig. 6C). Telomere dysfunction-induced foci (TIF) have been documented following siRNA-mediated TRF2 knockdown. In HeLa cells, ∼26% of cells were reported to contain four or more TIFs (Takai et al. 2003), some of which may be resolved as telomere fusion events. Thus, following TRF2 knockdown, fusion between any two chromosome ends is likely to be rare, and may be consistent with the frequency reported here. The fusion molecules observed following TRF2 knockdown were of a size consistent with the fusion of telomeres within the size range observed at these two ends in MCF7 cells (Fig. 6C). Sequence analysis revealed extensive TTAGGG repeats at both telomeres beyond the ability of DNA sequencing; these telomeric fusions were estimated to contain a mean of 288 TTAGGG repeats (Fig. 6D). We further tested the ability of our assays to detect telomere fusion events containing large tracts of TTAGGG repeats on either side of the fusion point by generating artificial fusion molecules. To do this, we included a restriction site within the linker used for STELA; following digestion and ligation, the products were subjected to the fusion assays. We could efficiently detect artificial fusions at the single-molecule level between the XpYp and 17p telomeres, with molecules of up to 10.8 kb being detected (Supplementary Fig. 4). These molecules contain >6 kb of telomere repeat sequences and are well within the size range of the fusion molecules observed in the various cells analyzed in this study (Figs. 1, 2). These data indicate that the fusion assays are capable of detecting the full range of fusion molecules between these two telomeres. However, these data also showed that artificial fusion molecules generated at the same chromosome end did not amplify efficiently (Supplementary Fig. 4). While we can detect sister chromatid-type fusion when there has been a deletion of least one of the telomere repeat arrays (Fig. 3F), sister chromatid-type fusions containing telomere repeats on either side of the fusion may not be detected in our assays. Together, the TRF2 knockdown and artificial fusion data show that the telomere fusion assays that we used are indeed capable of detecting fusion events between different chromosome ends that contain extensive TTAGGG repeats on either side of the fusion point. We conclude, therefore, that in the cells that we analyzed the absence of these types of telomere fusion events is of direct physiological relevance and relates to the mechanistic basis of telomeric fusion.
Figure 6.
siRNA-mediated knockdown of TRF2 generates telomere fusions containing extensive arrays of TTAGGG repeats. (A) Western blot analysis with TRF2 antibody (top panel) and actin loading control (bottom panel), with the various treatments indicated above. (B) Real-time PCR for TRF2; relative quantification using the comparative CT methods normalized to the untreated control. Error bars, ±SD. (C) Fusion PCR between the XpYp and 17p telomeres. (D) Sequence data from fusion molecules obtained from MCF7 cells subjected to siRNA-mediated TRF2 knockdown. The total number of telomere and variant repeats is estimated based on the sizes of the fragments sequenced.
Discussion
In human cells containing critically shortened telomeres, we observed a characteristic mutational profile at telomeric fusion points consisting of extensive telomeric deletion and microhomology. We show that the mutation profile of fusions arising from short telomeres is distinct from those observed following the knockdown of TRF2 that do not display telomeric deletion and that contain several kilobases of telomere repeat DNA on either side of the breakpoint. Thus, we hypothesize that the mutational profile may relate to the mechanistic basis of fusion between short telomeres, and that distinct mechanisms may be required for the fusion of short telomeres compared with those rendered dysfunctional by the experimental intervention with the telomeric protein complex (Smogorzewska et al. 2002; Celli and de Lange 2005). The mutational profile that we describe here is reminiscent of the Ku-independent end-joining process referred to as microhomology-mediated end-joining (MMEJ). Characterized in Saccharomyces cerevisiae and Xenopus cell-free extracts, MMEJ is an error-prone end-joining mechanism that requires microhomology at the point of fusion and results in extensive deletion events (Boulton and Jackson 1996; Gottlich et al. 1998; Ma et al. 2003; Yu and Gabriel 2003). MMEJ is independent of the Ku heterodimer and partially independent of Ligase IV, but dependent on Mre11, Rad50, and Rad1 (Ma et al. 2003). In S. cerevisiae, MMEJ appears to be the predominant mechanism for the end-joining of breaks that lack complementary sequences (Ma et al. 2003). MMEJ involves exonucleolytic end-processing that may be mediated via Mre11, possibly in conjunction with Exo1 (Moreau et al. 2001; Ma et al. 2003), creating extensive deletion events of >10 kb (Boulton and Jackson 1996; Feldmann et al. 2000; Yu and Gabriel 2003). The involvement of Mre11 in MMEJ is also consistent with biochemical data that reveal that the exonuclease activity is stimulated by the presence of nonhomologous ends, but is inhibited by the proximity of a second double-strand break that contains some degree of microhomology (Paull and Gellert 2000). It is interesting to note that, in addition to small patches of 100% micohomology observed at the fusion point (mean, 3.1 nt), we also observed lower levels of microhomology extending to one side of the fusion point only. This pattern appears characteristic of microhomology-mediated single-stranded annealing prior to ligation, such as that observed in Ku-deficient Xenopus and CHO cell-free extracts (Gottlich et al. 1998; Feldmann et al. 2000). The role of MMEJ in the fusion of short telomeres in human cells remains to be established. However, NHEJ-independent telomere fusion of short telomeres has been observed in other species (Baumann and Cech 2000; d’Adda di Fagagna et al. 2001; Heacock et al. 2004; Maser et al. 2007). In Arabidopsis, Mre11 has been implicated in the Ku-independent fusion of short telomeres, where the absence of Mre11 resulted in a reduction in the extent of microhomology at the fusion junctions (Heacock et al. 2004). In yeast, Exo1 has been implicated in the fusion of short telomeres (Hackett and Greider 2003) and in the generation of single-stranded subtelomeric DNA in the absence of Ku (Maringele and Lydall 2002). Thus, our data are consistent with the view that, as observed in other organisms, short dysfunctional telomeres in human cells may be subjected to error-prone end-processing mechanisms such as MMEJ. The relative contributions of error-prone end-joining and NHEJ in the processing of short dysfunctional telomeres in human cells remains to be established.
By using two independent assays—STELA and fusion PCR—we detected a critical telomere length threshold in telomerase-negative human cells. In clonal MRC5 fibroblast populations with ongoing cell division past the point of senescence, the gradual erosion of the shorter allele brought the telomere to within an apparent threshold telomere length, at which point the telomeres disappeared. This was not accompanied by further erosion into the telomere-adjacent DNA, despite the fact that the cells continued to proliferate and the larger telomeric allele continued to shorten. Instead, the disappearance of the shorter telomeric alleles was accompanied by the appearance of telomere fusion events. These data are consistent with the disappearance of telomeric alleles below a specific length threshold being a consequence of telomere fusion, without the need to invoke additional end-processing that may render the telomere undetectable with STELA; for example, the loss of the 3′ overhangs. From our sample of fusions that had been sequenced in telomerase-negative fibroblast cells, the threshold length at which telomere fusion occurs is 12-5/6 double-stranded TTAGGG repeats distal to the TVR region. Further sampling may yield fusion events involving more repeats. However, 47% of the fusion events did not contain any TTAGGG repeats at the fusion junction point, and of those that did, the mean telomere length was 4.7 repeats. Thus, we conclude that telomeres of <13 pure TTAGGG repeats distal to the TVR region are fusogenic, and that in the absence of telomerase fusogenic telomeres >13, repeats are likely to be rare. We speculate that telomeres of <13 repeats do not bind sufficient TRF1 and TRF2 to confer telomeric function. At this point, the telomere may be in an open and fusogenic state; if not involved in fusion immediately, the telomere may be subject to potentially extensive resection prior to fusion with a second fusogenic telomere or nontelomeric double-stranded DNA break.
We also observed that some epithelial cell lines displayed telomeric fusion despite the presence of telomerase. Fusion was only observed in those cell lines that displayed telomere length profiles in which the shorter telomeric molecules were close to or within the TVR region. These data were consistent with our observations in fibroblast cells that telomere length is an important determinant of telomeric stability and the presence of a lower telomere length threshold below which telomeres can become unstable. Importantly, these data show that, despite the expression of telomerase, telomeric instability can still occur. This instability may be related to the observation that some telomerase-expressing immortal cell lines—including 293 cells in which we observed high levels of telomeric instability—are comprised of clonal populations that maintain telomeres at varying lengths and exhibit varying levels of telomerase (Bryan et al. 1998). Some of these clones initially had no detectable telomerase but regained it in the later stages of growth (Bryan et al. 1998). We speculate that some of these clones that have no detectable telomerase activity may be subjected to significant telomere erosion and instability prior to reactivating telomerase.
We detected fusion events that involved long telomeres (>3.7 kb) that had, prior to fusion, suffered large deletion events that often included the telomere-adjacent DNA. This observation is of interest because we described previously that in normal cells and tissues, both in the presence or absence of telomerase, telomeres can be subjected to large-scale deletion events that can create telomeres within the fusogenic range we established here (Baird et al. 2003, 2006; Britt-Compton et al. 2006). Furthermore, we were also able to detect extremely rare fusion events in normal fibroblasts, at rates that were consistent with fusion between stochastically deleted telomeres. We conclude that normal cell populations that may appear to maintain the majority of telomeres at an apparently stable length can be subjected to telomere deletion events that lead to telomeric fusion. Thus, stochastic telomeric deletion provides a “cell-intrinsic” mutational mechanism that can result in the types of oncogenic rearrangements, such as nonreciprocal translocations, that can drive early-stage neoplasia in normal cells and tissues.
Materials and methods
Cell culture and tissue samples
We cultured IMR-90 (Nichols et al. 1977), MRC5 (Jacobs et al. 1970), and HCA2 (Bond et al. 1999) cells in EMEM (Invitrogen); HEK 293 (Graham et al. 1977) and MCF7 (Soule et al. 1973) cells in DMEM (Invitrogen); and CaSki (HPV 16) (Pattillo et al. 1977) cells in RPMI (Invitrogen), all supplemented with Earle’s salts containing, 2× nonessential amino acids (Sigma), 10% (v/v) fetal calf serum (15% for IMR-90) (Autogenbioclear), 1 × 105 IU/L penicillin, 100 mg/L streptomycin, and 2 mM glutamine (Sigma). HeLa (HPV 18) (Jones et al. 1971), SiHa (HPV 16) (Friedl et al. 1970), MS751 (HPV 18) (Pater and Pater 1985), and C33A (HPV-negative) (Auersperg 1964) were cultured in DMEM supplemented with 10% fetal bovine serum (Invitrogen), 100 U/mL penicillin, 100 mg/mL streptomycin (Invitrogen), 2 mM L-glutamine (Invitrogen), and 25 mM HEPES Buffer (Sigma). W12 cells (HPV 16) (Stanley et al. 1989) (kindly provided by Margaret Stanley, Cambridge University, Cambridge, UK) were cultured in Keratinocyte-SFM media (GIBCO-BRL) supplemented with bovine pituitary extract (25 μg/mL) and recombinant epidermal growth factor (0.1–0.2 ng/mL).
Retroviral gene transfer using amphotropic retroviral vectors expressing HPV 16 E6E7 and selected with G418 (0.4 mg/mL) was as described previously (Bond et al. 1999).
siRNA, RT–PCR, and Western blotting
siRNA duplexes against TRF2 and the “nontargeting” negative control were obtained from Dharmacon. Transfection was carried out in MCF7 cells using DharmaFECT (Dharmacon) according the protocol recommended by Dharmacon. TRF2 mRNA levels were analyzed by real-time PCR using a TaqMan probe directed against TRF2 (Applied Biosystems). TRF2 protein levels were analyzed by Western blot analysis of 20 μg of whole-cell lysate, with an anti-TRF2 antibody (Imgenex) at a concentration of 5 μg/mL, and ECL using Mouse IgG, and HRP-Linked Whole Ab (GE Healthcare).
DNA extraction and PCR
We extracted DNA from cells using standard proteinase K, RNase A, and phenol/chloroform protocols; solubilized the DNA by digestion with EcoRI; and quantified it in triplicate by Hoechst 33258 fluorometry (Bio-Rad) as detailed previously (Baird et al. 2003). XpYp telomere-adjacent SNPs were as described previously (Baird et al. 1995). The −13, −30, −146, −176, −297, and −298 positions were identified by sequence analysis of PCR products generated with the XpYpE2 and XpYpB2 primers, and the −415, −427, −540, −544, −554, −652, and −842 positions were identified using XpYpG and XpYpC primers. TVR distributions were determined in MRC5 cells using allele-specific TVR-PCR at the XpYp telomere as described previously (Baird et al. 1995). TVR-PCR was adapted to determine the distal extent of the extensive XpYp TVR regions in the other cell lines analyzed by using the XpYpE2 primer in conjunction with the TVR primers. Products were resolved by 0.5% TAE agarose gel electrophoresis, and were detected by Southern hybridization with a random-primed α-33P-labeled (GE Healthcare) probe generated by PCR between primers XpYpE2 and XpYpB2. For telomere length analysis at 2p, 11q, 12q, 17p, and XpYp, we used a modification of the STELA protocol (Baird et al. 2003; Britt-Compton et al. 2006). Briefly, solublized genomic DNA was diluted to 10 ng/μL in 10 mM Tris-HCl (pH 7.5). Ten nanograms of DNA were further diluted to 250 pg/μL in a volume of 40 μL containing 1 μM Telorette2 linker and 1 mM Tris-HCl (pH 7.5). Multiple PCRs (typically six reactions per sample) were carried out for each test DNA in 10-μL volumes containing 250 pg of diluted DNA, 0.5 μM telomere-adjacent and Teltail primers, 75 mM Tris-HCl (pH 8.8), 20 mM (NH4)2SO4, 0.01% Tween-20, 1.5 mM MgCl2, and 0.5 U of a 10:1 mixture of Taq (ABGene) and Pwo polymerase (Roche). The reactions were cycled with an MJ PTC-225 thermocycler (MJ research) as described previously (Baird et al. 2003; Britt-Compton et al. 2006). The DNA fragments were resolved by 0.5% TAE agarose gel electrophoresis, and were detected by Southern hybridization with a random-primed α-33P-labeled (GE Healthcare) telomere-adjacent probe and a probe to detect the 1-kb molecular weight marker (Stratagene). The hybridized fragments were detected by phosphorimaging with a Molecular Dynamics Storm 860 PhosphorImager (GE Healthcare). The molecular weights of the DNA fragments were calculated using the Phoretix 1D quantifier (Nonlinear Dynamics). For fusion PCR, the DNA was diluted to 50 ng/μL in 10 mM Tris-HCl (pH 7.5). We then carried out multiple PCRs (typically between 18 and 150 reactions per sample, depending on fusion frequency) for each test DNA in 10-μL volumes containing between 10 and 100 ng (depending on the frequency of fusion) of DNA, 0.5 μM telomere-adjacent primers, 1.2 mM NTPs, 75 mM Tris-HCl (pH 8.8), 20 mM (NH4)2SO4, 0.01% Tween-20, 1.5 mM MgCl2, and 0.5 U of a 10:1 mixture of Taq and Pwo polymerase. The reactions were cycled under the following conditions: 25 cycles of 15 sec at 94°C, 30 sec at 59°C, and 8 min at 68°C. We resolved the DNA fragments by 0.5% TAE agarose gel electrophoresis, and detected them by Southern hybridization with random-primed α-33P-labeled probes generated by PCR between primers XpYpO and XpYpB2 and 17p7 and 17p2. We detected the hybridized fragments by phosphorimaging with a Molecular Dynamics Storm 860 PhosphorImager. The total number of bands generated from each DNA analyzed was counted, and the number of input molecules in each analysis was estimated from the amount of DNA used in each reaction and then dividing by the size of a diploid human genome (6 pg). Thus, the frequency of fusion could be calculated directly from the number of positive molecules compared with the number of input molecules. Fusion products were reamplified for sequence analysis as follows: First-round PCR reactions containing fusions were diluted 1:20 in H2O, and 3 μL were used in a second PCR with nested primers (XpYpO and 17p7) using the same buffering system as described above with 3 mM MgCl2 and the same thermal-cycling conditions with 32 cycles. Reamplified DNA was gel-purified and sequenced using BigDye 3.1 or BigDye 3.1 dGTP in conjunction with XpYp and 17p subtelomeric primers (Baird et al. 1995; Britt-Compton et al. 2006). Artificial fusion molecules were generated by STELA as described above, using either MCF7 or MRC5 clone 1 E6E7 DNA diluted to 10 ng/μL with TelEcoR1 in place of the Telorette2 linker. The STELA PCR was carried out in 20-μL reactions for 25 cycles with 17p6 or XpYpM primers to amplify the 17p and XpYp telomeres, respectively. STELA products were digested with EcoRI using the manufacturer’s reaction conditions (Promega). The digested DNA was purified using a QIAquick PCR purification kit (Qiagen); the DNA was eluted in 50 μL of 10 mM Tris-HCl (pH 8.5). The DNA was then ligated in a 50-μL volume with 20 μL of purified DNA according to the manufacturer’s reaction conditions (Promega). Fusion products were detected as described above except using the 17p7 and XpYpO primers for 17p:XpYp fusion products; XpYpO alone for XpYp:XpYp fusion products; and XpYp-427G/415C or XpYp-427A/415T for allele-specific XpYp:XpYp fusion analysis.
Oligonucleotides
The oligonucleotide sequences were as follows (those marked with * were used for the fusion assay): *XpYpM, 5′-ACCAG GTTTTCCAGTGTGTT-3′; XpYpO, 5′-CCTGTAACGCTGT TAGGTAC-3′; XpYpB2, 5′-TCTGAAAGTGGACC(A/T)ATCA G-3′; XpYpE2, 5′-TTGTCTCAGGGTCCTAGTG-3′; XpYpG, 5′-AATTCCAGACACACTAGGACCCTGA-3′; XpYpC, 5′-CA GGGACCGGGACAAATAGAC-3′; *17p6, 5′-GGCTGAACTA TAGCCTCTGC-3′; 17p2, 5′-GAGTCAATGATTCCATTCCTA GC-3′; 17p7, 5′-CCTGGCATGGTATTGACATG-3′; 17pseqrev1, 5′-GAATCCACGGATTGCTTTGTGTAC-3′; TelEcoR1, 5′-TGCTCCGTGCATCTGGCATCGAATTCACCCTAA-3′; XpYp-427G/415C, 5′-GGTTATCGACCAGGTGCTCC-3′; XpYp427A/415T, 5′-GGTTATCAACCAGGTGCTCT-3′; *2p2, 5′-GA GCTGCGTTTTGCTGAGCAC-3′; *11q13B, 5′-CAGACCTTG GAGGCACGGCCTTCG-3′; *12q550, 5′-ACAGCCTTTTGGGG TACCGC-3′.
Acknowledgments
We thank David Wynford-Thomas and David Kipling for helpful discussions on the manuscript, and Barrie Francis for sequence analysis. This work was funded by Cancer Research UK and the MRC. D.M.B. is a Cancer Research UK Senior Cancer Research Fellow.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.439107
References
- Artandi S.E., Chang S., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Chang S., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Alson S., Gottlieb G.J., Chin L., DePinho R.A., Gottlieb G.J., Chin L., DePinho R.A., Chin L., DePinho R.A., DePinho R.A. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Atkin N.B. Lack of reciprocal translocations in carcinomas. Cancer Genet. Cytogenet. 1986;21:275–278. doi: 10.1016/0165-4608(86)90009-9. [DOI] [PubMed] [Google Scholar]
- Auersperg N. Long-term cultivation of hypodiploid human tumor cells. J. Natl. Cancer Inst. 1964;32:135–163. [PubMed] [Google Scholar]
- Bailey S.M., Cornforth M.N., Kurimasa A., Chen D.J., Goodwin E.H., Cornforth M.N., Kurimasa A., Chen D.J., Goodwin E.H., Kurimasa A., Chen D.J., Goodwin E.H., Chen D.J., Goodwin E.H., Goodwin E.H. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293:2462–2465. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
- Baird D.M., Jeffreys A.J., Royle N.J., Jeffreys A.J., Royle N.J., Royle N.J. Mechanisms underlying telomere repeat turnover, revealed by hypervariable variant repeat distribution patterns in the human Xp/Yp telomere. EMBO J. 1995;14:5433–5443. doi: 10.1002/j.1460-2075.1995.tb00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird D.M., Rowson J., Wynford-Thomas D., Kipling D., Rowson J., Wynford-Thomas D., Kipling D., Wynford-Thomas D., Kipling D., Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- Baird D.M., Britt-Compton B., Rowson J., Amso N.N., Gregory L., Kipling D., Britt-Compton B., Rowson J., Amso N.N., Gregory L., Kipling D., Rowson J., Amso N.N., Gregory L., Kipling D., Amso N.N., Gregory L., Kipling D., Gregory L., Kipling D., Kipling D. Telomere instability in the male germline. Hum. Mol. Genet. 2006;15:45–51. doi: 10.1093/hmg/ddi424. [DOI] [PubMed] [Google Scholar]
- Baumann P., Cech T.R., Cech T.R. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell. 2000;11:3265–3275. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P., Cech T.R., Cech T.R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- Bianchi A., Stansel R.M., Fairall L., Griffith J.D., Rhodes D., de Lange T., Stansel R.M., Fairall L., Griffith J.D., Rhodes D., de Lange T., Fairall L., Griffith J.D., Rhodes D., de Lange T., Griffith J.D., Rhodes D., de Lange T., Rhodes D., de Lange T., de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J.A., Haughton M.F., Rowson J.M., Smith P.J., Gire V., Wynford-Thomas D., Wyllie F.S., Haughton M.F., Rowson J.M., Smith P.J., Gire V., Wynford-Thomas D., Wyllie F.S., Rowson J.M., Smith P.J., Gire V., Wynford-Thomas D., Wyllie F.S., Smith P.J., Gire V., Wynford-Thomas D., Wyllie F.S., Gire V., Wynford-Thomas D., Wyllie F.S., Wynford-Thomas D., Wyllie F.S., Wyllie F.S. Control of replicative life span in human cells: Barriers to clonal expansion intermediate between M1 senescence and M2 crisis. Mol. Cell. Biol. 1999;19:3103–3114. doi: 10.1128/mcb.19.4.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S.J., Jackson S.P., Jackson S.P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Britt-Compton B., Rowson J., Locke M., Mackenzie I., Kipling D., Baird D.M., Rowson J., Locke M., Mackenzie I., Kipling D., Baird D.M., Locke M., Mackenzie I., Kipling D., Baird D.M., Mackenzie I., Kipling D., Baird D.M., Kipling D., Baird D.M., Baird D.M. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans. Hum. Mol. Genet. 2006;15:725–733. doi: 10.1093/hmg/ddi486. [DOI] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska A., Chong L., de Lange T., Smogorzewska A., Chong L., de Lange T., Chong L., de Lange T., de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- Bryan T.M., Englezou A., Dunham M.A., Reddel R.R., Englezou A., Dunham M.A., Reddel R.R., Dunham M.A., Reddel R.R., Reddel R.R. Telomere length dynamics in telomerase-positive immortal human cell populations. Exp. Cell Res. 1998;239:370–378. doi: 10.1006/excr.1997.3907. [DOI] [PubMed] [Google Scholar]
- Celli G.B., de Lange T., de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Chadeneau C., Siegel P., Harley C.B., Muller W.J., Bacchetti S., Siegel P., Harley C.B., Muller W.J., Bacchetti S., Harley C.B., Muller W.J., Bacchetti S., Muller W.J., Bacchetti S., Bacchetti S. Telomerase activity in normal and malignant murine tissues. Oncogene. 1995;11:893–898. [PubMed] [Google Scholar]
- Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T., Hanish J., Tempst P., de Lange T., Tempst P., de Lange T., de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- di d’Adda Fagagna F., Hande M.P., Tong W.M., Roth D., Lansdorp P.M., Wang Z.Q., Jackson S.P., Hande M.P., Tong W.M., Roth D., Lansdorp P.M., Wang Z.Q., Jackson S.P., Tong W.M., Roth D., Lansdorp P.M., Wang Z.Q., Jackson S.P., Roth D., Lansdorp P.M., Wang Z.Q., Jackson S.P., Lansdorp P.M., Wang Z.Q., Jackson S.P., Wang Z.Q., Jackson S.P., Jackson S.P. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- di d’Adda Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P., Saretzki G., Carter N.P., Jackson S.P., Carter N.P., Jackson S.P., Jackson S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- Feldmann E., Schmiemann V., Goedecke W., Reichenberger S., Pfeiffer P., Schmiemann V., Goedecke W., Reichenberger S., Pfeiffer P., Goedecke W., Reichenberger S., Pfeiffer P., Reichenberger S., Pfeiffer P., Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: Implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl F., Kimura I., Osato T., Ito Y., Kimura I., Osato T., Ito Y., Osato T., Ito Y., Ito Y. Studies on a new human cell line (SiHa) derived from carcinoma of uterus. I. Its establishment and morphology. Proc. Soc. Exp. Biol. Med. 1970;135:543–545. doi: 10.3181/00379727-135-35091a. [DOI] [PubMed] [Google Scholar]
- Gisselsson D., Jonson T., Petersen A., Strombeck B., Dal Cin P., Hoglund M., Mitelman F., Mertens F., Mandahl N., Jonson T., Petersen A., Strombeck B., Dal Cin P., Hoglund M., Mitelman F., Mertens F., Mandahl N., Petersen A., Strombeck B., Dal Cin P., Hoglund M., Mitelman F., Mertens F., Mandahl N., Strombeck B., Dal Cin P., Hoglund M., Mitelman F., Mertens F., Mandahl N., Dal Cin P., Hoglund M., Mitelman F., Mertens F., Mandahl N., Hoglund M., Mitelman F., Mertens F., Mandahl N., Mitelman F., Mertens F., Mandahl N., Mertens F., Mandahl N., Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc. Natl. Acad. Sci. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D., Gorunova L., Hoglund M., Mandahl N., Elfving P., Gorunova L., Hoglund M., Mandahl N., Elfving P., Hoglund M., Mandahl N., Elfving P., Mandahl N., Elfving P., Elfving P. Telomere shortening and mitotic dysfunction generate cytogenetic heterogeneity in a subgroup of renal cell carcinomas. Br. J. Cancer. 2004;91:327–332. doi: 10.1038/sj.bjc.6601803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlich B., Reichenberger S., Feldmann E., Pfeiffer P., Reichenberger S., Feldmann E., Pfeiffer P., Feldmann E., Pfeiffer P., Pfeiffer P. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eur. J. Biochem. 1998;258:387–395. doi: 10.1046/j.1432-1327.1998.2580387.x. [DOI] [PubMed] [Google Scholar]
- Graham F.L., Smiley J., Russell W.C., Nairn R., Smiley J., Russell W.C., Nairn R., Russell W.C., Nairn R., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Greider C.W., Greider C.W. End resection initiates genomic instability in the absence of telomerase. Mol. Cell. Biol. 2003;23:8450–8461. doi: 10.1128/MCB.23.23.8450-8461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C.B., Futcher A.B., Greider C.W., Futcher A.B., Greider C.W., Greider C.W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Heacock M., Spangler E., Riha K., Puizina J., Shippen D.E., Spangler E., Riha K., Puizina J., Shippen D.E., Riha K., Puizina J., Shippen D.E., Puizina J., Shippen D.E., Shippen D.E. Molecular analysis of telomere fusions in Arabidopsis: Multiple pathways for chromosome end-joining. EMBO J. 2004;23:2304–2313. doi: 10.1038/sj.emboj.7600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Strong M.A., Hao L.Y., Greider C.W., Strong M.A., Hao L.Y., Greider C.W., Hao L.Y., Greider C.W., Greider C.W. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- Jacobs J.P., Jones C.M., Baille J.P., Jones C.M., Baille J.P., Baille J.P. Characteristics of a human diploid cell designated MRC-5. Nature. 1970;227:168–170. doi: 10.1038/227168a0. [DOI] [PubMed] [Google Scholar]
- Jones H.W., McKusick V.A., Harper P.S., Wuu K.D., McKusick V.A., Harper P.S., Wuu K.D., Harper P.S., Wuu K.D., Wuu K.D. George Otto Gey (1899–1970). The HeLa cell and a reappraisal of its origin. Obstet. Gynecol. 1971;38:945–949. [PubMed] [Google Scholar]
- Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W., Wright W.E., Weinrich S.L., Shay J.W., Weinrich S.L., Shay J.W., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kolquist K.A., Ellisen L.W., Counter C.M., Meyerson M., Tan L.K., Weinberg R.A., Haber D.A., Gerald W.L., Ellisen L.W., Counter C.M., Meyerson M., Tan L.K., Weinberg R.A., Haber D.A., Gerald W.L., Counter C.M., Meyerson M., Tan L.K., Weinberg R.A., Haber D.A., Gerald W.L., Meyerson M., Tan L.K., Weinberg R.A., Haber D.A., Gerald W.L., Tan L.K., Weinberg R.A., Haber D.A., Gerald W.L., Weinberg R.A., Haber D.A., Gerald W.L., Haber D.A., Gerald W.L., Gerald W.L. Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nat. Genet. 1998;19:182–186. doi: 10.1038/554. [DOI] [PubMed] [Google Scholar]
- Kyo S., Kanaya T., Ishikawa H., Ueno H., Inoue M., Kanaya T., Ishikawa H., Ueno H., Inoue M., Ishikawa H., Ueno H., Inoue M., Ueno H., Inoue M., Inoue M. Telomerase activity in gynecological tumors. Clin. Cancer Res. 1996;2:2023–2028. [PubMed] [Google Scholar]
- Lei M., Baumann P., Cech T.R., Baumann P., Cech T.R., Cech T.R. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560– 14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- Ma J.L., Kim E.M., Haber J.E., Lee S.E., Kim E.M., Haber J.E., Lee S.E., Haber J.E., Lee S.E., Lee S.E. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L., Lydall D., Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes & Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser R.S., Wong K.K., Sahin E., Xia H., Naylor M., Hedberg H.M., Artandi S.E., DePinho R.A., Wong K.K., Sahin E., Xia H., Naylor M., Hedberg H.M., Artandi S.E., DePinho R.A., Sahin E., Xia H., Naylor M., Hedberg H.M., Artandi S.E., DePinho R.A., Xia H., Naylor M., Hedberg H.M., Artandi S.E., DePinho R.A., Naylor M., Hedberg H.M., Artandi S.E., DePinho R.A., Hedberg H.M., Artandi S.E., DePinho R.A., Artandi S.E., DePinho R.A., DePinho R.A. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol. Cell. Biol. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M., Counter C.M., Eaton E.N., Ellisen L.W., Steiner P., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., Counter C.M., Eaton E.N., Ellisen L.W., Steiner P., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., Eaton E.N., Ellisen L.W., Steiner P., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., Ellisen L.W., Steiner P., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., Steiner P., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., Beijersbergen R.L., Davidoff M.J., Liu Q., Davidoff M.J., Liu Q., Liu Q., et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Mitelman F., Johansson B., Mandahl N., Mertens F., Johansson B., Mandahl N., Mertens F., Mandahl N., Mertens F., Mertens F. Clinical significance of cytogenetic findings in solid tumors. Cancer Genet. Cytogenet. 1997;95:1–8. doi: 10.1016/s0165-4608(96)00252-x. [DOI] [PubMed] [Google Scholar]
- Moreau S., Morgan E.A., Symington L.S., Morgan E.A., Symington L.S., Symington L.S. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols W.W., Murphy D.G., Cristofalo V.J., Toji L.H., Greene A.E., Dwight S.A., Murphy D.G., Cristofalo V.J., Toji L.H., Greene A.E., Dwight S.A., Cristofalo V.J., Toji L.H., Greene A.E., Dwight S.A., Toji L.H., Greene A.E., Dwight S.A., Greene A.E., Dwight S.A., Dwight S.A. Characterization of a new human diploid cell strain, IMR-90. Science. 1977;196:60–63. doi: 10.1126/science.841339. [DOI] [PubMed] [Google Scholar]
- O’Hagan R.C., Chang S., Maser R.S., Mohan R., Artandi S.E., Chin L., DePinho R.A., Chang S., Maser R.S., Mohan R., Artandi S.E., Chin L., DePinho R.A., Maser R.S., Mohan R., Artandi S.E., Chin L., DePinho R.A., Mohan R., Artandi S.E., Chin L., DePinho R.A., Artandi S.E., Chin L., DePinho R.A., Chin L., DePinho R.A., DePinho R.A. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- Olovnikov A.M. Principle of marginotomy in template synthesis of polynucleotides. Dokl. Akad. Nauk SSSR. 1971;201:1496–1499. [PubMed] [Google Scholar]
- Pater M.M., Pater A., Pater A. Human papillomavirus types 16 and 18 sequences in carcinoma cell lines of the cervix. Virology. 1985;145:313–318. doi: 10.1016/0042-6822(85)90164-3. [DOI] [PubMed] [Google Scholar]
- Pattillo R.A., Hussa R.O., Story M.T., Ruckert A.C., Shalaby M.R., Mattingly R.F., Hussa R.O., Story M.T., Ruckert A.C., Shalaby M.R., Mattingly R.F., Story M.T., Ruckert A.C., Shalaby M.R., Mattingly R.F., Ruckert A.C., Shalaby M.R., Mattingly R.F., Shalaby M.R., Mattingly R.F., Mattingly R.F. Tumor antigen and human chorionic gonadotropin in CaSki cells: A new epidermoid cervical cancer cell line. Science. 1977;196:1456–1458. doi: 10.1126/science.867042. [DOI] [PubMed] [Google Scholar]
- Paull T.T., Gellert M., Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl. Acad. Sci. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett M.R., Alazawi W.O., Roberts I., Dowen S., Smith D.I., Stanley M.A., Coleman N., Alazawi W.O., Roberts I., Dowen S., Smith D.I., Stanley M.A., Coleman N., Roberts I., Dowen S., Smith D.I., Stanley M.A., Coleman N., Dowen S., Smith D.I., Stanley M.A., Coleman N., Smith D.I., Stanley M.A., Coleman N., Stanley M.A., Coleman N., Coleman N. Acquisition of high-level chromosomal instability is associated with integration of human papillomavirus type 16 in cervical keratinocytes. Cancer Res. 2004;64:1359–1368. doi: 10.1158/0008-5472.can-03-3214. [DOI] [PubMed] [Google Scholar]
- Rudolph K.L., Chang S., Lee H.W., Blasco M., Gottlieb G.J., Greider C., DePinho R.A., Chang S., Lee H.W., Blasco M., Gottlieb G.J., Greider C., DePinho R.A., Lee H.W., Blasco M., Gottlieb G.J., Greider C., DePinho R.A., Blasco M., Gottlieb G.J., Greider C., DePinho R.A., Gottlieb G.J., Greider C., DePinho R.A., Greider C., DePinho R.A., DePinho R.A. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- Rudolph K.L., Millard M., Bosenberg M.W., DePinho R.A., Millard M., Bosenberg M.W., DePinho R.A., Bosenberg M.W., DePinho R.A., DePinho R.A. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- Shay J.W., Wright W.E., Brasiskyte D., Van der Haegen B.A., Wright W.E., Brasiskyte D., Van der Haegen B.A., Brasiskyte D., Van der Haegen B.A., Van der Haegen B.A. E6 of human papillomavirus type 16 can overcome the M1 stage of immortalization in human mammary epithelial cells but not in human fibroblasts. Oncogene. 1993;8:1407–1413. [PubMed] [Google Scholar]
- Shih I.M., Zhou W., Goodman S.N., Lengauer C., Kinzler K.W., Vogelstein B., Zhou W., Goodman S.N., Lengauer C., Kinzler K.W., Vogelstein B., Goodman S.N., Lengauer C., Kinzler K.W., Vogelstein B., Lengauer C., Kinzler K.W., Vogelstein B., Kinzler K.W., Vogelstein B., Vogelstein B. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 2001;61:818–822. [PubMed] [Google Scholar]
- Smogorzewska A., Karlseder J., Holtgreve-Grez H., Jauch A., de Lange T., Karlseder J., Holtgreve-Grez H., Jauch A., de Lange T., Holtgreve-Grez H., Jauch A., de Lange T., Jauch A., de Lange T., de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- Soule H.D., Vazguez J., Long A., Albert S., Brennan M., Vazguez J., Long A., Albert S., Brennan M., Long A., Albert S., Brennan M., Albert S., Brennan M., Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- Stanley M.A., Browne H.M., Appleby M., Minson A.C., Browne H.M., Appleby M., Minson A.C., Appleby M., Minson A.C., Minson A.C. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int. J. Cancer. 1989;43:672–676. doi: 10.1002/ijc.2910430422. [DOI] [PubMed] [Google Scholar]
- Takai H., Smogorzewska A., de Lange T., Smogorzewska A., de Lange T., de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T., Smogorzewska A., de Lange T., de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Yu X., Gabriel A., Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]