Abstract
The ribonuclease inhibitor protein (RI) binds to members of the bovine pancreatic ribonuclease (RNase A) superfamily with an affinity in the femtomolar range. Here, we report on structural and energetic aspects of the interaction between human RI (hRI) and human pancreatic ribonuclease (RNase 1). The structure of the crystalline hRI·RNase 1 complex was determined at a resolution of 1.95 Å, revealing the formation of 19 intermolecular hydrogen bonds involving 13 residues of RNase 1. In contrast, only 9 such hydrogen bonds are apparent in the structure of the complex between porcine RI and RNase A. hRI, which is anionic, also appears to use its horseshoe-shaped structure to engender long-range Coulombic interactions with RNase 1, which is cationic. In accordance with the structural data, the hRI·RNase 1 complex was found to be extremely stable (t½ = 81 days; Kd = 2.9 × 10-16 M). Site-directed mutagenesis experiments enabled the identification of two cationic residues in RNase 1—Arg39 and Arg91—that are especially important for both the formation and stability of the complex, and are thus termed “electrostatic targeting residues”. Disturbing the electrostatic attraction between hRI and RNase 1 yielded a variant of RNase 1 that maintained ribonucleolytic activity and conformational stability but had a 2.8 × 103-fold lower association rate for complex formation and 5.9 × 109-fold lower affinity for hRI. This variant of RNase 1, which exhibits the largest decrease in RI affinity of any engineered ribonuclease, is also toxic to human erythroleukemia cells. Together, these results provide new insight into an unusual and important protein-protein interaction, and could expedite the development of human ribonucleases as chemotherapeutic agents.
Keywords: cancer, cytotoxin, protein-protein interaction, tight-binding inhibitor, X-ray crystallography
Introduction
The stability of a protein·protein complex is governed by intermolecular forces that mediate the rates at which the proteins associate and the complex dissociates. The rate of dissociation is affected largely by forces that act over short distances, including hydrophobic forces, hydrogen bonds, and van der Waals interactions. The rate of association, however, depends primarily on diffusion but can be increased by Coulombic forces.1-7 Although a large energetic penalty is incurred upon desolvation of charged amino acids,8,9 the rate of association and, consequently, the stability of a complex can be increased by optimizing Coulombic interactions.5,6,10
The ribonuclease inhibitor protein forms a tight (Kd = kd/ka ≈ 10-15 M) complex with multiple members of the bovine pancreatic ribonuclease (RNase A11; EC 3.1.27.5) superfamily, as shown in equation (1) for the human inhibitor (hRI) and enzyme (RNase 1).12,13
| (1) |
RI achieves its high affinity for ribonucleases through the burial of a large surface area (2908 Å2 for the hRI·angiogenin complex14), along with one of the largest known electrostatic energies of interaction (ΔU = -12.3 kcal/mol for the hRI·angiogenin complex15). Indeed, among 68 heterodimeric protein·protein complexes, the hRI·angiogenin complex ranked behind only the karyopherin β2-Ran complex in the relative contribution of electrostatic energy to complex formation.15
The evasion of RI by ribonucleases has medicinal implications, as variants of RNase A that evade RI are toxic to cancer cells.16,17 By using the structure of the complex between porcine RI (pRI) and RNase A,18 we designed variants of RNase A that are more toxic to human leukemic cells in vitro than is Onconase® (ONC), a naturally cytotoxic homologue from Rana pipiens that is now in Phase III clinical trials as a cancer chemotherapeutic agent.17 Disruption of the pRI·RNase A interface was accomplished by designing RNase A variants with amino-acid substitutions that disturbed regions of high shape-complementarity.17 These substitutions targeted short-range pRI·RNase A interactions by instilling steric hindrance or excising hydrogen bonds. We also applied this strategy to bovine seminal ribonuclease (BS-RNase, 87% sequence similarity), another homologue of RNase A that is dimeric in its native state.19,20 A BS-RNase variant with substitutions in the same high shape-complementarity regions was also more cytotoxic than ONC in vitro.20
Designing proteins that have diminished affinity for a cognate protein could be accomplished by targeting either component of the equilibrium dissociation constant: kd or ka. Previous studies of the RI·ribonuclease interface have focused on short-range intermolecular contacts between the proteins, thereby raising the dissociation rate.21,22,17 Diminishing the affinity of RNase 1, the human homologue of RNase A, by modulating short-range interactions has, however, proven to be difficult.23-25 Although RNase 1 and RNase A share 70% sequence identity, mutagenesis studies have indicated substantial variation in how each is recognized by RI.23-25 We sought to elaborate how RNase 1 is recognized by hRI.
Here, we report the atomic structure of the crystalline hRI·RNase 1 complex. We use this structure to design RNase 1 variants that reveal the contribution of specific residues to the affinity for hRI and to design a variant that has micromolar (rather than femtomolar) affinity for hRI. This variant is toxic to human erythroleukemia cells. Our findings cause us to reevaluate the stability of the wild-type hRI·RNase 1 complex, which we find to have a Kd value that is nearly 103-fold lower than any reported previously. Overall, this work highlights the structural basis for intraspecies regulation of ribonucleolytic activity as well as facilitates the development of chemotherapeutic agents based on human ribonucleases.
Results
Important interactions between hRI and RNase 1
The three-dimensional crystal structure of the hRI·RNase 1 complex was refined to an R cryst value of 0.175 (Rfree = 0.236) at a resolution of 1.95 Å (Table 1). The asymmetric unit of the crystal of the hRI·RNase 1 complex resembles that of the hRI·angiogenin complex in its containing two molecular complexes (Figure 1).14 The two complexes are held together by 24 residue-to-residue hydrogen bonds formed between the N-terminal β-strand of the two hRI molecules. This dimerization of the hRI molecules buries an additional 1700 Å2 of surface area.
Table 1.
Crystallographic, data processing, and refinement statistics; values in parentheses refer to the highest resolution shell
| Data Collection Statistics | |
| Space group | P212121 |
| Unit cell parameters | a=71.338, b=107.546, c=155.036 |
| alpha beta gamma | 90.00 90.00 90.00 |
| X-ray energy (keV) | 12.399 |
| X-ray wavelength (Å) | 0.99997 |
| Overall resolution range (Å) | 47.17-1.95 (2.00-1.95) |
| Number of reflections | measured 573,939, unique 84,446 |
| Completeness (%) | 97.0 (72.6) |
| Rmerge a | 0.078 (0.424) |
| Redundancy | 6.8 (3.6) |
| Mean I/σ (I) | 16.96 (2.94) |
| Phasing | |
| MR correlation coefficient (MOLREP)d | 0.223 |
| MR modeld | 1DFJ |
| Refinement and Model Statistics from REFMAC 5.2.0005 | |
| Number of reflections (total) | 80,141 |
| Rcryst b (Rfree c) | 0.175 (0.236) |
| RMSD bonds (Å) | 0.016 |
| RMSD angles (°) | 1.515 |
| ESU based on Rfree (Å)d | 0.166 |
| Average B factor (Å2) | 28.04 |
| Number of water molecules | 854 |
| Ramachandran plot | |
| Residues in most favorable region | 86.8% |
| Residues in additional allowed region | 12.8% |
| Residues in generously allowed region | 0.4% |
| Residues in disallowed region | 0.0% |
, where Ii(h) is the intensity of an individual measurement of the reflection and <I(h)> is the mean intensity of the reflection.
, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
Rfree was calculated as Rcryst using 5.0% of the randomly selected unique reflections that were omitted from structure refinement.
Abbreviations used: MR, molecular replacement; ESU, estimated standard uncertainty.
Figure 1.
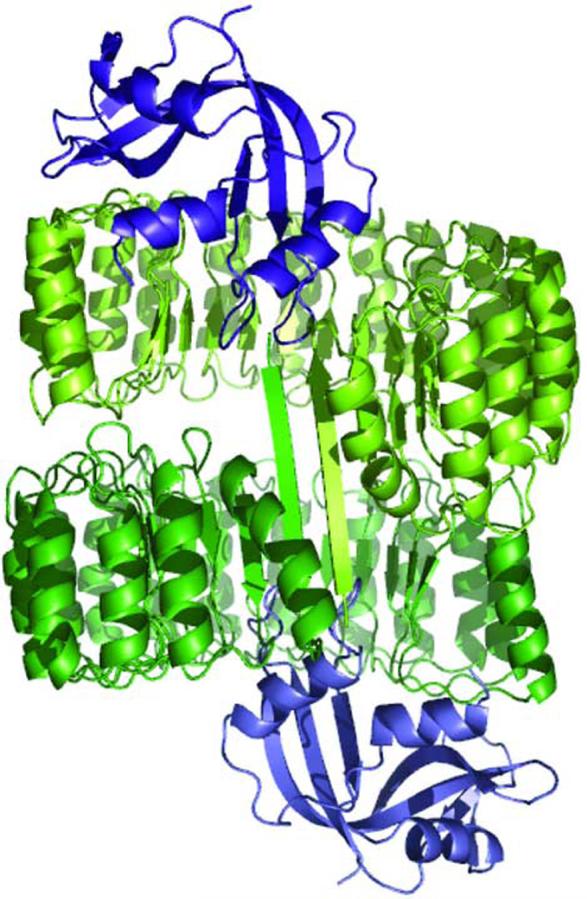
Structure of the crystalline complex of hRI (green) and RNase 1 (blue). Ribbon diagram of the contents of the asymmetric unit in which the N-terminal β-strands of hRI from two molecular complexes form an anti-parallel β-sheet. The image was created with the program PyMOL (DeLano Scientific, South San Francisco, CA).
In chain X of RNase 1 (Figure 2a), a bound citrate molecule forms hydrogen bonds to all three of the key catalytic residues in the enzymic active site (His12, Lys41, and His119). The bound citrate perturbs the substrate binding cleft of RNase 1, causing Arg10 and Lys66 to undergo conformational changes. To exclude the effect of citrate, the complex between chain Z of RNase 1 (without citrate bound) and chain Y of hRI will serve herein for comparisons to the structure of the pRI·RNase A complex.26
Figure 2.
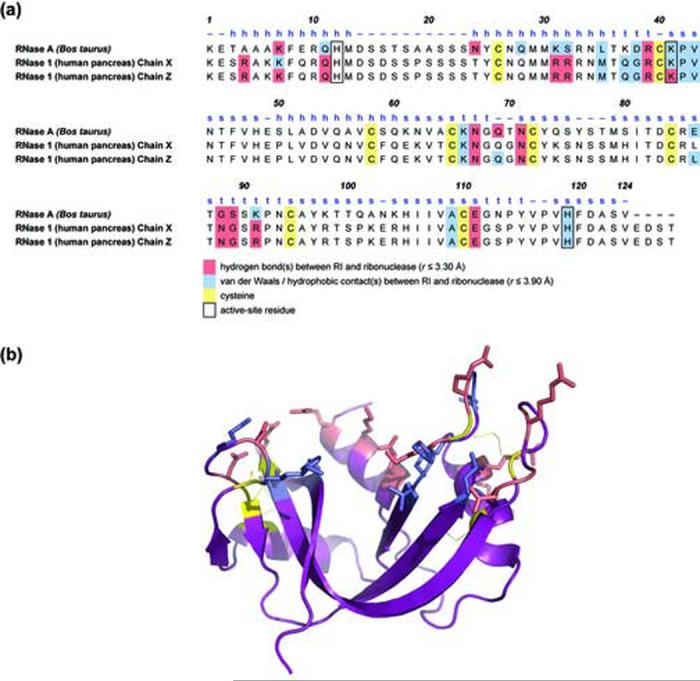
Contact residues from the crystal structures of hRI·RNase 1 and pRI·RNase A. (a) Amino acid sequence alignment of RNase A and RNase 1. hRI contacts for both chains of RNase 1 in the unit cell are given. The secondary structure of RNase A is identified with h (α-helix), s (β-strand), or t (turn). Residues in van der Waals or hydrophobic contact with RI are in blue. Residues with a hydrogen bond to RI are in red. Conserved cysteine residues are in yellow. Catalytic residues are enclosed in black boxes. (b) Three-dimensional structure of RNase 1, chain Z. Residues are colored using the same scheme as in panel a, except that active-site residues are not highlighted. The image was created with the program PyMOL.
The root-mean-square deviation (rmsd) between the alpha carbons of hRI·RNase 1 and pRI·RNase A is 2.8 Å.26 Much of the deviation between the complexes originates from the structural variation between hRI and pRI (rmsd = 1.6 Å) because pRI, unlike hRI, was observed to undergo a conformational change upon ribonuclease binding. The alpha carbons of RNase 1 and RNase A have less deviation (rmsd = 0.6 Å). In contrast, angiogenin and eosinophil-derived neurotoxin (EDN), the other human ribonucleases that have been co-crystallized with hRI, have rmsd values of 7.4 and 6.3 Å from RNase 1, respectively,14,27 underscoring the similarity between RNase 1 and RNase A.
The conservation of contact residues between the hRI·RNase 1 and pRI·RNase A complexes is shown in Figure 2a. The location of hRI-contact residues on the substrate-binding face of RNase 1 is shown in Figure 2b. The manner in which RI recognizes RNase 1 and RNase A is similar. This similarity is evident from the superposition of the β4-β5 loop of RNase 1 and RNase A in the two complexes, as shown in Figure 3. Moreover, the total number of RI-contact residues (23) is conserved in RNase 1 and RNase A.26 In RNase 1, however, 13 residues form 19 hydrogen bonds with hRI, whereas 9 residues in RNase A form 11 hydrogen bonds with pRI (Table 2). Overall, the 19 hydrogen bonds observed between hRI and RNase 1 are shorter than the analogous ones between pRI and RNase A (Table 2). Likewise, 375 Å2 more surface area is buried in the hRI·RNase 1 complex (2800 Å2 versus 2425 Å2), indicative of a more intimate complex.
Figure 3.
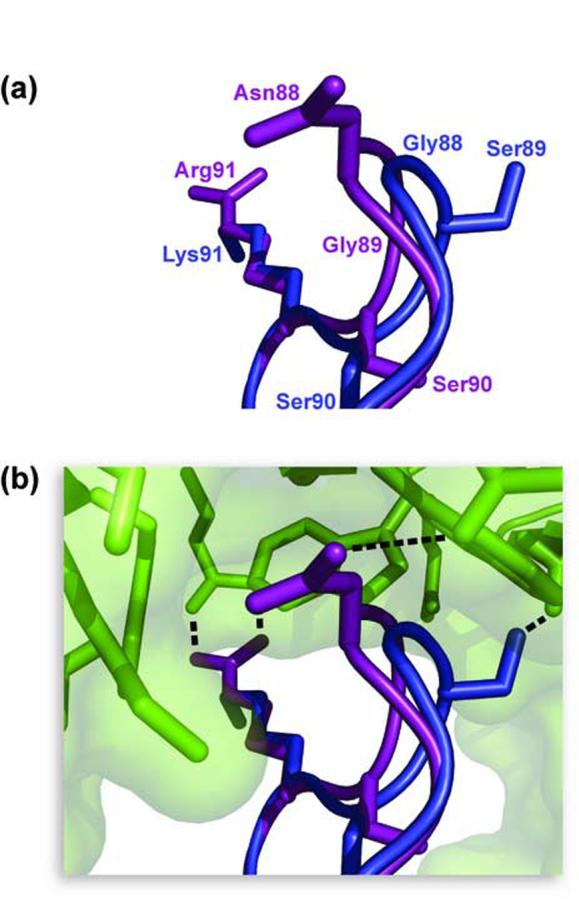
Comparison of the β4-β5 loop of RNase 1 (purple) and RNase A (blue) when bound to RI (green). α-Carbon atoms of RNase 1 and RNase A were superimposed with the program Sequoia, and images were created with the program PyMOL. (a) Structure of the β4-β5 loops. Sidechains of residues 88-91 are shown as sticks. Amino acids are labeled with the color corresponding to the color of the ribonuclease. (b) Orientation of the β4-β5 loops bound to RI. A model of RNase A (PDB entry 1DFJ, chain E) bound to hRI (chain Y) was created based on its alignment to RNase 1 (chain Z). Hydrogen bonds are shown as dotted lines. Hydrogen bonds between hRI and RNase A are hypothesized based on the alignment of hRI (chain Y) and pRI (chain I).
Table 2.
Intermolecular hydrogen bonds (rX···X < 3.35 Å) in the structures of the crystalline hRI·RNase 1 and pRI·RNase A complexesa
| PDB Entry 1Z7X (Chains Y and Z) |
PDB Entry 1DFJ (Chains E and I) |
||||
|---|---|---|---|---|---|
| Residue |
Residue |
||||
| RNase 1 | hRI | r (Å) | RNase A | pRI | r (Å) |
| Arg4 | Trp438 | 2.74 | — | — | — |
| Arg4 | Trp438 | 2.91 | — | — | — |
| Lys7 | Ser460 | 2.80 | Lys7 | Ser456 | 3.20b |
| Gln11 | Ser460 | 2.94 | — | — | — |
| — | — | — | Asn24 | Asp117 | 2.72 |
| Arg31 | Gln10 | 2.79 | — | — | — |
| Arg31 | Asp35 | 2.82 | — | — | — |
| Arg32 | Asp36 | 2.67 | — | — | — |
| Arg32 | Asp36 | 2.74 | — | — | — |
| Arg39 | Tyr434 | 2.74 | — | — | — |
| Arg39 | Glu401 | 2.76 | Arg39 | Glu397 | 2.32 |
| Arg39 | Glu401 | 3.03 | — | — | — |
| Lys41 | Asp435 | 2.57 | — | — | — |
| Asn67 | Tyr437 | 2.87 | Asn67 | Val405 | 2.81 |
| — | — | — | Asn67 | Val405 | 2.83 |
| — | — | — | Gln69 | Asp407 | 3.19 |
| Asn71 | Tyr437 | 2.75 | Asn71 | Tyr437 | 2.36 |
| Asn88 | Glu264 | 2.71 | Gly88 | Trp257 | 3.24 |
| Gly89 | Trp261 | 2.97 | Ser89 | Glu202 | 2.97 |
| Arg91 | Glu287 | 2.61 | — | — | — |
| Arg91 | Glu287 | 2.93 | — | — | — |
| — | — | — | Glu111 | Glu436 | 2.89 |
| Glu111 | Tyr437 | 2.58 | Glu111 | Tyr433 | 3.00 |
Hydrogen bonds were identified using the program PDBsum.72
No definitive electron density.
Previous studies on the interaction of RNase A and BS-RNase with RI focused on three structural regions that are remote from the active site: residues 38/39, residue 67, and residues in the β4-β5 loop.20,17 The hydrogen bonding network and electron density of these regions in the hRI·RNase 1 complex are depicted in Figures 3 and 4. Based on this information, two variants of RNase 1 were designed in which residues were replaced in all three of these regions. One variant (G38R/R39G/N67R/N88R RNase 1) mimics the most cytotoxic of known RNase A variants (D38R/R39D/N67R/G88R RNase A) by swapping the amino acids in positions 38/39 and installing arginine residues at positions 67 and 88. The other RNase 1 variant (R39D/N67D/N88A/G89D/R91D RNase 1) is focused on the same regions, but instead uses Coulombic repulsion to suppress the binding of RI.
Figure 4.
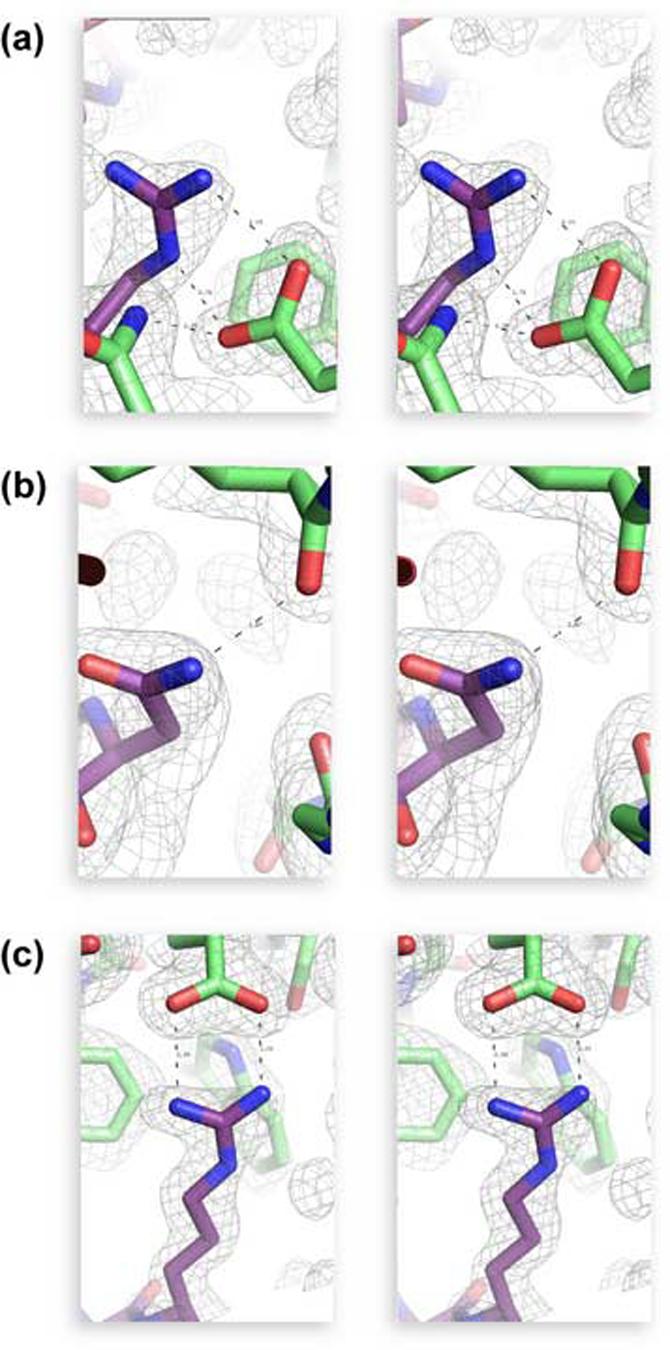
Electron density at 1σ (2Fobs - Fcalc) of key contact residues between hRI (green) and RNase 1 (purple). Specific residues shown in detail are (a) Arg39, (b) Asn67, and (c) Arg91. Highlighted regions are shown in wall-eyed stereo; interprotein hydrogen bonds are displayed by black dotted lines. Images were created with the program PyMOL.
Ribonucleolytic activity
The ability of a ribonuclease to cleave RNA in the presence of RI correlates closely with its cytotoxicity in vitro.28 For a ribonuclease variant to achieve its full cytotoxic potential, an amino-acid substitution that decreases RI binding must not be detrimental to catalytic activity.29 Consequently, variants of RNase 1 were assayed for their catalytic activity toward a tetranucleotide substrate in buffer that lacks oligo(vinylsulfonic acid), a potent inhibitor of ribonucleolytic activity.30,31 Values of kcat/KM for RNase A, RNase 1, and their variants are given in Table 3. The kcat/KM value for wild-type RNase 1 is 10-fold higher than that reported previously.24 A similar increase in the catalytic activity was observed for RNase A when contaminating oligo(vinylsulfonic acid) was removed from the reaction buffer.31 The kcat/KM values for the RNase 1 variants are within 6-fold of that for the wild-type enzyme.
Table 3.
Biochemical parameters of RNase 1, RNase A, and their variants
| Ribonuclease | Tm a (°C) | kcat/KMb (106 M-1s-1) | Kdc (nM) | ΔGd (kcal/mol) | ΔΔGe (kcal/mol) | IC50f (μM) | Z |
|---|---|---|---|---|---|---|---|
| Wild-type RNase A | 64g | 52 ± 4g | 44×10-6h | >25 | +4 | ||
| D38R/R39D/N67R/G88R RNase A | 56g | 38 ± 6g | 510 ± 30g | 0.15 ± 0.01 | +6 | ||
| Wild-type RNase 1 | 57 | 21 ± 2 | 29×10-8i | >25 | +6 | ||
| G38R/R39G/N67R/N88R RNase 1 | 61 | 4.2 ± 0.4 | 0.032 ± 0.016 | 6.9 | >25 | +8 | |
| R39D/N67D/N88A/G89D/R91D RNase 1 | 58 | 6.3 ± 0.5 | (1.7 ± 0.5)×103 | 13.3 | 13.3 ± 1.7 | 0 | |
| R39L/N67L/N88A/G89L/R91L RNase1 | 65 | 30 ± 3 | 30 ± 1 | 10.9 | 2.4 | >25 | +4 |
| N67D/N88A/G89D/R91D RNase 1 | 51 | 16 ± 6 | 45 ± 15 | 11.2 | 2.1 | >25 | +2 |
| R39D/N88A/G89D/R91D RNase 1 | 57 | 10 ± 3 | 68 ± 8 | 11.4 | 1.9 | >25 | +1 |
| R39D/N67D/G89D/R91D RNase 1 | 54 | 3.3 ± 0.5 | (1.0 ± 0.1)×103 | 13.0 | 0.3 | >25 | 0 |
| R39D/N67D/N88A/R91D RNase 1 | 51 | 10 ± 1 | 278 ± 50 | 12.2 | 1.1 | >25 | +1 |
| R39D/N67D/N88A/G89D RNase 1 | 57 | 5 ± 1 | 16 ± 3 | 10.6 | 2.7 | >25 | +2 |
Values of Tm (±2 °C) for RNase 1 and its variants were determined in PBS by UV spectroscopy.
Values of kcat/KM (±SE) were determined for catalysis of 6-FAM-dArU(dA)2-6-TAMRA cleavage at 25 °C in 0.10 M MES-NaOH buffer (OVS-free), pH 6.0, containing 0.10 M NaCl (9).
Values of Kd (±SE) were determined for the complex with hRI at 25 °C (10).
Values of ΔG were calculated with the equation: ΔG = -RTln(Kd wild-type/Kd variant).
Values of ΔΔG = ΔΔGR39D/N67D/N88A/G89D/R91D RNase 1 - ΔΔGRNase 1 variant.
Values for IC50 (±SE) are for the incorporation of [methyl-3 H]thymidine into the DNA of K-562 cells treated with the ribonuclease, and were calculated with equation (4).
From ref 17.
From ref 12.
Conformational stability
The conformational stability of a ribonuclease is linked to its susceptibility to proteolysis and, consequently, its cytotoxicity.32 The Tm values for all RNase 1 variants are shown in Table 3. The Tm value of wild-type RNase 1 is close to that reported previously.24 In agreement with previous studies, incorporation of charged patches on the surface of RNase 1 does not reduce the Tm value by more than 6 °C.6 Neither arginine nor aspartate substitutions at residues 38/39, residue 67, or residues in the β4-β5 loop disturb the conformational stability significantly, as G38R/R39G/N67R/N88R RNase 1 and R39D/N67D/N88A/G89D/R91D RNase 1 have Tm values comparable to that of wild-type RNase 1 (61 and 58 °C, respectively). Thus, all of the variants have high conformational stability at physiological temperature.
Evasion of ribonuclease inhibitor
RI binds multiple members of the RNase A superfamily with equilibrium dissociation constants in the femtomolar range,12 forming one of the tightest known noncovalent interactions among biomolecules. Replacing residues 38, 39, 67, and 88 in RNase A (D38R/R39D/N67R/G88R RNase A) increased the equilibrium dissociation constant of the hRI·RNase A complex by seven orders of magnitude (Table 3).17 In marked contrast, the analogous variant in RNase 1 (G38R/R39G/N67R/N88R RNase 1) had an affinity for RI near that reported previously for wild-type RNase 1 (Table 3).13,33,23
This surprising observation led us to reinvestigate the affinity of wild-type RNase 1 for hRI. Previously, the stability of the hRI·RNase 1 complex had been determined by measuring the inhibition of catalytic activity. The Ki values (2 × 10-11, 5.2 × 10-12, or 2 × 10-13 M (ref 13,33,23)) obtained by this method are lower than the concentration of wild-type RNase 1 used in the experiment itself. Accordingly, these values can only be an upper limit to the true Kd value.34,24
We determined the dissociation rate of the hRI·RNase 1 complex by monitoring the release of fluorescently-labeled RNase 1 over time (Figure 5). We found that the hRI·RNase 1 complex (Table 4) has a half-life (t½ = ln2/kd) of 81 days, which puts its value of kd closer to that of the hRI·angiogenin complex (t½ = 62 days) than that of the hRI·RNase A complex (t½ = 13 h).12 To calculate the value of Kd of the hRI·RNase 1 complex, the value of ka was assumed to be similar to that for the association of hRI with angiogenin or RNase A. These ka values are within twofold of each other, and are close to the diffusion limit.34,12 The ka value of RNase A was assumed herein for wild-type RNase 1 due to the similarity of the sequence and structure of RNase A with those of RNase 1. Using k = 3.4 × 108 M-1 -1 35 as, the value of Kd (= kd/ka) for the hRI·RNase 1 complex was then calculated to be 2.9 × 10-16 M, which is at least 690-fold lower than any reported previously for the hRI·RNase 1 complex,13,33,23 150-fold lower than that for the hRI·RNase A complex and comparable to that of the hRI·angiogenin complex (7.1 × 10-16 M).12
Figure 5.
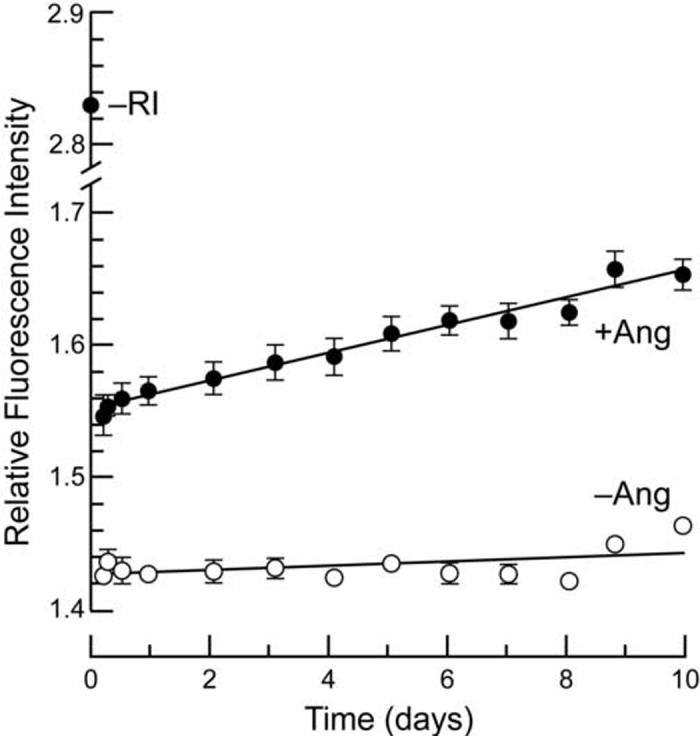
Dissociation rate of the complex between wild-type RNase 1 and hRI. The release of diethylfluorescein-labeled RNase 1 (100 nM) from hRI (100 nM) was followed over time after addition of a 50-fold molar excess of angiogenin (5 μM) (•) or after addition of an identical volume of PBS (○). The initial fluorescence of unbound RNase 1 was used as the end-point for complete RNase 1 release. Data points are the mean (±SE) of six separate measurements and are normalized for the average fluorescence of rhodamine 110 (10 nM).
Table 4.
Rate constants for the binding of hRI to RNase 1 and its variants
| RNase 1 | kd (s-1) | ka (M-1 s-1) |
|---|---|---|
| wild-type | (9.9 ± 0.7) ×10-8 a | 3.4×108 b |
| R39D/N67D/N88A/G89D/R91D | 0.22 ± 0.03 c (2.2×106) | 1.2 × 105 d (2.8×103) |
| R39L/N67L/N88A/G89L/R91L | 0.092 ± 0.003 c (9.3×105) | 3.1 × 106 d (1.1×102) |
The value of kd (±SE) was determined by following the release of diethylfluorescein- labeled RNase 1 from hRI over time and fitting the curve to equation (3).
The value of ka is from ref 35. Both angiogenin and RNase A have association rates within twofold of each other,35 and so the association rate of RNase A was chosen for comparison due to the greater similarity of its sequence and structure with those of RNase 1.
Values of kd (±SE) were determined by following the release of a fluorescein-labeled RNase 1 variant from hRI over time and fitting the curves to equation (3). Values in parentheses represent the fold increase from wild-type RNase 1.
Values of ka were calculated with the equation: Kd = kd/ka. Numbers in parentheses represent the fold decrease from wild-type RNase 1.
Designing a variant to overcome the extraordinary stability of the hRI·RNase 1 complex required a new strategy. Instead of inserting sterically bulky residues at the complementarity regions, multiple aspartate residues and one alanine residue were substituted in the same regions. The value of Kd = 1.7 μM for the resulting variant, R39D/N67D/N88A/G89D/R91D RNase 1, is close to the highest measured for any RNase A variant (2.9 μM),17 despite the high stability of the wild-type complex. These substitutions reduced the affinity of hRI for RNase 1 by 5 × 109-fold, which is the greatest evasion yet reported for a mammalian ribonuclease. When the aspartate substitutions in R39D/N67D/N88A/G89D/R91D RNase 1 are replaced with an isologous amino acid, leucine (R39L/N67L/N88A/G89L/R91L RNase 1), the variant loses 50-fold in RI evasion.
The influence of electrostatics on the binding of hRI to RNase 1 variants was analyzed by determining the value of kd for the complexes formed by hRI and these two RNase 1 variants (Table 4). The value of kd increases by 9 × 105-fold upon substitution of R39L/N67L/N88A/G89L/R91L. Yet, the value of kd increases by only twofold upon aspartate substitution (R39D/N67D/N88A/G89D/R91D RNase 1). The values of ka for these two variants were calculated by using values of Kd from Table 3, values of kd from Table 4, and equation (1). The association rate constant is affected more substantially by both leucine substitution (110-fold decrease) and aspartate substitutions (25-fold decrease). The substantial change in the value of ka with both leucine and aspartate substitution demonstrates a contribution gained by both the loss of attractive forces (leucine substitution) and gain of repulsive forces (aspartate substitution). Overall, the 5 × 109-fold decrease in binding affinity of hRI for R39D/N67D/N88A/G89D/R91D RNase 1 contains significant contributions from both an increase in the value of kd (2.2 × 106-fold) and a decrease in that of k (2.7 × 103 a -fold).
The impact of individual substitutions in R39D/N67D/N88A/G89D/R91D RNase 1 to its overall binding constant for hRI was elucidated by the reversion of each substitution in R39D/N67D/N88A/G89D/R91D RNase 1 to the residue in the wild-type enzyme. In this analysis, single substitutions with little impact on RI affinity had small values of ΔΔG (Table 3). The energetic contributions of the substitutions increased in the order: N88A < G89D < N67D < R39D < R91D. In this analysis, installing an aspartate residue at position 91 contributed 2.7 kcal/mol to evasion, whereas an alanine residue at position 88 contributed only 0.3 kcal/mol.
Molecular charge
The cytotoxicity of a ribonuclease is modulated by its molecular charge.36,20,17,37 This interplay is apparent in the data shown in Figure 6 and listed in Table 3. D38R/R39D/N67R/G88R RNase A (Z = +6) and R39D/N67D/N88A/G89D/R91D RNase 1 (Z = 0) have similar conformational stability, ribonucleolytic activity, and affinity for RI, but their IC50 values for K-562 cells differ by 87-fold (Table 3). R39D/N67D/N88A/G89D/R91D RNase 1 has an IC50 of only 13.3 μM, making it more toxic than wild-type RNase 1, but twofold less toxic than G88R RNase A.21
Figure 6.
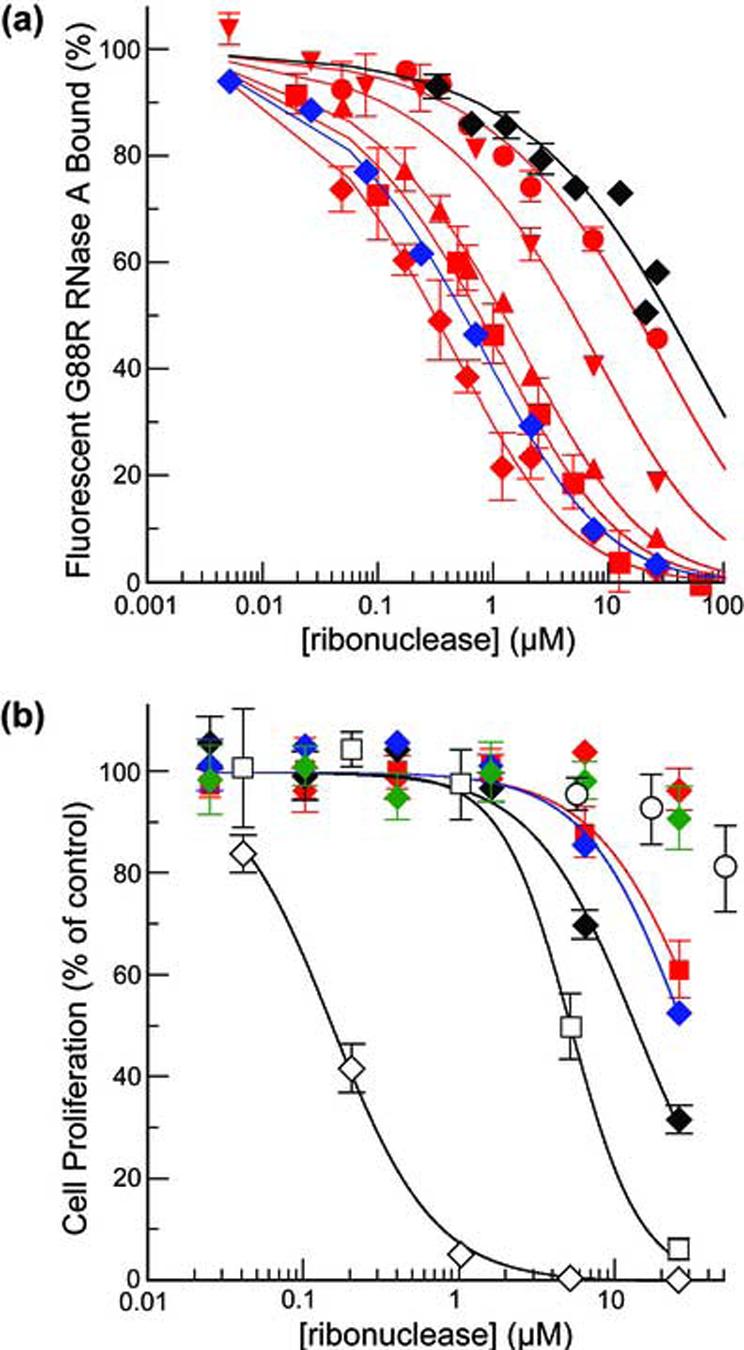
Affinity for hRI and cytotoxic activity of ribonucleases. (a) Affinity of hRI for RNase 1 variants was determined by using a competition assay with fluorescein-labeled A19C/G88R RNase A (50 nM). Data points are the mean (±SE) of at least three separate measurements. RNase 1 variants in order of decreasing affinity for hRI: R39D/N67D/N88A/G89D (red ◆), R39L/N67L/N88A/G89L/R91L (blue ◆), N67D/N88A/G89D/R91D (red ■), R39D/N88A/G89D/R91D (red ▲), R39D/N67D/N88A/R91D (red ▼), R39D/N67D/G89D/R91D (red •), and R39D/N67D/N88A/G89D/R91D (◆). (b) Effect of ribonucleases on the proliferation of K-562 cells was determined by monitoring the incorporation of [methyl-3H]thymidine into cellular DNA in the presence of ribonucleases. Data points are the mean (±SE) of at least three separate experiments carried out in triplicate. Ribonucleases in order of decreasing cytotoxic activity: D38R/R39D/N67R/G88R RNase A (◇), G88R RNase A (□), R39D/N67D/N88A/G89D/R91D RNase 1 (◆), R39L/N67L/N88A/G89L/R91L RNase 1 (blue ◆), and N67D/N88A/G89D/R91D RNase 1 (red ■). R39D/N67D/N88A/G89D RNase 1 (red ◆), G38R/R39G/N67R/N88R RNase 1 (green ◆), wild-type RNase 1 (○), and other variants of RNase 1 (not shown) have negligible cytotoxic activity.
The IC50 values for all of the other variants of RNase 1 listed in Table 3 fall outside the measurable range of the assay (IC50 > 25 μM). R39L/N67L/N88A/G89L/R91L RNase 1 and N67D/N88A/G89D/R91D RNase 1 killed approximately 60% of the K-562 cells at 25 μM (Figure 6b), indicative of IC50 values only slightly above 25 μM. The lack of cytotoxicity for RNase 1 variants other than R39D/N67D/N88A/G89D/R91D RNase 1 is likely due to an increased affinity for RI when compared to R39D/N67D/N88A/G89D/R91D RNase 1 (Table 3).
Discussion
Ribonuclease inhibitor is a 50-kDa resident of the cytosol that comprises 0.1% of all cellular proteins.38,39 RI serves as a sentry for rogue ribonucleases and through its horseshoe-shaped binding surface, inhibits the ribonucleolytic activity of members of the RNase A superfamily.26,14,27 RI manifests this control despite the low sequence identity between RNase A family members,40 which are evolving rapidly.41 The structural basis for the interaction of RI with four RNase A family members (RNase 1, RNase A, angiogenin, and EDN) has been determined by Deisenhofer, Acharya, and their coworkers.26,14,27 A thorough comparison of the recognition by RI of structurally diverse ribonucleases (angiogenin and EDN) has been reported previously.27 Here, we focus on recognition of RNase 1 and RNase A, two ribonucleases with high sequence identity.
Recognition of RNase 1 by RI
The fast atomic density evaluator (FADE) algorithm revealed regions of high shape-complementarity in the pRI·RNase A complex.42,17 By inserting disruptive substitutions in those regions, D38R/R39D/N67R/G88R RNase A (K = 510 nM)17 d and C31A/C32A/G38R/K39D/G88R BS-RNase (K = 110 nM)20d were developed, and each was found to have significantly decreased affinity for hRI. Using the same reasoning, we designed G38R/R39G/N67R/N88R RNase 1 (Table 3). This variant of RNase 1, however, retained subnanomolar affinity for hRI and was not cytotoxic to K-562 cells (Figure 6). Consequently, we sought to reveal the distinguishing aspects of the recognition of RNase 1 and RNase A by hRI.
Hydrogen bonds play an important role in the stability of protein·protein complexes, as well as the conformational stability of individual proteins.43-46 Hydrogen bonds between functional groups of opposite charge (i.e., salt bridges) are especially strong.47 Overall, the hRI·RNase 1 complex has both more intermolecular hydrogen bonds than does the pRI·RNase A complex and more between residues of opposite charge (Table 2). Moreover, those hydrogen bonds that are conserved are shorter in the hRI·RNase 1 complex. The hRI·RNase 1 complex also buries an additional 375 Å2 of surface area, which could also enhance its stability.48,45,49 In line with the more intimate complex formed by hRI and RNase 1, RNase 1 dissociates from hRI 150-fold more slowly than does RNase A from hRI (Figure 5; Table 3).12 The slower dissociation rate (t½ = 81 days) for the hRI·RNase 1 complex is in the range of that for the hRI·angiogenin complex (t½ = 62 days).12 Accordingly, hRI seems to have coevolved with human ribonucleases in a manner that would enable hRI to abrogate more effectively any lethal ribonucleolytic activity. The greater stability of the hRI·RNase 1 complex also explains why the design of RI-evasive RNase 1 variants has been more difficult than the design of RI-evasive RNase A variants.23-25
To characterize differential binding within the hRI·RNase 1 and pRI·RNase A complexes, we investigated the energetic contribution of the residues in RNase 1 that are analogous to the residues in high shape-complementarity regions of RNase A.17 These residues are Arg39, Asn67, and residues in the β4-β5 loop.
Residue 39
Arg39 of RNase A had the highest shape complementarity score of any RNase A residue in the pRI·RNase A complex17 and was proposed to be a secondary anchor residue.50 When Arg39 was replaced with an aspartate residue in G88R RNase A to create R39D/G88R RNase A, the R39D substitution instilled 725-fold lower RI affinity. In RNase 1, Arg39 interacts even more extensively with hRI, forming 2 hydrogen bonds that are absent in the pRI·RNase A complex (Figure 4a). Specifically, Arg39 of RNase 1 makes a bidentate hydrogen bond with the side chain of Glu401 and a main-chain hydrogen bond with the main chain of Tyr434. Consequently, the energetic consequence of the R39D substitution (ΔΔG = 2.1 kcal/mol; Table 3) is the second highest among the residues studied herein.
Residue 67
Interactions with Asn67 had been exploited previously to develop RI variants that bind to angiogenin but not to RNase 1 or RNase A. Incorporating a tryptophan at positions 408 and 410 hindered the interaction of hRI with Asn67 of RNase 1 and RNase A, yielding a variant of hRI that bound only to angiogenin.22 Asn67 had been proposed to be a primary anchor residue in the pRI-RNase A interface, due to its burial of surface area and its lack of molecular motion.50 Asn67 in RNase 1 forms a main-chain hydrogen bond to Tyr437 (Table 2) and is in van der Waals contact with Leu409 and Gly410 of hRI. We find, however, that replacing Asn67 to hinder interaction with residues 408-410 in hRI does not produce a large effect on binding, as an aspartate residue at position 67 only destabilizes the complex by ΔΔG = 1.9 kcal/mol (Table 3). Hence, Arg39 and Arg91 provide more energetic stabilization than does Asn67 to the hRI·RNase 1 complex.
β4-β5 loop
Replacing Gly88 with an arginine residue in the β4-β5 loop of RNase A had been found previously to decrease the affinity of pRI for RNase A by 104-fold21 and the affinity of BS-RNase for hRI by 250-fold.21,20 In the crystalline hRI·RNase 1 complex, the β4-β5 loop adopts a similar conformation to that of RNase A in the pRI·RNase A complex (Figure 3). One major difference is with residue 88, as Asn88 of RNase 1 forms a hydrogen bond with Glu264 instead of residing in the pocket formed by Trp261 and Trp263. (The numbering of RI residues herein refers to their position in hRI, which has four more N-terminal residues than does pRI.) Asn88 in RNase 1 is located on the outer surface of the hRI-RNase 1 interface. This location seems to be able to accommodate the bulk of an arginine residue23 (or even a carbohydrate chain51) while maintaining high affinity for RI.
Gly89 of RNase 1 has been proposed to constitute the structural analogue of Gly88 in RNase A.52,53 Yet, replacing Gly89 with arginine in RNase 1 does not yield a cytotoxic variant.21,23,25 Gly89 in RNase 1 overlays more closely with Ser89 in RNase A (Figure 3), but Gly89 is unable to form a hydrogen bond with Glu206, as does Ser89 in RNase A.26 Gly89 in RNase 1 is within van der Waals distance of Trp261 and Trp263 in hRI·RNase 1, but due to the increased stability of the hRI·RNase 1 complex, a single G89R substitution does not lower the affinity enough to make this variant cytotoxic.23
Among the five residues investigated, Arg91 had the greatest influence on the stability of the hRI·RNase 1 complex (ΔΔG = 2.7 kcal/mol, Table 3). Arg91 makes contact with hRI in the concave anionic surface of hRI (Figure 7), forming two hydrogen bonds with Glu287. Lys91 has been proposed to play a secondary role in anchoring RNase A to pRI,50 but Arg91 of RNase 1 could serve as a primary targeting residue in binding to hRI. Replacing Arg91 with an aspartate residue severs the hydrogen bonds formed with Glu287 of hRI, and replaces a favorable Coulombic interaction with an unfavorable one.
Figure 7.
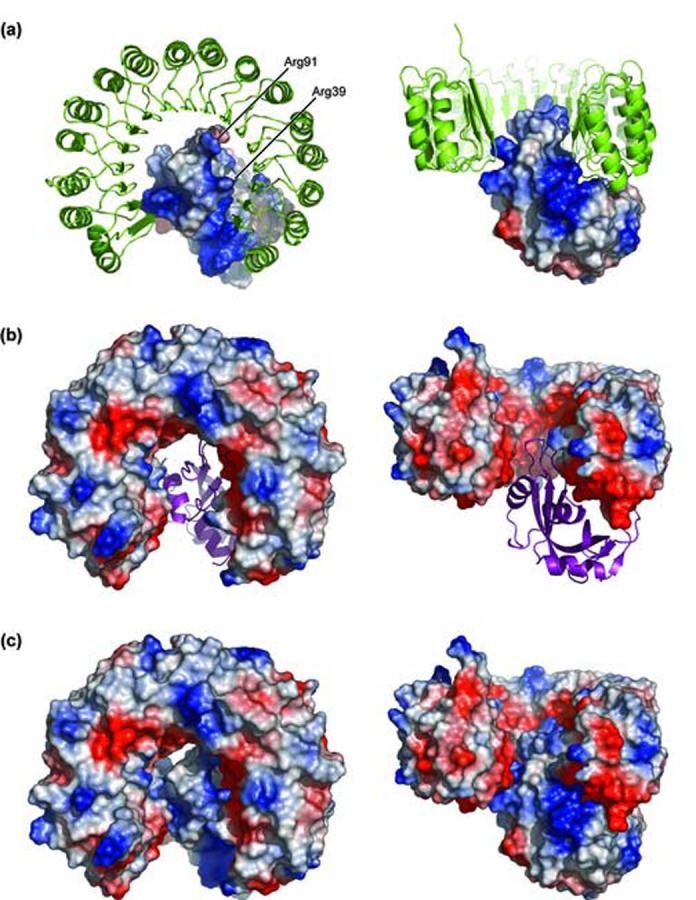
Electrostatic representation of the interaction between hRI (green) and RNase 1 (purple). Protein contact potential of RNase 1 (a), hRI (b), and the hRI·RNase 1 complex (c) are shown. Residues 39 and 91 are labeled in panel a. The intensity of the blue (positive) and red (negative) coloration is indicative of the local electrostatic environment. Vacuum electrostatics were calculated and images were created with the program PyMOL.
Energetics of RI-evasion
Charged amino acids comprise 19% of all exposed amino acids on a protein surface.54 Fewer charged residues are exposed, however, in the average protein-protein interface. Charge-charge interactions in protein-protein interfaces are disfavored by the large energetic penalty incurred by desolvation of the exposed charge upon binding.8,9 The energetic penalty of desolvation can be circumvented by leaving key charged residues partially exposed to solvent upon complex formation.9 In Figure 4, the electron density for multiple solvent molecules is visible surrounding favorable Coulombic interactions between hRI and RNase 1. This residual exposure to solvent diminishes the energetic desolvation penalty and allows electrostatics to remain a driving force to complex formation.
The cationicity of the RNase 1 surface (Figure 7) facilitates the binding of its anionic substrates.55 RI exploits this cationicity to inhibit RNase 1 rapidly and strongly using long-range Coulombic interactions.15 In a theoretical study of 14 enzyme·inhibitor complexes, all were found to be suffer a net destabilization by electrostatic interactions, due to the energetic penalty of breaking hydrogen bonds to water in the unbound state.8 RI·ribonuclease complexes are, however, atypical.50 The hRI·angiogenin complex has ΔU = -12.3 kcal/mol and a calculated rate increase due to electrostatics of 106 M-1s-1.15 Figure 7 highlights the distribution of positive and negative charges on hRI and RNase 1. In these highly charged proteins, the favorable electrostatic interaction of key solvent-exposed charged residues like Arg39 and Arg91 of RNase 1 appear to drive complex formation (Figures 4 and 7).
Arg39 and Arg91 contribute more free energy to complex formation than do Asn67, Asn88, or Gly89 (Table 3). Because Arg39 and Arg91 are the only charged residues changed in R39L/N67L/N88A/G89L/R91L RNase 1 and electrostatics determine the association rate,5,6 data with R39L/N67L/N88A/G89L/R91L RNase 1 can be used to estimate the influence of Arg39 and Arg91 on the association rate. Replacing Arg39 and Arg91 with leucine residues to give R39L/N67L/N88A/G89L/R91L RNase 1 decreases the association rate by 110-fold (Table 4). Hence, Arg39 and Arg91 serve a special role in the hRI·RNase 1 complex, one that we define as “electrostatic targeting residues”. A residue that directs the formation of a protein-protein complex could also provide the major energetic force to maintain a stable complex. Arg39 and Arg91 fit these criteria, as they strongly affect the association rate of the complex (Table 4). Then, Arg39 and Arg91 keep RNase 1 bound to hRI through multiple hydrogen bonds with anionic residues (Figure 4), allowing other contacts in the complex to form.
Rates of association and dissociation
Electrostatics can guide the formation of protein-protein complexes through space and can increase the rate of association beyond that of diffusion alone.5-7 We measured the association and dissociation rates for two of the RNase 1 variants listed in Table 3, thereby dissecting equation (1) into its component parts.15 Changes in the values of both kd (2.2 × 106-fold increase) and ka (2.8 × 103-fold decrease) make a substantial contribution to the ability of R39D/N67D/N88A/G89D/R91D RNase 1 to evade hRI (Table 4). The effect of the electrostatic forces that originate from charged residues 39, 67, 89, and 91 in this aspartate variant can be discerned with a comparison to the isologous leucine variant, R39L/N67L/N88A/G89L/R91L RNase 1. The decrease to the value of ka upon leucine substitution (110-fold from the loss of attractive forces) and then aspartate substitution (25-fold from the gain of repulsive forces) highlights the contribution of electrostatic forces to the stability of the hRI·RNase 1 complex and reinforces previous calculations on the importance of electrostatics in the affinity of ribonucleases for RI.15
RNase 1 as a chemotherapeutic agent
Ribonucleases show great promise as cancer chemotherapeutic agents.16 ONC, a homologue of RNase 1 from the Northern leopard frog, is currently in phase III clinical trials for the treatment of malignant mesothelioma.56 A chemotherapeutic agent based on a human ribonuclease have multiple advantages over ONC, including enhanced catalytic activity,17 decreased renal toxicity,57,58 and decreased immunogenicity.59 To endow ribonucleases that are not naturally cytotoxic with cytotoxic activity requires the consideration of multiple biochemical attributes, including ribonucleolytic activity, cationic charge, conformational stability, and (especially) RI evasion.29,17
Variants of RNase 1, unlike those of RNase A, have been difficult to engineer with decreased affinity for hRI.23,25 The data reported herein reveal the origin of this difficulty. The hRI·RNase 1 complex has a more extensive and tighter network of hydrogen bonds than does the pRI·RNase A complex (Table 2). This structural data is manifested in the stability of the hRI·RNase 1 complex, which has a subfemtomolar Kd value (Table 3), which is 150-fold less than that of the hRI·RNase A complex.12 Accordingly, cytotoxic variants of RNase 1 must overcome greater RI inhibition to become cytotoxic. We have overcome this barrier by designing a variant of RNase 1 with an affinity for RI in the micromolar range. R39D/N67D/N88A/G89D/R91D RNase 1 has a 5 × 109-fold decrease in affinity for RI, making it the most RI-evasive engineered ribonuclease reported to date. This RNase 1 variant, which has nearly wild-type ribonucleolytic activity and conformational stability, is cytotoxic (Figure 6; Table 3). Its cytotoxic activity is, however, likely compromised by its overall neutrality (Z = 0), as net charge is known to correlate with cytotoxic activity among ribonucleases with similar ribonucleolytic activity, conformational stability, and affinity for RI.17,37 The cytotoxicity of a ribonuclease can be increased, however, by adding nine arginine residues to the C-terminus.60 Accordingly, liberating RNase 1 from inhibition by hRI overcomes the major barrier to endowing RNase 1 with cytotoxic activity.
Conclusions
RI and its cognate ribonucleases represent a unique system for characterizing a protein·protein complex. Toward this end, we have examined the interaction of RNase 1, which is the human homologue of RNase A, and hRI. We find that the affinity of RNase 1 for hRI is subfemtomolar, indicative of the imperative of regulating ribonucleolytic activity in humans. By determining the three-dimensional structure of the hRI·RNase 1 complex at atomic resolution, we were able to reveal those residues that are responsible for its extraordinary stability. We were also able to design an RNase 1 variant that retains its ribonucleolytic activity and conformational stability but has only micromolar affinity for hRI. Arg39 and Arg91 of RNase 1 are especially important in this context. These two “electrostatic targeting residues” (1) substantially increase the association rate of the complex, and (2) form tight hydrogen bonds that maintain the complex. Together, these data provide detailed insight into one of the most stable protein·protein complexes, and represent a key step in the development of human ribonucleases as chemotherapeutic agents.
Materials and Methods
Materials
Escherichia coli strain BL21(DE3) was from Novagen (Madison, WI). 6-FAM-dArU(dA)2-6-TAMRA, a fluorogenic ribonuclease substrate, was from Integrated DNA Technologies (Coralville, IA). Enzymes were from Promega (Madison, WI). K-562 cells, which are an erythroleukemia cell line derived from a chronic myeloid leukemia patient, were from the American Type Culture Collection (Manassas, VA). Cell culture medium and supplements were from Invitrogen (Carlsbad, CA). [methyl-3H]thymidine (6.7 Ci/mmol) was from Perkin-Elmer (Boston, MA). Protein purification columns were from Amersham Biosciences (Piscataway, NJ). RNase A Type III-A was from Sigma-Aldrich (St. Louis, MO). Human angiogenin was prepared as described previously.61
MES buffer (Sigma-Aldrich, St. Louis, MO) was purified by anion-exchange chromatography to remove trace amounts of oligomeric vinylsuflonic acid.31,60 Costar 96-well NBS microtiter plates were from Corning Life Sciences (Acton, MA). Rhodamine 110 (sold as Rhodamine 560) was from Exciton (Dayton, OH). All other chemicals were of commercial grade or better, and were used without further purification.
Terrific Broth (TB) contained (in 1.00 L) tryptone (12 g), yeast extract (24 g), glycerol (4 mL), KH2PO4 (2.31 g), and K2HPO4 (12.54 g). Phosphate-buffered saline (PBS; pH 7.4) contained (in 1.00 L) NaCl (8.0 g), KCl (2.0 g), Na2HPO4·7H2O (1.15 g), KH2PO4 (2.0 g), and NaN3 (0.10 g).
Instrumentation
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry was performed with a Voyager-DE-PRO Biospectrometry Workstation (Applied Biosystems, Foster City, CA) at the Biophysics Instrumentation Facility. Fluorescence spectroscopy was performed with a QuantaMaster1 photon-counting fluorimeter equipped with sample stirring (Photon Technology International, South Brunswick, NJ). The fluorescence intensity in microtiter plates was recorded with a Perkin-Elmer EnVison 2100 plate reader equipped with an FITC filter set (excitation at 485 nm with 14 nm bandwidth; emission at 535 nm with 25 nm bandwidth; dichroic mirror cutoff at 505 nm) in the W.M. Keck Center for Chemical Genomics. Thermal denaturation data were collected using a Cary 3 double-beam spectrophotometer equipped with a Cary temperature-controller (Varian, Palo Alto, CA). [methyl-3H]thymidine incorporation into genomic DNA was quantified by liquid scintillation counting using a Microbeta TriLux liquid scintillation and luminescence counter (Perkin-Elmer, Wellesley, MA).
RNase 1 purification
DNA encoding variants of RNase 1 was created by using plasmid pHP-RNase24 and the Quikchange site-directed mutagenesis kit or Quikchange Multi site-directed mutagenesis kit (Stratagene, La Jolla, CA). RNase 1 and its variants were purified from inclusion bodies using the same oxidative folding procedure described previously.24 These proteins all lacked the C-terminal threonine residue (Thr128), which was absent from the initial report of the amino acid sequence.62
Variants of RNase 1 with a free cysteine residue at position 19 were initially protected by reaction with 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB).63,64 Immediately before fluorophore attachment, TNB-protected variants were deprotected using a three-fold molar excess of dithiothreitol (DTT) and desalted by chromatography using a PD-10 desalting column (Amersham Biosciences, Piscataway, NJ). Deprotected RNase 1 variants were reacted for 4-6 h at 25 °C with a ten-fold molar excess of 5-iodoacetamido fluorescein (Sigma-Aldrich, St. Louis, MO). Fluorescein-labeled RNase 1 variants were purified by chromatography using a HiTrap SP FF column.
The molecular mass of RNase 1, its variants, and conjugates was confirmed by MALDI-TOF mass spectrometry.
hRI purification
hRI was purified by procedures similar to those described previously.65,20,17 Briefly, a plasmid20,17 that directs the expression of hRI was transformed into E. coli BL21(DE3) cells, and a single colony was used to inoculate LB medium (25 mL) containing ampicillin (150 μg/mL). A starter culture was grown for 16 h at 37 °C and 250 rpm and was used to inoculate cultures of TB medium (1.00 L) containing ampicillin (200 μg/mL). These cultures were grown at 37 °C and 225 rpm until OD600 ≥ 3.0. Expression of the hRI cDNA was induced by adding IPTG (0.5 mM) and growing for 16 h at 18 °C and 225 rpm. Bacteria were collected by centrifugation (12,000g for 10 min) and resuspended in 30 mL of 50 mM Tris-HCl buffer, pH 7.5, containing EDTA (10 mM) and DTT (10 mM). Bacteria were lysed by two passes through a French pressure cell, and the cellular debris was removed by ultracentrifugation. RNase A was attached covalently to the resin in two 5-mL HiTrap NHS-ester columns, following the manufacturer’s protocol. The supernatant from the ultracentrifuge tubes was loaded onto the RNase A-affinity resin. hRI was eluted in 100 mM sodium acetate-NaOH buffer, pH 6.0, containing NaCl (3.0 M), DTT (5 mM), and EDTA (1 mM), after extensive washing with 50 mM sodium phosphate-NaOH buffer, pH 6.4, containing NaCl (1.0 M), DTT (5 mM), and EDTA (1 mM). The peak eluted from the RNase A-affinity resin was dialyzed for 16 h against 4 L of 20 mM Tris-HCl buffer, pH 7.5, containing DTT (10 mM) and EDTA (1 mM) and purified further by cation-exchange chromatography using a HiTrap Q column.65 The purity of the eluted hRI was shown to be >99% by SDS-PAGE (data not shown).
hRI·RNase 1 complex purification
Purified RNase 1 (50 mg/mL) and hRI (10 mg/mL) were mixed at a 1.2:1.0 molar ratio, and this solution was incubated at 25 °C for 60 min to allow for complex formation. The solution was then loaded onto a 5-mL HiTrap Q column that was pre-equilibrated with 20 mM Hepes-NaOH buffer, pH 7.5, containing DTT (10 mM) and glycerol (2% v/v). The complex was eluted with a linear gradient of NaCl (0-0.4 M) over 30 column volumes. Free RNase 1 eluted with the flowthrough, and the hRI·RNase 1 complex eluted around 0.15 M NaCl. Purified complex was dialyzed for 16 h at 4 °C against 20 mM Hepes-NaOH buffer, pH 7.5, containing DTT (10 mM) and glycerol (2% v/v). Purified complex was concentrated to a final concentration of 10 mg/mL in a Vivaspin 20 mL centrifugal concentrator (Vivascience AG, Hannover, Germany). Aliquots were flash frozen and stored at -80 °C.
Crystallization
Crystals of the hRI·RNase 1 complex were obtained by hanging-drop vapor diffusion in 20 mM sodium citrate-HCl buffer, pH 4.2, containing methyl ether PEG 2000 (10% w/v), ammonium sulfate (1 mM), and DTT (25 mM) with the hanging-drop solution containing a mixture of purified hRI·RNase 1 complex (0.9 μL) and crystallization solution (5.1 μL), which was composed of the citrate buffer described above. Diffraction-quality crystals grew within a week at 25 °C. Protein crystals were soaked in reservoir solutions containing increasing amounts of ethylene glycol up to 25% (v/v), and were flash-cooled in a stream of cryogenic N2(g).
Structure Determination
Diffraction data were collected at SER-CAT Sector 22 at Argonne National Laboratories. The crystal was maintained at 100 K during data collection, and X-rays were tuned to a wavelength of 0.99997 Å. The diffraction images were integrated and scaled using HKL2000.66 The initial phases were determined through molecular replacement using MOLREP from the CCP4 suite67,68 with PDB entry 1DFJ26 as the starting model. Arp-Warp software69 was used to build the initial model, which was then completed by alternating cycles of model building and refinement using the programs Xfit70 and REFMAC,71 respectively (Table 1). The coordinates have been deposited in the PDB with accession code 1Z7X. To enable a more precise comparison, the structure of the pRI·RNase A complex was refined using ten rounds of refinement in REFMAC with the same parameters (Rcryst = 21.4; Rfree = 28.1). Reflections used for the refinement were from PDB entry 1DFJ (ref 26), and 5% of the reflections were chosen randomly to define as Rfree. Both the structure of the hRI·RNase 1 complex and the refined structure of the pRI·RNase A complex were analyzed with the program PDBsum72 to identify intermolecular hydrogen bonds and van der Waals contacts. PDBsum uses the algorithm HBPLUS to identify hydrogen bonds (rX···X < 3.35 Å).23
Ribonucleolytic activity
The ribonucleolytic activity of RNase 1 and its variants was determined by quantitating their ability to cleave 6-FAM-dArU(dA)2-6-TAMRA, cleavage of this substrate at the uridine ribonucleotide leads to a 180-fold increase in fluorescence.30 Assays were carried out at 23 (±2) °C in 2 mL of 0.10 M Mes-NaOH buffer, pH 6.0, containing NaCl (0.10 M). Fluorescence data were fitted to equation (2), wherein ΔI/Δt represents the initial reaction velocity, I0 is the fluorescence intensity before the addition of a ribonuclease, If corresponds to the final fluorescence intensity after complete substrate hydrolysis, and [E] is the total ribonuclease concentration.
| (2) |
Conformational stability
The conformational stability was determined by following the change in UV absorbance of a solution of ribonuclease at 287 nm with increasing temperature.32 The temperature of PBS containing a ribonuclease (0.1-0.2 mg/mL) was raised from 20 to 80 °C at 0.15 °C/min. The A287 was followed at 1-°C intervals, and the absorbance was fitted to a two-state model of denaturation, in which the temperature at the midpoint of the transition curve corresponds to the value of Tm.
RI evasion
The affinity of hRI for variants of RNase 1 was determined by using a competition assay reported previously.74 Briefly, 2.0 mL of PBS containing DTT (5 mM), fluorescein-labeled G88R RNase A (50 nM), and an unlabeled RNase 1 variant was incubated at 23 (±2) °C for 20 min. The initial fluorescence intensity of the unbound fluorescein-labeled G88R RNase A was monitored for 3 min (excitation: 491 nm; emission: 511 nm). hRI was then added to 50 nM and the final fluorescence intensity was measured. Values for Kd were obtained by nonlinear least-squares analysis of the binding isotherm using the program DELTAGRAPH 5.5 (Red Rock Software, Salt Lake City, UT). The Kd value for the complex of hRI and fluorescein-labeled G88R RNase A was assumed to be 1.4 nM.17
Dissociation rate
The dissociation rate constant for the complex between hRI and variants of RNase 1 was determined by a procedure similar to that described previously.74 Briefly, equimolar concentrations of hRI and a fluorescein-labeled RNase 1 variant were allowed to reach equilibrium in PBS containing DTT (5 mM). The equimolar concentrations were 20-fold greater than the Kd value previously determined for each hRI·RNase 1 complex. After equilibrium was reached, wild-type RNase A (100-fold molar excess) was added to the reaction to scavenge free hRI. The increase in fluorescence upon irreversible complex disassociation was followed by fluorescence spectroscopy (excitation: 491 nm; emission: 511 nm). To calculate the value of kd, the fluorescence data were fitted to equation (3), wherein F0 is the fluorescence before the addition of wild-type RNase A and F∞ is the fluorescence after complete dissociation of the complex.
| (3) |
The dissociation rate of the complex between hRI and wild-type RNase 1 was determined using a procedure similar to above except that the experiment was performed using a 96-well microtiter-plate format (L.D. Lavis, T.J. Rutkoski, and R.T. Raines, unpublished results). Briefly, diethylfluorescein-labeled RNase 1 (100 nM) in PBS containing tris(2-carboxyethyl)phosphine (TCEP; 100μM) was added to a 96-well microtiter plate and the initial fluorescence was measured. hRI was then added at equimolar concentrations and incubated with labeled RNase 1 at 25 °C for 5 min. A 50-fold molar excess of human angiogenin (5 μM) was added to scavenge dissociated complex and the change in fluorescence was measured at various time points. To insure that the protein stability was maintained over the duration of the experiment, additional data points were monitored under the same conditions only without the addition of the 50-fold molar excess of angiogenin. Also, to account for drift in the instrument, data are the mean (± SE) from six solutions normalized for the fluorescence of four solutions of rhodamine 110 (10 nM). Fluorescence data were fitted to equation (3) to determine the dissociation rate constant. Initial fluorescence data (<4 h) were not included in the analysis, as these values showed a rapid burst in fluorescence similar to that observed in previous dissociation rate determinations.12
Cytotoxicity
The effect of RNase 1 and its variants on the proliferation of K-562 cells was assayed as described previously.24,75,17 Briefly, after a 44-h incubation with ribonuclease, K-562 cells were treated with [methyl-3H]thymidine for 4-h and the incorporation of radioactive thymidine into the cellular DNA was quantified by liquid scintillation counting. Results are shown as the percentage of [methyl-3H]thymidine incorporated into the DNA as compared to the incorporation into control K-562 cells where only PBS was added. Data are the average of three measurements for each concentration, and the entire experiment was repeated in triplicate. Values for IC50 were calculated by fitting the curves using nonlinear regression to equation (4), wherein y is the total DNA synthesis following the [methyl-3H]thymidine pulse, and h is the slope of the curve.
| (4) |
Acknowledgements
We are grateful to B.D. Smith for production systems for hRI and human angiogenin, to L.D. Lavis for diethylfluorescein derivatives, and to L.D. Lavis, T.J. Rutkoski, K.A. Dickson, R.F. Turcotte, and B.D. Smith for contributive discussions. R.J.J. was supported by Biotechnology Training grant GM08349 (NIH). J.G.M. was supported by Computational and Informatics in Biology and Medicine Training Grant LM007359 (NIH) and Genome Sciences Training Grant HG002760 (NIH). This work was supported by grants CA73808, P50 GM64598, and U54 GM074901 (NIH). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract No. W-31-109-Eng-38. Data were collected at Southeast Regional Collaborative Access Team (SERCAT) 22-BM, beamline at the Advance Photon Source, Argonne National Laboratory. Supporting institutions can be found at http://www.ser-cat.org/members.html. The Biophysics Instrumentation Facility was established with grants BIR-9512577 (NSF) and RR13790 (NIH). The W.M. Keck Center for Chemical Genomics was established by a grant from the W.M. Keck Foundation.
Glossary
Abbreviations used:
- BS-RNase
bovine seminal ribonuclease
- DTNB
5,5′-dithio-bis(2-nitrobenzoic acid)
- DTT
dithiothreitol
- EDN
eosinophil-derived neurotoxin
- FADE
fast atomic density evaluator
- 6-FAM
6-carboxyfluorescein
- hRI
human ribonuclease inhibitor
- MALDI-TOF
matrix-assisted laser desorption/ionization-time-of-flight
- ONC
Onconase® (a registered trademark of Alfacell Inc.)
- PBS
phosphate-buffered-saline
- PDB
Protein Data Bank
- pRI
porcine ribonuclease inhibitor
- RI
ribonuclease inhibitor
- RNase A
bovine pancreatic ribonuclease
- RNase 1
human pancreatic ribonuclease
- rmsd
root-mean-square deviation
- 6-TAMRA
6-carboxytetramethylrhodamine
- TB
terrific broth
- TCEP
tris(2-carboxyethyl)phosphine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shoup D, Szabo A. Role of diffusion in ligand binding to macromolecules and cell-bound receptors. Biophys. J. 1982;40:33–39. doi: 10.1016/S0006-3495(82)84455-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharp K, Fine R, Honig B. Computer simulations of the diffusion of a substrate to an active site of an enzyme. Science. 1987;236:1460–1463. doi: 10.1126/science.3589666. [DOI] [PubMed] [Google Scholar]
- 3.Getzoff ED, Cabelli DE, Fisher CL, Parge HE, Viezzoli MS, Banci L, Hallewell RA. Faster superoxide dismutase mutants designed by enhancing electrostatic guidance. Nature. 1992;358:347–351. doi: 10.1038/358347a0. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber G, Fersht AR. Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 5.Selzer T, Schreiber G. Predicting the rate enhancement of protein complex formation from the electrostatic energy of interaction. J. Mol. Biol. 1999;287:409–419. doi: 10.1006/jmbi.1999.2615. [DOI] [PubMed] [Google Scholar]
- 6.Selzer T, Albeck S, Schreiber G. Rational design of faster associating and tighter binding protein complexes. Nat. Struct. Biol. 2000;7:537–541. doi: 10.1038/76744. [DOI] [PubMed] [Google Scholar]
- 7.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 8.Norel R, Sheinerman F, Petrey D, Honig B. Electrostatic contributions to protein-protein interactions: Fast energetic filters for docking and their physical basis. Protein Sci. 2001;10:2147–2161. doi: 10.1110/ps.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheinerman FB, Honig B. On the role of electrostatic interactions in the design of protein-protein interfaces. J. Mol. Biol. 2002;318:161–177. doi: 10.1016/S0022-2836(02)00030-X. [DOI] [PubMed] [Google Scholar]
- 10.Lee LP, Tidor B. Barstar is electrostatically optimized for tight binding to barnase. Nat. Struct. Biol. 2001;8:73–76. doi: 10.1038/83082. [DOI] [PubMed] [Google Scholar]
- 11.Raines RT. Ribonuclease A. Chem. Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 12.Lee FS, Shapiro R, Vallee BL. Tight-binding inhibition of angiogenin and ribonuclease A by placental ribonuclease inhibitor. Biochemistry. 1989;28:225–230. doi: 10.1021/bi00427a031. [DOI] [PubMed] [Google Scholar]
- 13.Boix E, Wu Y, Vasandani VM, Saxena SK, Ardelt W, Ladner J, Youle RJ. Role of the N-terminus in RNase A homologues: Differences in catalytic activity, ribonuclease inhibitor interaction and cytotoxicity. J. Mol. Biol. 1996;257:992–1007. doi: 10.1006/jmbi.1996.0218. [DOI] [PubMed] [Google Scholar]
- 14.Papageorgiou A, Shapiro R, Acharya K. Molecular recognition of human angiogenin by placental ribonuclease inhibitor—an X-ray crystallographic study at 2.0 Å resolution. EMBO J. 1997;16:5162–5177. doi: 10.1093/emboj/16.17.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaul Y, Schreiber G. Exploring the charge space of protein-protein association: A proteomic study. Proteins. 2005;60:341–352. doi: 10.1002/prot.20489. [DOI] [PubMed] [Google Scholar]
- 16.Leland PA, Raines RT. Cancer chemotherapy—ribonucleases to the rescue. Chem. Biol. 2001;8:405–413. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutkoski TJ, Kurten EL, Mitchell JC, Raines RT. Disruption of shape-complementarity markers to create cytotoxic variants of ribonuclease A. J. Mol. Biol. 2005;354:41–54. doi: 10.1016/j.jmb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J. Mol. Biol. 1996;264:1028–1043. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
- 19.Kim J-S, Soucek J, Matousek J, Raines RT. Structural basis for the biological activities of bovine seminal ribonuclease. J. Biol. Chem. 1995;270:10525–10530. doi: 10.1074/jbc.270.18.10525. [DOI] [PubMed] [Google Scholar]
- 20.Lee JE, Raines RT. Cytotoxicity of bovine seminal ribonuclease: Monomer versus dimer. Biochemistry. 2005;44:15760–15767. doi: 10.1021/bi051668z. [DOI] [PubMed] [Google Scholar]
- 21.Leland PA, Schultz LW, Kim B-M, Raines RT. Ribonuclease A variants with potent cytotoxic activity. Proc. Natl. Acad. Sci. U.S.A. 1998;98:10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar K, Brady M, Shapiro R. Selective abolition of pancreatic RNase binding to its inhibitor protein. Proc. Natl. Acad. Sci. U.S.A. 2004;101:53–58. doi: 10.1073/pnas.0307268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaur D, Swaminathan S, Batra JK. Interaction of human pancreatic ribonuclease with human ribonuclease inhibitor. J. Biol. Chem. 2001;276:24978–24984. doi: 10.1074/jbc.M102440200. [DOI] [PubMed] [Google Scholar]
- 24.Leland PA, Staniszewski KE, Kim B-M, Raines RT. Endowing human pancreatic ribonuclease with toxicity for cancer cells. J. Biol. Chem. 2001;276:43095–43102. doi: 10.1074/jbc.M106636200. [DOI] [PubMed] [Google Scholar]
- 25.Bosch M, Benito A, Ribó M, Puig T, Beaumelle B, Vilanova M. A nuclear localization sequence endows human pancreatic ribonuclease with cytotoxic activity. Biochemistry. 2004;43:2167–2177. doi: 10.1021/bi035729+. [DOI] [PubMed] [Google Scholar]
- 26.Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 27.Iyer S, Holloway DE, Kumar K, Shapiro R, Acharya KR. Molecular recognition of human eosinophil-derived neurotoxin (RNase 2) by placental ribonuclease inhibitor. J. Mol. Biol. 2005;347:637–655. doi: 10.1016/j.jmb.2005.01.035. [DOI] [PubMed] [Google Scholar]
- 28.Bretscher LE, Abel RL, Raines RT. A ribonuclease A variant with low catalytic activity but high cytotoxicity. J. Biol. Chem. 2000;275:9893–9896. doi: 10.1074/jbc.275.14.9893. [DOI] [PubMed] [Google Scholar]
- 29.Dickson KA, Dahlberg CL, Raines RT. Compensating effects on the cytotoxicity of ribonuclease A variants. Arch. Biochem. Biophys. 2003;415:172–177. doi: 10.1016/s0003-9861(03)00214-5. [DOI] [PubMed] [Google Scholar]
- 30.Kelemen BR, Klink TA, Behlke MA, Eubanks SR, Leland PA, Raines RT. Hypersensitive substrate for ribonucleases. Nucleic Acids Res. 1999;27:3696–3701. doi: 10.1093/nar/27.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith BD, Soellner MB, Raines RT. Potent inhibition of ribonuclease A by oligo(vinylsulfonic acid) J. Biol. Chem. 2003;278:20934–20938. doi: 10.1074/jbc.M301852200. [DOI] [PubMed] [Google Scholar]
- 32.Klink TA, Raines RT. Conformational stability is a determinant of ribonuclease A cytotoxicity. J. Biol. Chem. 2000;275:17463–17467. doi: 10.1074/jbc.M001132200. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki M, Saxena SK, Boix E, Prill RJ, Vasandani VM, Ladner JE, Sung C, Youle RJ. Engineering receptor-mediated cytotoxicity into human ribonucleases by steric blockage of inhibitor interaction. Nat. Biotechnol. 1999;17:265–270. doi: 10.1038/7010. [DOI] [PubMed] [Google Scholar]
- 34.Stone SR, Hofsteenge J. Kinetics of the inhibition of thrombin by hirudin. Biochemistry. 1986;25:4622–4628. doi: 10.1021/bi00364a025. [DOI] [PubMed] [Google Scholar]
- 35.Lee FS, Auld DS, Vallee BL. Tryptophan fluorescence as a probe of placental ribonuclease inhibitor binding to angiogenin. Biochemistry. 1989;28:219–224. doi: 10.1021/bi00427a030. [DOI] [PubMed] [Google Scholar]
- 36.Futami J, Maeda T, Kitazoe M, Nukui E, Tada H, Seno M, Kosaka M, Yamada H. Preparation of potent cytotoxic ribonucleases by cationization: Enhanced cellular uptake and decreased interaction with ribonuclease inhibitor by chemical modification of carboxyl groups. Biochemistry. 2001;26:7518–7524. doi: 10.1021/bi010248g. [DOI] [PubMed] [Google Scholar]
- 37.Notomista E, Mancheno JM, Crescenzi O, Di Donato A, Gavilanes J, D′Alessio G. The role of electrostatic interactions in the antitumor activity of dimeric RNases. FEBS J. 2006;273:3687–97. doi: 10.1111/j.1742-4658.2006.05373.x. [DOI] [PubMed] [Google Scholar]
- 38.Haigis MC, Kurten EL, Raines RT. Ribonuclease inhibitor as an intracellular sentry. Nucleic Acids Res. 2003;31:1024–1032. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: Structure and function. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:349–374. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beintema JJ. Structure, properties and molecular evolution of pancreatic-type ribonucleases. Life Chem. Rep. 1987;4:333–389. [Google Scholar]
- 41.Rosenberg HF, Dyer KD, Tiffany HL, Gonzalez M. Rapid evolution of a unique family of primate ribonuclease genes. Nature Genet. 1995;10:219–223. doi: 10.1038/ng0695-219. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell JC, Kerr R, Ten Eyck LF. Rapid atomic density methods for molecular shape characterization. J. Mol. Graph. Model. 2001;19:325–330. doi: 10.1016/s1093-3263(00)00079-6. [DOI] [PubMed] [Google Scholar]
- 43.Jeffrey GA, Saenger W. Hydrogen Bonding in Biological Structures. Springer-Verlag; New York: 1991. [Google Scholar]
- 44.Pace CN, Shirley BA, McNutt M, Gajiwala K. Forces contributing to the conformational stability of proteins. FASEB J. 1996;10:75–83. doi: 10.1096/fasebj.10.1.8566551. [DOI] [PubMed] [Google Scholar]
- 45.Pace CN. Polar group burial contributes more to protein stability than nonpolar group burial. Biochemistry. 2001;40:310–313. doi: 10.1021/bi001574j. [DOI] [PubMed] [Google Scholar]
- 46.Rose GD, Fleming PJ, Banavar JR, Maritan A. A backbone-based theory of protein folding. Proc. Natl. Acad. Sci. USA. 2006;103:16623–16633. doi: 10.1073/pnas.0606843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fersht AR, Shi J-P, Knill-Jones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Arter P, Waye MMY, Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 48.Vallone B, Miele AE, Vecchini P, Chiancone E, Brunori M. Free energy of burying hydrophobic residues in the interface between protein subunits. Proc. Natl. Acad. Sci. USA. 1998;95:6103–6107. doi: 10.1073/pnas.95.11.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parsegian VA. van der Waals Forces. Cambridge University Press; New York: 2006. [Google Scholar]
- 50.Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ribó M, Beintema JJ, Osset M, Fernández E, Bravo J, de Llorens R, Cuchillo CM. Heterogeneity in the glycosylation pattern of human pancreatic ribonuclease. Biol. Chem. Hoppe-Seyler. 1994;375:357–363. [PubMed] [Google Scholar]
- 52.Pous J, Canals A, Terzyan SS, Guasch A, Benito A, Ribó M, Vilanova M, Coll M. Three-dimensional structure of a human pancreatic ribonuclease variant, a step forward in the design of cytotoxic ribonucleases. J. Mol. Biol. 2000;303:49–60. doi: 10.1006/jmbi.2000.4506. [DOI] [PubMed] [Google Scholar]
- 53.Pous J, Mallorquí-Fernández G, Peracaula R, Terzyan SS, Futami J, Tada H, Yamada H, Seno M, de Llorens R, Gomis-Rüth FX, Coll M. Three-dimensional structure of human RNase 1ΔN7 at 1.9 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 2001;57:498–505. doi: 10.1107/s0907444901001147. [DOI] [PubMed] [Google Scholar]
- 54.Miller S, Janin J, Lesk AM, Chothia C. Interior and surface of monomeric proteins. J. Mol. Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 55.Park C, Raines RT. Catalysis by ribonuclease A is limited by the rate of substrate association. Biochemistry. 2003;42:3509–3518. doi: 10.1021/bi026076k. [DOI] [PubMed] [Google Scholar]
- 56.Mikulski SM, Costanzi JJ, Vogelzang NJ, McCachren S, Taub RN, Chun H, Mittelman A, Panella T, Puccio C, Fine R, Shogen K. Phase II trial of a single weekly intravenous dose of ranpirnase in patients with unresectable malignant mesothelioma. J. Clin. Oncol. 2002;20:274–281. doi: 10.1200/JCO.2002.20.1.274. [DOI] [PubMed] [Google Scholar]
- 57.Vasandani VM, Wu Y-N, Mikulski SM, Youle RJ, Sung C. Molecular determinants in the plasma clearance and tissue distribution of ribonucleases of the ribonuclease A superfamily. Cancer Res. 1996;56:4180–4186. [PubMed] [Google Scholar]
- 58.Vasandani VM, Burris JA, Sung C. Reversible nephrotoxicity of onconase and effect of lysine pH on renal onconase uptake. Cancer Themother. Pharmacol. 1999;44:164–169. doi: 10.1007/s002800050962. [DOI] [PubMed] [Google Scholar]
- 59.Matousek J, Soucek J, Slavik T, Tomanek M, Lee JE, Raines RT. Comprehensive comparison of the cytotoxic activities of onconase and bovine seminal ribonuclease. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2003;136:343–356. doi: 10.1016/j.cca.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Fuchs SM, Raines RT. Polyarginine as a multifunctional fusion tag. Protein Sci. 2005;14:1538–1544. doi: 10.1110/ps.051393805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leland PA, Staniszewski KE, Park C, Kelemen BR, Raines RT. The ribonucleolytic activity of angiogenin. Biochemistry. 2002;41:1343–1350. doi: 10.1021/bi0117899. [DOI] [PubMed] [Google Scholar]
- 62.Beintema JJ, Wietzes P, Weickmann JL, Glitz DG. The amino acid sequence of human pancreatic ribonuclease. Anal. Biochem. 1984;136:48–64. doi: 10.1016/0003-2697(84)90306-3. [DOI] [PubMed] [Google Scholar]
- 63.Riddles PW, Blakeley RL, Zerner B. Reassessment of Ellman′s reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 64.Lavis L, Chao T-Y, Raines RT. Fluorogenic label for biomolecular imaging. ACS Chem. Biol. 2006;1:252–260. doi: 10.1021/cb600132m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klink TA, Vicentini AM, Hofsteenge J, Raines RT. High-level soluble production and characterization of porcine ribonuclease inhibitor. Protein Expr. Purif. 2001;22:174–179. doi: 10.1006/prep.2001.1422. [DOI] [PubMed] [Google Scholar]
- 66.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 67.Bailey S. The CCP4 suite: Programs for protein crystallography. Acta. Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 68.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 69.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 70.McRee DE. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 71.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 72.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: New summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 74.Abel RL, Haigis MC, Park C, Raines RT. Fluorescence assay for the binding of ribonuclease A to the ribonuclease inhibitor protein. Anal. Biochem. 2002;306:100–107. doi: 10.1006/abio.2002.5678. [DOI] [PubMed] [Google Scholar]
- 75.Haigis MC, Kurten EL, Abel RL, Raines RT. KFERQ sequence in ribonuclease A-mediated cytotoxicity. J. Biol. Chem. 2002;277:11576–11581. doi: 10.1074/jbc.M112227200. [DOI] [PubMed] [Google Scholar]


