Abstract
The formation of many forms of long-term memory requires several molecular mechanisms including regulation of gene expression. The mechanisms directing transcription require not only activation of individual transcription factors but also recruitment of transcriptional coactivators. CBP and p300 are transcriptional coactivators that interact with a large number of transcription factors and regulate transcription through multiple mechanisms, including an intrinsic histone acetyltransferase (HAT) activity. HAT activity mediates acetylation of lysine residues on the amino-terminal tails of histone proteins, thereby increasing DNA accessibility for transcription factors to activate gene expression. CBP has been shown to play an important role in long-term memory formation. We have investigated whether p300 is also required for certain forms of memory. p300 shares a high degree of homology with CBP and has been shown to interact with transcription factors known to be critical for long-term memory formation. Here we demonstrate that conditional transgenic mice expressing an inhibitory truncated form of p300 (p300Δ1), which lacks the carboxy-terminal HAT and activation domains, have impaired long-term recognition memory and contextual fear memory. Thus, our study demonstrates that p300 is required for certain forms of memory and that the HAT and carboxy-terminal domains play a critical role.
Information is first stored as a short-term memory lasting minutes to hours and can then be stabilized into long-term memory lasting days to lifetime. These forms of memory differ in that the formation of long-term memory requires activation of transcription (for review, see Korzus 2003). Transcriptional activation requires recruitment of a large number of proteins in addition to individual transcription factors. Cyclic AMP-responsive element binding protein (CREB) binding protein (CBP) and its homolog E1A binding protein (p300) are transcriptional coactivators (Chrivia et al. 1993; Eckner et al. 1994) that interact with multiple transcriptional factors to facilitate gene-specific transcription (for review, see Vo and Goodman 2001).
Several studies have shown that CBP plays an important role in long-term memory formation (Oike et al. 1999; Bourtchouladze et al. 2003; Alarcon et al. 2004; Korzus et al. 2004; Wood et al. 2005, 2006). The first demonstration that CBP may play a role in memory formation came from a study in which genetically modified mice exhibited long-term memory deficits as well as developmental defects that resembled the phenotype observed in Rubinstein Taybi syndrome (RTS) patients. In these mice, a single Cbp allele is truncated (truncated protein contains residues 1–1084), and this truncated form is expressed throughout developmental and adult stages (Oike et al. 1999). In our laboratory, transgenic mice that express the same truncation form (CBPΔ1) only in adulthood and in forebrain neurons were generated to study the role of CBP in memory independently of its role in development (Wood et al. 2005). CBPΔ1 transgenic mice exhibit deficits in specific forms of hippocampal synaptic plasticity and long-term memory formation. In the present study we have investigated whether p300 is also required for long-term memory formation.
Recently, a screen for mutations in RTS patients showed that only 40% of the patients carried mutations in the CBP gene, suggesting that mutations in other genes could also be the cause of this syndrome (Roelfsema et al. 2005). A potential candidate is the EP300 gene, encoding the coactivator p300, because of its high degree of homology with CBP. Indeed, some RTS patients carry mutations in the EP300 gene that lead to proteins that do not contain the HAT domain (Roelfsema et al. 2005). Although all RTS patients have varying degrees of cognitive impairment and mental retardation, the phenotypes of patients with mutations in either the CBP or EP300 genes do not overlap completely; RTS patients with mutations in the EP300 do not have the skeletal abnormalities that are usually observed in patients with mutations in the CBP gene (Bartholdi et al. 2007). The phenotypic differences between RTS patients with mutations in the CBP gene and EP300 gene and the observation that CBP and p300 have different functions during embryogenesis and hematopoiesis (Tanaka et al. 1997; Yao et al. 1998; Kasper et al. 2002, 2006; for review, see also Kalkhoven 2004) suggest that CBP and p300, despite their high degree of homology, also have unique functions in vivo. In support of this idea, we have recently found that CBP and p300 have distinct roles in motor skill learning (Oliveira et al. 2006).
CBP and p300 regulate transcription through multiple mechanisms. CBP and p300 function as scaffolds that form macromolecular regulatory complexes, linking gene-specific transcription factors to the basal transcription machinery. Furthermore, CBP and p300 contain intrinsic histone acetyltransferase (HAT) activity in the carboxy-terminal domain that mediates acetylation of lysine residues on the amino-terminal tails of histone proteins (for review, see Chan and La Thangue 2001). Acetylation neutralizes the positively charged lysine residues in histones and disrupts the interaction between histones and DNA, increasing DNA accessibility for transcription factors to activate gene expression (Grunstein 1997). In addition to HAT activity, CBP and p300 have also been shown to acetylate other proteins, including transcription factors (Gu and Roeder 1997; Furia et al. 2002). p300 HAT activity has been shown to be highest in the adult mouse brain compared with other tissues, suggesting that this protein may play an important role in the regulation of gene transcription in the brain (Li et al. 2002). p300 has also been shown to interact in vitro with transcription factors that are known to play a role in learning and memory, such as CREB, NF-κB, Elk-1, and C/EBPβ (Lundblad et al. 1995; Li et al. 2003; Romano et al. 2006; Cesena et al. 2007).
Thus, p300 functions in vitro and cell culture as a transcriptional coactivator for transcription factors known to be critical in memory. p300 is highly homologous to CBP, and mutations in CBP produce memory deficits in mice. Mutations in CBP or EP300 produce similar cognitive impairments in RTS patients. Therefore, we have directly investigated whether p300 plays a role in long-term memory formation using genetically modified mice expressing a truncated form of p300 (p300Δ1). p300Δ1 lacks the carboxy-terminal portion of the protein and therefore does not contain the HAT domain. Transgenic mice were generated using the tetracycline system to allow for temporal and regional control of the expression of the transgene. We show that expression of p300Δ1 is associated with deficits in long-term recognition memory and contextual fear memory, suggesting that p300 is involved in these forms of memory.
Results
p300 expression in the mouse brain
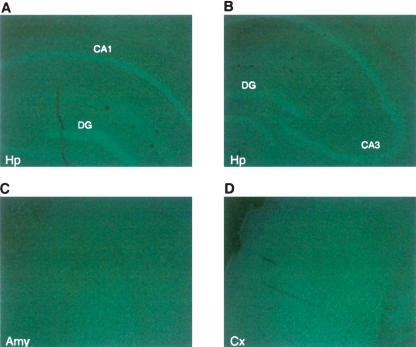
We first examined the expression of p300 in the mouse brain as this has not been established. We used antibodies shown to be specific for p300 in previous studies (Kasper et al. 2006). Kasper et al. (2006) used the same antibodies to show by immunohistochemistry that p300 is absent in T-cells derived from p300 conditional knockout mice. We show that p300 is expressed ubiquitously in the mouse brain, including areas that are critical for long-term memory formation such as hippocampus, amygdala, and cortex (Fig. 1).
Figure 1.
p300 expression in the brain. Fluorescent immunohistochemistry of p300 in brain coronal sections from naïve C57Bl/6J mice. p300 is ubiquitously expressed in the mouse forebrain including the CA1, CA3, and DG regions of the hippocampus (Hp) (A,B), amygdala (Amy) (C), and cortex (Cx) (D).
Generation and characterization of p300Δ1 transgenic mice
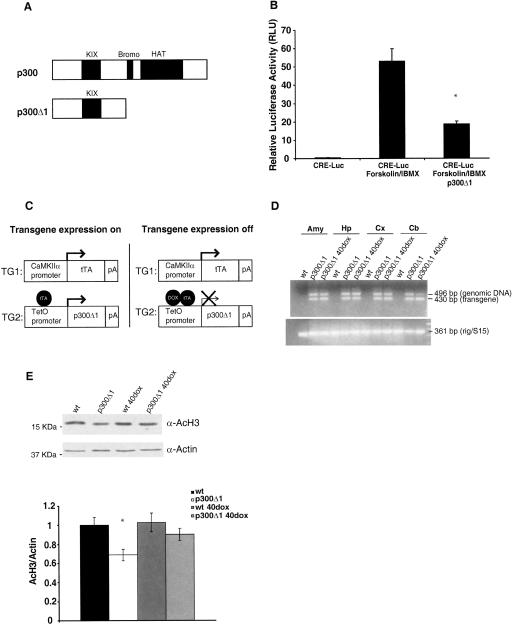
Many lines of evidence suggest that CBP plays a critical role in the formation of certain types of long-term memory (Oike et al. 1999; Bourtchouladze et al. 2003; Alarcon et al. 2004; Korzus et al. 2004; Wood et al. 2005, 2006). It is not known, however, whether its homolog p300 is also required for long-term memory formation. To address this question, we generated transgenic mice expressing a truncated form of p300 (p300Δ1) that lacks the carboxy-terminal part of p300 (Fig. 2A). We chose a conditional transgenic system in which expression of the transgene could be spatially and temporally restricted.
Figure 2.
Generation and characterization of p300Δ1 transgenic mice. (A) Schematic representation of the truncated form of p300 (p300Δ1). p300Δ1 lacks the bromo and histone acetyltransferase (HAT) domains contained within the carboxy-terminal portion of this protein. (KIX) Kinase inducible interaction domain. (B) Transient transfection of p300Δ1 and a CRE-luciferase reporter in HEK293 cells showed that p300Δ1 significantly reduces CRE-dependent transcription induced by forskolin and IBMX. (C) Schematic representation of the tetracycline system used to regionally and temporally control the expression of p300Δ1 transgene in mice. p300Δ1 transgene is under the control of a tetracycline operator sequence (tetO) and a minimal promoter and mice carrying this transgene were generated and crossed to mice that contain the tTA transactivator driven by the CaMKIIα promoter (line B, Mayford et al. 1996) giving rise to bitransgenic mice (referred to in this paper as p300Δ1 transgenic mice). In the absence of doxycycline (dox), p300Δ1 is expressed in postnatal forebrain neurons and in the presence of doxycycline p300Δ1 expression should be turned off. (D) RT-PCR analyses of cDNA synthesized from RNA isolated from amygdala (Amy), hippocampus (Hp), cortex (Cx), and cerebellum (Cb) of wild-type and p300Δ1 transgenic mice on and off dox. (E) Western blot analysis of levels of acetylated lysines K9 and K14 in histone H3 (AcH3) in forebrain of wild-type (wt) and p300Δ1 transgenic mice on and off dox (n = 6 per group) normalized to the levels of actin in each sample.
To spatially restrict the expression of the transgene to forebrain neurons, we used one transgenic mouse line (TG1 in Fig. 2C) that contains the tetracycline responsive transactivator (tTA) under the control of the CaMKIIα promoter (CaMKIIα-tTA line B) (Mayford et al. 1996). To temporally restrict the expression of the transgene, TG1 was combined with a second transgenic mouse line (TG2 in Fig. 2C) that contains the p300Δ1 transgene driven by the tetracycline operator sequence (tetO) and a minimal promoter (Mayford et al. 1996) (tetOp300Δ1). Crossing both transgenic lines generates bitransgenic mice (referred to in this paper as p300Δ1 transgenic mice) in which expression of the transgene should be restricted to excitatory neurons in the adult mouse forebrain and suppressed by administration of doxycycline (dox) in the animals’ food (Fig. 2C). We generated three lines of tetOp300Δ1 transgenic mice and the line with higher copy number (determined by Southern blot, data not shown) was used in the present study as this line showed the strongest impairment in contextual fear conditioning in an initial behavioral screening of all three lines.
CREB is one of the many transcription factors that interacts with p300 through the kinase inducible interaction domain (KIX) (Vo and Goodman 2001), and it has been shown to require the HAT activity of p300 for transcriptional activation (Lundblad et al. 1995; Yuan and Gambee 2001). Therefore, we predicted that p300Δ1 expression would impair CRE-dependent transcription. We observed that CRE-dependent transcription induced by forskolin and 3-isobutyl-1-methylxanthine (IBMX) is significantly inhibited (t(5) = 4.95, P < 0.05) when HEK293 cells are cotransfected with p300Δ1 and a CRE-luciferase reporter (Fig. 2B). p300Δ1 is likely acting in an inhibitory fashion by competing with endogenous p300 for p300-specific binding sites, thus compromising the function of proteins, such as CREB, that interact with the amino-terminal part of p300 and require its HAT and/or other carboxy-terminal domain.
p300Δ1 transgenic mice do not show gross morphological abnormalities or reduced size compared with wild-type or single transgenic littermates. Similarly, p300Δ1 transgenic mice on dox appear normal (data not shown), suggesting that p300Δ1 transgenic mice are developmentally normal. To assess the expression of the transgene, we performed reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of cDNA prepared from RNA from amygdala, hippocampus, cortex, and cerebellum of p300Δ1 transgenic and wild-type mice (Fig. 2D). We chose PCR primers that span an intron to discriminate between transgene genomic DNA and transgene cDNA. We observed the presence of the p300Δ1 transgene after PCR analysis of cDNA from all brain regions analyzed in p300Δ1 transgenic mice both on and off dox (Fig. 2D). The identity of the bands was confirmed by restriction enzyme analysis. These observations suggest that transgene expression is not restricted to the forebrain and is not suppressed by dox treatment. However, because this approach is not quantitative, we could not determine whether dox regulation is totally absent or just incomplete. Therefore, we examined how transgene expression affects acetylation of histone H3 in the forebrain of p300Δ1 transgenic mice and their wild-type littermates (Fig. 2E) by Western blot. We chose to measure acetylation of the lysines K9 and K14 in histone H3 because it has been previously shown to be enhanced in the hippocampus after contextual fear conditioning (Levenson et al. 2004). Furthermore, lysines K9 and K14 in histone H3 are p300 targets (McManus and Hendzel 2003). We observed a significant effect of genotype (F(1,20) = 7.94, P < 0.05). A direct comparison of the groups showed that p300Δ1 transgenic mice off dox have significantly lower levels of acetylated histone H3 (AcH3) in the forebrain than their wild-type littermates (P < 0.05). Wild-type animals on dox show similar levels of AcH3 as compared with wild-type animals off dox, showing that dox does not affect histone H3 acetylation. p300Δ1 transgenic mice on dox show slightly reduced levels of AcH3 compared with wild-type mice, but this reduction was not statistically significant (P = 0.38). This finding suggests that p300Δ1 transgenic mice on dox still express the transgene in the forebrain but at lower levels than in p300Δ1 transgenic mice off dox and that these levels are not enough to produce a significant reduction of acetylation of lysines K9 and K14 in histone H3.
The presence of the transgene in the cerebellum (Fig. 2D) is perhaps not surprising because CaMKIIα is expressed in Purkinje cells of the cerebellum; however, the level of CaMKIIα in the cerebellum is much lower overall than in the forebrain (Burgin et al. 1990; Hanson and Schulman 1992; Zou et al. 2002). We observed that the levels of AcH3 in the cerebellum of wild-type and p300Δ1 transgenic mice are not different (data not shown). However, it remains possible that histone acetylation is reduced in Purkinje cells, but, since the levels of CaMKIIα in these cells are very low and the majority of cells in the cerebellum are granule cells, we do not observe a significant difference when analyzing the levels of AcH3 in the whole cerebellum of p300Δ1 transgenic mice when compared with wild-type littermates by Western blot. Also, p300Δ1 transgenic mice show normal locomotor activity (see below) despite expression of the transgene in the cerebellum.
Overall, p300Δ1 transgene expression seems to be incompletely regulated by dox and not fully restricted to the forebrain, but the levels of leaky expression in the presence of dox are not sufficient to cause a significant effect on acetylation of lysines K9 and K14 of histone H3.
p300Δ1 transgenic mice show normal spatial memory but impaired recognition memory
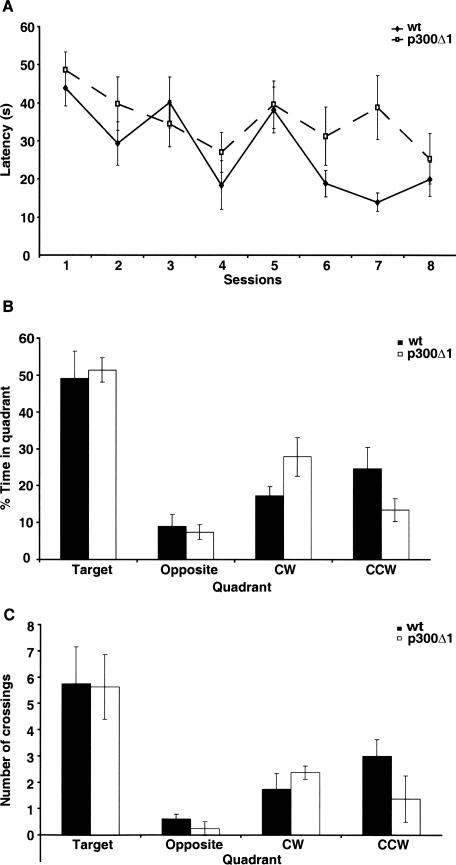
To assess spatial memory, p300Δ1 transgenic mice and wild-type littermates were tested in the hidden platform version of the Morris water maze task (Fig. 3). The ability of rodents to acquire this task has been shown to depend on an intact hippocampus (Morris et al. 1982). p300Δ1 transgenic mice and wild-type littermates showed a significant improvement across the acquisition phase (effect of training day F(7,98) = 6.1, P < 0.0001), showing that both groups acquired this task. However, no effect of genotype (F(1,98) = 2.3, P = NS) or genotype × day interaction (F(7,98) = 1.6, P = NS) was observed. Overall, p300Δ1 transgenic mice exhibited similar latencies to find the hidden platform as their wild-type littermates (Fig. 3A). To assess spatial memory, two probe trials were performed: the first, one day after session four, and the second, one day after session eight. No differences were observed between p300Δ1 transgenic mice and wild-type littermates in the time spent in target quadrant or number of crossings over the area where the platform was placed during acquisition in either probe trail (data not shown; Fig. 3B,C). Swim speed, thigmotaxis, and total swimming distance were also not different between genotypes, showing that p300Δ1 transgenic mice have normal locomotor activity despite expression of the transgene in the cerebellum. Transgenic mice also showed normal motor coordination and motor skill learning when tested in the rotarod task (data not shown).
Figure 3.
p300Δ1 transgenic mice show normal spatial memory in the Morris water maze. (A) Wild-type (n = 8) and p300Δ1 transgenic (n = 8) mice do not show a significant difference in the latency to find the hidden platform during the acquisition phase of the task. (B) During probe trial (performed after session 8), p300Δ1 transgenic mice and wild-type littermates showed a similar percentage of time spent swimming in the target quadrant. (C) During the probe trial (performed after session 8) the number of crossings over the area where the platform was located during acquisition was not different between wild-type and p300Δ1 transgenic mice.
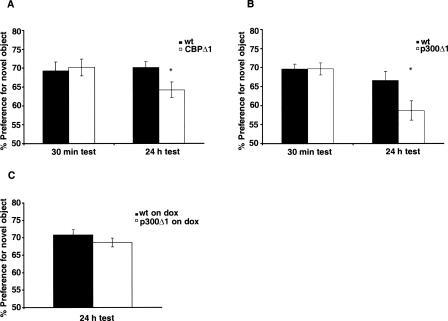
Several studies have demonstrated a role for CBP in recognition memory (Bourtchouladze et al. 2003; Alarcon et al. 2004; Korzus et al. 2004; Wood et al. 2006). In agreement with these studies, we show here that CBPΔ1 transgenic mice generated in our laboratory (Wood et al. 2005) have a significant deficit in long-term memory for novel object recognition (Fig. 4A). We observed that CBPΔ1 transgenic mice had a significantly lower preference for the novel object when tested 24 h after training (t(14) = 2.34, P < 0.05). In contrast, when tested 30 min after training, the preference for the novel object was similar between CBPΔ1 transgenic and wild-type animals (Fig. 4A). This observation further confirms that CBP is important for long-term recognition memory. We then determined if p300 also plays a role in this type of memory (Fig. 4B,C). We observed that p300Δ1 transgenic mice show significantly lower preference for the novel object when tested 24 h after training, as compared with wild-type littermates (t(28) = 2.20, P < 0.05) (Fig. 4B). Short-term recognition memory, tested 30 min after training, was normal in p300Δ1 transgenic mice (Fig. 4B), showing that p300Δ1 is disrupting consolidation of the learning experience without affecting acquisition of the task. The long-term recognition memory phenotype was reversed in a different set of animals that were kept on dox food from conception (Fig. 4C), indicating that dox presumably reduced the expression of p300Δ1 transgene to levels that were not sufficient to cause an impairment. The observation that p300Δ1 transgenic mice on dox do not show a significant impairment supports the idea that the phenotype of p300Δ1 transgenic mice off dox is not due to a transgene insertion effect. The total time spent exploring both objects during training and testing phases among the different conditions tested was not different between the two genotypes (data not shown). Together, these results suggest that p300 plays a role in the formation of long-term recognition memory but not spatial memory.
Figure 4.
CBPΔ1 and p300Δ1 transgenic mice show impaired long-term recognition memory. (A) CBPΔ1 transgenic mice (n = 5) and wild-type littermates (n = 6) show similar preference for the novel object when tested 30 min after training. However, when tested 24 h after training, CBPΔ1 transgenic mice (n = 9) show significant lower preference for the novel object than their wild-type littermates (n = 8). (B) p300Δ1 transgenic mice (n = 16) and wild-type littermates (n = 16) show similar short-term recognition memory tested 30 min after training. However, p300Δ1 transgenic mice (n = 15) show impaired novel object recognition tested 24 h after training compared with their wild-type littermates (n = 15). (C) p300Δ1 transgenic mice (n = 10) and wild-type littermates (n = 12) fed with doxycycline show similar recognition memory tested 24 h after object familiarization.
p300Δ1 transgenic mice show impaired contextual fear memory but normal cued fear memory
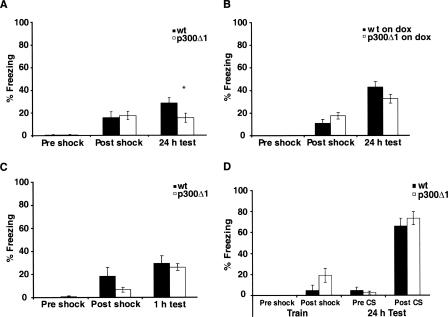
We assessed the role of p300 in associative memory by using contextual and cued fear conditioning (Fig. 5). Contextual and cued fear conditioning partially differ in the neural systems required. Lesions in the hippocampus only affect contextual fear, but both tasks are sensitive to lesions in the amygdala (for review, see Maren and Quirk 2004). p300Δ1 transgenic mice showed significantly lower freezing compared with their wild-type littermates (t(35) = 2.12, P < 0.05) when tested 24 h after conditioning (Fig. 5A). In the case of mice treated with dox, we still observed a small impairment in the p300Δ1 transgenic mice, but this decrease was not statistically significant (Fig. 5B). This pattern is consistent with the levels of histone H3 acetylation in the forebrain of p300Δ1 transgenic and wild-type mice on and off dox (Fig. 2E). We also found that p300Δ1 transgenic mice had normal cued fear memory at 24 h (Fig. 5D), normal short-term contextual fear memory at 1 h (Fig. 5C), and normal freezing during the 30 sec immediately after shock. Together, these results suggest that p300 is required for consolidation of contextual fear memory but is not necessary for acquisition of contextual fear memory or for long-term cued fear memory.
Figure 5.
p300Δ1 transgenic mice show impaired long-term contextual fear memory. (A) p300Δ1 transgenic mice (n = 18) show significant lower percentage of freezing compared with their wild-type littermates (n = 19) when tested 24 h after training in the contextual fear conditioning. (B) p300Δ1 transgenic mice fed with doxycycline (n = 20) show a reduced (but not statistically significant) percentage of freezing compared with their wild-type littermates (n = 20). (C) p300Δ1 transgenic mice (n = 10) and wild-type littermates (n = 10) show comparable contextual fear memory tested 1 h after training. (D) p300Δ1 transgenic mice (n = 7) and wild-type littermates (n = 7) show comparable long-term cued fear memory tested 24 h after training.
Discussion
This study is the first demonstration that the transcriptional coactivator p300 plays a role in long-term memory formation. We have investigated the role of p300 in memory using genetically modified mice that express a truncated form of p300. p300Δ1 transgenic mice show deficits in long-term recognition memory and long-term memory for contextual fear conditioning, with no deficits in short-term memory, hippocampus-independent cued fear conditioning, or spatial learning in the Morris water maze. These results suggest that p300 is required during learning-induced transcription activation that is critical for the consolidation of memory (for review, see Korzus 2003).
We generated transgenic mice that express a truncated form of p300 (p300Δ1) under the control of the tetracycline system. We observed that treating p300Δ1 transgenic mice with dox did not completely suppress expression of the transgene. Previous studies have also shown incomplete suppression of expression of the transgene in a doxycycline-regulated system (Tremblay et al. 1998). Despite incomplete suppression of the transgene expression, p300Δ1 transgenic mice on dox did not show significant behavioral impairments, and extracts from p300Δ1 transgenic mice on dox had similar levels of acetylation of lysines K9 and K14 in histone H3 as extracts from wild-type littermates. In contrast, the expression of the p300Δ1 transgene in p300Δ1 transgenic mice off dox lead to significant impairments in long-term recognition memory and contextual fear memory, as well as significant reduction in H3 acetylation in the forebrain. Our results suggest that p300 is specifically required for these types of memory. Morris water maze and contextual fear conditioning are both hippocampus-dependent tasks (Morris et al. 1982; Phillips and LeDoux 1992); however, increasing evidence suggests that different molecules are required for each type of memory (for review, see Mizuno and Giese 2005). Thus, it is not surprising that p300 may be involved in the molecular mechanisms involved in long-term contextual fear memory formation but not in mechanisms underlying spatial memory.
The transcriptional coactivator functions of p300 appear to be exerted through multiple mechanisms: p300 functions as a scaffold, forming a multiprotein transcriptional regulatory complex, and through its acetyltranferase enzymatic activity can influence histone and nonhistone proteins activity (for review, see Chan and La Thangue 2001). Similar to CBPΔ1 (Wood et al. 2005), p300Δ1 binds to molecules that interact with the amino-terminal domain of p300. Thus, factors that bind to p300Δ1 will not recruit HAT activity and carboxy-terminal activation domains of p300. p300 binds to transcription factors that have been shown to be involved in memory formation (e.g., CREB and family members, c-Fos, Elk-1, C/EBP, NF-κB) (see Kasper et al. 2006). The memory impairments observed in this study may be a result of impaired CRE-dependent transcription, as we observed that p300Δ1 inhibits CRE-dependent transcription in cell culture. However, we cannot rule out the idea that other transcription factors that interact with p300 may also be involved. Regulation of gene expression is a multifactorial and combinatorial process that involves a set of transcription factors and coactivators. Previous studies have implicated CBP as a critical transcriptional coactivator in long-term memory formation (Oike et al. 1999; Bourtchouladze et al. 2003; Alarcon et al. 2004; Korzus et al. 2004; Wood et al. 2005, 2006), and the present study constitutes the first evidence that p300 may also play a role in this process. Besides CREB, other transcription factors have been shown to respond to the neuronal activation triggered by learning. It is now well established that NF-κB is required for memory formation and that NF-κB-dependent transcription requires coactivators possessing HAT activity (for review, see Quivy and Van Lint 2004; Romano et al. 2006). Zhong and colleagues showed that PKA regulates the transcriptional activity of NF-κB in cultured cells by modulating its interaction with CBP and p300 (Zhong et al. 1998). Also, the transcription factor Elk-1 has been implicated in associative memory formation (Cammarota et al. 2000). Interestingly, a study using fibroblasts showed that p300 is important in Elk-1 activation of transcription and that the interactions between Elk-1 and p300 change following Elk-1 phosphorylation by MAPK, resulting in increased p300 HAT activity (Li et al. 2003). Moreover, it was shown that p300 acetylates C/EBPβ at K39 and that this post-translational modification is required for transcription of c-fos (Cesena et al. 2007). However, regulation of NF-κB, Elk-1, and C/EBPβ activities by p300 in neuronal cells remains to be investigated.
A comparison of memory phenotypes in CBPΔ1 (Wood et al. 2005) and p300Δ1 transgenic mice suggests that CBP, but not p300, is required for spatial memory, revealing a differential role for these highly homologous coactivators. However, such a conclusion should be made with caution because different transgenic models may give rise to different levels and patterns of transgene expression (Furth et al. 1994), and this difference may account for the distinct behavioral phenotype. Indeed, we observed that CBPΔ1 transgenic mice show much lower levels of forebrain acetylation of histone H3 than p300Δ1 transgenic mice (data not shown), suggesting higher levels of expression of the CBPΔ1 transgene than the p300Δ1 transgene. On the other hand, both mice showed impaired long-term contextual fear conditioning and long-term recognition memory. Investigation of whether CBP and p300 are recruited interchangeably after activation triggered by learning or whether they are differentially recruited by different signaling pathways that lead to transcription activation will be the subject of further studies. Increasing evidence has shown that CBP and p300 have unique functions. In vivo studies found that during embryogenesis, hematopoiesis, and motor skill learning, CBP and p300 have distinct functions (Tanaka et al. 1997; Yao et al. 1998; Kasper et al. 2002, 2006; Oliveira et al. 2006). Different substrate specificity and interaction with distinct molecules may account for these differences. McManus and Hendzel (2003) have shown that CBP has a preference for acetylating K12 on histone H4, whereas p300 preferentially acetylates K8 on histone H4 in vivo. Also, as mentioned above, Li et al. (2003) showed that in fibroblasts p300 is important in Elk-1 activation of transcription and that the interactions between Elk-1 and p300, but not CBP, change following Elk-1 phosphorylation by MAPK, resulting in increased p300 HAT activity. In neuronal cells, the activation of CaMKII, CaMKIV, or PKA, but not MAPK, can lead to induction of CBP-mediated transcription (Hu et al. 1999). In agreement with this study, Impey et al. (2002) showed that in hippocampal neurons, CaMKIV signaling activates CBP- but not p300-dependent transcription, and that MAPK signaling is dispensable for activation of CBP (Impey et al. 2002). However, it is not known whether MAPK signaling activates p300 in neuronal cells.
Interestingly, neither CBPΔ1 (Wood et al. 2005) nor p300Δ1 transgenic mice showed impairments in cued fear conditioning. These findings may be explained by low levels of transgene expression in the amygdala in those mice or it may suggest that transcriptional coactivators other than CBP or p300 play a role in cued fear conditioning. In agreement with our observations, other recent studies that investigated a role for CBP in memory also found no significant deficits in cued fear conditioning in three different genetically modified CBP mutant mice (Alarcon et al. 2004; Korzus et al. 2004; Wood et al. 2006). As it is known that activation of transcription in the amygdala is essential for this form of associative memory (Bailey et al. 1999), it would be interesting to investigate whether other transcriptional coactivators with HAT activity, such as p300/CBP associated factor (P/CAF) and GCN5, play a role in this form of memory.
Transcriptional coactivators play critical roles in coordinating and integrating multiple signal-dependent events with the transcriptional apparatus allowing gene activation to occur in response to diverse stimuli. The present study suggests that the transcriptional coactivator p300 is required in the process of gene activation induced by certain forms of learning. It remains to be investigated which signaling pathways recruit p300 during memory formation. A recent study showed that some RTS patients, who have cognitive abnormalities, have mutations in the EP300 gene, some of which lead to proteins that lack the HAT domain (Roelfsema et al. 2005). Our study is the first to suggest that the impaired p300 activity may partially contribute to the cognitive impairments observed in these patients.
Materials and Methods
Immunohistochemistry
C57Bl/6J mice were anesthetized with isoflurane and transcardially perfused with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde in PBS using a peristaltic perfusion pump. Fixed brains were dissected, post-fixed overnight, and then cryoprotected in 30% sucrose at 4°C. Brains were flash frozen in 2-methylbutane on dry ice and mounted on cryostat chucks using OCT (Optimal Cutting Temperature compound; Sakura). Coronal sections were cut at a thickness of 30 μm and collected in 1× PBS. Floating sections were washed with 1× PBS. Permeabilization was done with 0.1% Triton X-100 (Fisher), and sections were blocked in 8% normal goat serum (NGS, Vector Laboratories) with 0.3% Triton-X for 50 min at room temperature. Incubation with p300 antibodies (C-20 and N-15, 1:500, Santa Cruz Biotechnology) was done overnight at 4°C in 2% NGS, 0.3% Triton X-100 in PBS and for two hours at room temperature with goat anti-rabbit IgG-FITC secondary antibody (1:500, Santa Cruz Biotechnology). Sections were washed three times for 5 min each in PBS before and after each incubation step. Sections were mounted on slides in 50% glycerol and imaged using a Leica DMRBE fluorescent microscope using a 10× objective and a Hamamatsu CCD camera.
Generation of p300Δ1 transgenic mice
The full-length wild-type cDNA for human p300 was kindly provided by Dr. Richard Eckner (University of New Jersey, Newark). Amino acids 1–1031 (p300Δ1) were cloned into pBluescript II SK (+/−) using NotI and ScaI sites. The truncated p300 cDNA was then FLAG-epitope tagged by Muta-Gene Phagemid In Vitro Mutagenesis (Bio-Rad). FLAG-p300Δ1 was cloned into the EcoRV site of MM400, which contains the tetracycline operator sequence (tetO) and a minimal promoter (Mayford et al. 1996). All cloning junctions were verified by DNA sequencing. The FLAG-p300Δ1 transgene construct was excised from MM400 using NotI sites and purified by CsCl gradient centrifugation. Transgenic mice were generated by injecting purified TetO-FLAG-p300Δ1 into pronuclei of B6-SJL/F1 zygotes (Transgenic and Chimeric Mouse Facility, University of Pennsylvania, Philadelphia). Founders were backcrossed to C57BL/6J mice for at least 5 generations. CaMKIIα-tTA (line B) mice were obtained from Dr. Mark Mayford (Mayford et al. 1996). Mice were maintained and bred under standard conditions, consistent with National Institutes of Health guidelines and approved by the Institutional Animal Care and Use Committee. Mice were maintained on a 12 h light/12 h dark cycle with the behavioral testing occurring during the light phase. For experiments subjects were 8–16 wk old at the time of testing and had free access to food and water. Littermate mice were used for controls in all experiments. For reversal experiments, animals were fed with food supplemented with 40 mg/kg doxycycline (Bio-Serv) during development and adulthood and throughout the experiment. For genotyping, tail DNA was prepared and analyzed by Southern blotting and/or PCR using transgene-specific probe and/or primers respectively (Abel et al. 1997; forward PCR primer 5′-CTTTCCCAGCCAGCTCTAAG-3′; reverse PCR primer 5′-GGAAAGTCCTTGGGGTCTTC-3′).
Cell culture
HEK 293 cells (ATCC) were maintained in Minimum Essential Media with Earle’s Salts (GIBCO) supplemented with 10% horse serum (ATCC) and 1% penicillin/streptomycin (LTI) and grown at 37°C, 5% CO2. Before transfection, cells at 60%–70% confluence were incubated in serum-free media with 1% penicillin/streptomycin for 3 h. Cells were transfected with a total of 375 ng plasmid DNA using FuGene 6 Transfection Reagent (Roche) according to manufacturer’s protocol. The following plasmids were used: pßgal-Control (BD Biosciences), CRE-Luciferase (Oh et al. 2003), and p300Δ1-MM400. pßgal-Control contains the SV40 early promoter and enhancer sequences inserted upstream and downstream, respectively, of the lacZ gene and was used to normalize the transfection efficacy. CRE-luciferase expresses firefly luciferase under the control of a CRE and functions as a reporter of CRE-mediated transcription. p300Δ1-MM400 expresses p300Δ1 (residues 1–1031) from the CMV promoter. MM400 was added to keep the amount of DNA in each transfection equal. One day after transfection, the cells were treated with 10 μM of forskolin (Sigma) and 10 μM of 3-isobutyl-1-methylxanthine (IBMX) (Sigma) and incubated for 6 h. At the end of incubation, cells were harvested and assayed for luciferase activity with the Luciferase Assay System with Reporter Lysis Buffer (Promega) and β-Galactosidase activity with β-Gal Reporter Gene Assay (Roche).
RT-PCR
Preparation of mRNA and cDNA from amygdala, hippocampi, cortex, and cerebellum was done as described in Wood et al. (2006). Preparation of cDNA was performed with 3 μg of total RNA. Two sets of primers were used for nested PCR. The primers recognize part of the p300Δ1 transgene and the SV40 polyA tail (set 1: forward 5′-CTTTCCCAGCCAGCTCTAAG-3′; reverse 5′-GGAAAGTCCTTGGGGTCTTC-3′; set 2: forward 5′-TTCCCAGG AAGTGAAGATGG-3′; reverse 5′-AGATGGCATTTCTTCTGAG CA-3′) and span one intron. Control primers amplify a 361-bp fragment of a constitutively expressed housekeeping gene, rig/S15, which encodes a small ribosomal subunit protein (RETROscript kit, Ambion).
AcH3 Western blotting
Forebrain from one mouse was homogenized in hypotonic lysis buffer (10 mM Tris at pH 8.0, 1 mM EDTA, 2.5 mM sodium pyrophosphate, 1 mM sodium ortovanadate, 1 mM PMSF, 1% protease inhibitor cocktail [Sigma]). The nuclear pellet was resuspended in 5 volumes of 0.2 M HCl/10% glycerol and the acid supernatant was mixed with 10 volumes of ice cold acetone. The histone pellet was then resuspended in 9 M Urea. The protein quantification was done using the Bradford reagent (Bio-Rad). Samples were loaded in a 15% acrylamide gel and blotted onto a nitrocellulose membrane (Bio-Rad). The membrane was blocked in Tris-buffered saline (TBS) and 5% nonfat dry milk and probed with Actin (Chemicon) and AcH3 (Upstate) antibodies (1:5000 dilution in TBS containing 0.05% Tween 20 [TBST] and 2% milk). Blots were then incubated with AP-conjugated anti-rabbit or anti-mouse secondary antibodies (1:5000 dilution in TBST and 2% milk, Jackson ImmunoResearch). Blots were then incubated in enhanced chemifluorescent substrate (Amersham) and exposed to a fluorescence scanner (Storm, Amersham).
Water maze
The hidden platform experiment was performed using the method previously described (Wood et al. 2005).
Fear conditioning
The fear conditioning experiments were performed using the methods previously described (Wood et al. 2005).
Novel object recognition
The experimental apparatus consisted of a white rectangular open field (60 cm × 50 cm × 26 cm). Prior to training, mice were handled for one minute a day for two days and habituated to the experimental apparatus for five minutes in the absence of objects. During the training phase, mice were placed in the experimental apparatus in the presence of two identical objects and allowed to explore for 15 min. After a retention interval of 30 min or 24 h, mice were placed again in the apparatus where this time one of the objects was replaced by a novel one. Mice were allowed to explore for 15 min. Objects were rinsed with ethanol between trials and before first trial. All testing and training sessions were videotaped and analyzed by an experimenter blind to the genotype of the animals. It was considered exploration of the objects when mice were facing the objects and/or touching it. Preference for the novel object was expressed as the percent time spent exploring the novel object relative to the total time spent exploring both objects. The identity of the objects—which one was novel or familiar—as well as the spatial locations where they were located were balanced between subjects.
Statistical analyses
Statistical analyses were performed using Student’s t-test, with the exception of the Western blot analysis of acetyl histone H3 levels in forebrain of wild-type and transgenic mice and the Morris water maze experiments. In the Western blot experiment, two-way analyses of variance (ANOVA) were performed using Statistica 7. In the Morris water maze experiment, two-way repeated-measures ANOVA were performed using SigmaStat. To compare groups, we used the Student-Newman-Keuls multiple comparisons post-hoc test. All experiments were done by an individual blind to genotype.
Acknowledgments
We thank Dr. Jean Richa at the Transgenic and Chimeric Mouse Facility at the University of Pennsylvania for generating tetOp300Δ1 transgenic animals, Dr. Mark Mayford for the CaMKIIα-tTA line B mice, and Dr. Richard Eckner for providing us p300 cDNA. We thank Christopher Vecsey for comments on the manuscript. This research was supported by a predoctoral fellowship from Fundação para a Ciência e Tecnologia, Portugal (A.M.M.O.), a SFN MNFP postdoctoral fellowship (M.A.W.), and an Alavi-Dabiri postdoctoral fellowship (M.A.W.), and by grants from the National Institutes of Health, the Human Frontiers Science Program, and the Packard Foundation (T.A.).
Footnotes
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.656907
References
- Abel T., Nguyen P.V., Barad M., Deuel T.A., Kandel E.R., Bourtchouladze R. Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long-term memory. Cell. 1997;88:615–626. doi: 10.1016/s0092-8674(00)81904-2. [DOI] [PubMed] [Google Scholar]
- Alarcon J.M., Malleret G., Touzani K., Vronskaya S., Ishii S., Kandel E.R., Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Bailey D.J., Kim J.J., Sun W., Thompson R.F., Helmstetter F.J. Acquisition of fear conditioning in rats requires the synthesis of mRNA in the amygdala. Behav. Neurosci. 1999;113:276–282. doi: 10.1037//0735-7044.113.2.276. [DOI] [PubMed] [Google Scholar]
- Bartholdi D., Roelfsema J.H., Papadia F., Breuning M.H., Niedrist D., Hennekam R.C., Schinzel A., Peters D.J. Genetic heterogeneity in Rubinstein-Taybi Syndrome: Delineation of the phenotype of the first patients carrying mutations in EP300. J. Med. Genet. 2007; 44:327–333. doi: 10.1136/jmg.2006.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchouladze R., Lidge R., Catapano R., Stanley J., Gossweiler S., Romashko D., Scott R., Tully T. A mouse model of Rubinstein-Taybi syndrome: Defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc. Natl. Acad. Sci. 2003;100:10518–10522. doi: 10.1073/pnas.1834280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin K.E., Waxham M.N., Rickling S., Westgate S.A., Mobley W.C., Kelly P.T. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J. Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarota M., Bevilaqua L.R., Ardenghi P., Paratcha G., de Levi Stein M., Izquierdo I., Medina J.H. Learning-associated activation of nuclear MAPK, CREB and Elk-1, along with Fos production, in the rat hippocampus after a one-trial avoidance learning: Abolition by NMDA receptor blockade. Brain Res. Mol. Brain Res. 2000;76:36–46. doi: 10.1016/s0169-328x(99)00329-0. [DOI] [PubMed] [Google Scholar]
- Cesena T.I., Cardinaux J.R., Kwok R., Schwartz J. CCAAT/enhancer-binding protein (C/EBP) β is acetylated at multiple lysines: Acetylation of C/EBPβ at lysine 39 modulates its ability to activate transcription. J. Biol. Chem. 2007;282:956–967. doi: 10.1074/jbc.M511451200. [DOI] [PubMed] [Google Scholar]
- Chan H.M., La Thangue N.B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- Chrivia J.C., Kwok R.P., Lamb N., Hagiwara M., Montminy M.R., Goodman R.H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Eckner R., Ewen M.E., Newsome D., Gerdes M., DeCaprio J.A., Lawrence J.B., Livingston D.M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes & Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- Furia B., Deng L., Wu K., Baylor S., Kehn K., Li H., Donnelly R., Coleman T., Kashanchi F. Enhancement of nuclear factor-κB acetylation by coactivator p300 and HIV-1 Tat proteins. J. Biol. Chem. 2002;277:4973–4980. doi: 10.1074/jbc.M107848200. [DOI] [PubMed] [Google Scholar]
- Furth P.A., St Onge L., Boger H., Gruss P., Gossen M., Kistner A., Bujard H., Hennighausen L. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc. Natl. Acad. Sci. 1994;91:9302–9306. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Gu W., Roeder R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Schulman H. Neuronal Ca2+/calmodulin- dependent protein kinases. Annu. Rev. Biochem. 1992;61:559–601. doi: 10.1146/annurev.bi.61.070192.003015. [DOI] [PubMed] [Google Scholar]
- Hu S.C., Chrivia J., Ghosh A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- Impey S., Fong A.L., Wang Y., Cardinaux J.R., Fass D.M., Obrietan K., Wayman G.A., Storm D.R., Soderling T.R., Goodman R.H. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- Kasper L.H., Boussouar F., Ney P.A., Jackson C.W., Rehg J., van Deursen J.M., Brindle P.K. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419:738–743. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
- Kasper L.H., Fukuyama T., Biesen M.A., Boussouar F., Tong C., de Pauw A., Murray P.J., van Deursen J.M., Brindle P.K. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol. Cell. Biol. 2006;26:789–809. doi: 10.1128/MCB.26.3.789-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E. The relation of transcription to memory formation. Acta Biochim. Pol. 2003;50:775–782. [PubMed] [Google Scholar]
- Korzus E., Rosenfeld M.G., Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson J.M., O'Riordan K.J., Brown K.D., Trinh M.A., Molfese D.L., Sweatt J.D. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Li Q., Xiao H., Isobe K. Histone acetyltransferase activities of cAMP-regulated enhancer-binding protein and p300 in tissues of fetal, young, and old mice. J. Gerontol. A Biol. Sci. Med. Sci. 2002;57:B93–B98. doi: 10.1093/gerona/57.3.b93. [DOI] [PubMed] [Google Scholar]
- Li Q.J., Yang S.H., Maeda Y., Sladek F.M., Sharrocks A.D., Martins-Green M. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 2003;22:281–291. doi: 10.1093/emboj/cdg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad J.R., Kwok R.P., Laurance M.E., Harter M.L., Goodman R.H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- Maren S., Quirk G.J. Neuronal signalling of fear memory. Nature Rev. Neurosci. 2004;5:844–852. doi: 10.1038/nrn1535. [DOI] [PubMed] [Google Scholar]
- Mayford M., Bach M.E., Huang Y.Y., Wang L., Hawkins R.D., Kandel E.R. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- McManus K.J., Hendzel M.J. Quantitative analysis of CBP- and P300-induced histone acetylations in vivo using native chromatin. Mol. Cell. Biol. 2003;23:7611–7627. doi: 10.1128/MCB.23.21.7611-7627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K., Giese K.P. Hippocampus-dependent memory formation: Do memory type-specific mechanisms exist? J. Pharmacol. Sci. 2005;98:191–197. doi: 10.1254/jphs.crj05005x. [DOI] [PubMed] [Google Scholar]
- Morris R.G., Garrud P., Rawlins J.N., O'Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- Oh D.Y., Wang L., Ahn R.S., Park J.Y., Seong J.Y., Kwon H.B. Differential G protein coupling preference of mammalian and nonmammalian gonadotropin-releasing hormone receptors. Mol. Cell. Endocrinol. 2003;205:89–98. doi: 10.1016/s0303-7207(03)00204-1. [DOI] [PubMed] [Google Scholar]
- Oike Y., Hata A., Mamiya T., Kaname T., Noda Y., Suzuki M., Yasue H., Nabeshima T., Araki K., Yamamura K. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: Implications for a dominant-negative mechanism. Hum. Mol. Genet. 1999;8:387–396. doi: 10.1093/hmg/8.3.387. [DOI] [PubMed] [Google Scholar]
- Oliveira A.M., Abel T., Brindle P.K., Wood M.A. Differential role for CBP and p300 CREB-binding domain in motor skill learning. Behav. Neurosci. 2006;120:724–729. doi: 10.1037/0735-7044.120.3.724. [DOI] [PubMed] [Google Scholar]
- Phillips R.G., LeDoux J.E. Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav. Neurosci. 1992;106:274–285. doi: 10.1037//0735-7044.106.2.274. [DOI] [PubMed] [Google Scholar]
- Quivy V., Van Lint C. Regulation at multiple levels of NF-κB-mediated transactivation by protein acetylation. Biochem. Pharmacol. 2004;68:1221–1229. doi: 10.1016/j.bcp.2004.05.039. [DOI] [PubMed] [Google Scholar]
- Roelfsema J.H., White S.J., Ariyurek Y., Bartholdi D., Niedrist D., Papadia F., Bacino C.A., den Dunnen J.T., van Ommen G.J., Breuning M.H., et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: Mutations in both the CBP and EP300 genes cause disease. Am. J. Hum. Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A., Freudenthal R., Merlo E., Routtenberg A. Evolutionarily-conserved role of the NF-κB transcription factor in neural plasticity and memory. Eur. J. Neurosci. 2006;24:1507–1516. doi: 10.1111/j.1460-9568.2006.05022.x. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Naruse I., Maekawa T., Masuya H., Shiroishi T., Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: A partial similarity with Rubinstein-Taybi syndrome. Proc. Natl. Acad. Sci. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay P., Meiner Z., Galou M., Heinrich C., Petromilli C., Lisse T., Cayetano J., Torchia M., Mobley W., Bujard H., et al. Doxycycline control of prion protein transgene expression modulates prion disease in mice. Proc. Natl. Acad. Sci. 1998;95:12580–12585. doi: 10.1073/pnas.95.21.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N., Goodman R.H. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Wood M.A., Kaplan M.P., Park A., Blanchard E.J., Oliveira A.M., Lombardi T.L., Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn. Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M.A., Attner M.A., Oliveira A.M., Brindle P.K., Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn. Mem. 2006;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T.P., Oh S.P., Fuchs M., Zhou N.D., Ch'ng L.E., Newsome D., Bronson R.T., Li E., Livingston D.M., Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- Yuan L.W., Gambee J.E. Histone acetylation by p300 is involved in CREB-mediated transcription on chromatin. Biochim. Biophys. Acta. 2001;1541:161–169. doi: 10.1016/s0167-4889(01)00141-0. [DOI] [PubMed] [Google Scholar]
- Zhong H., Voll R.E., Ghosh S. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- Zou D.J., Greer C.A., Firestein S. Expression pattern of αCaMKII in the mouse main olfactory bulb. J. Comp. Neurol. 2002;443:226–236. doi: 10.1002/cne.10125. [DOI] [PubMed] [Google Scholar]