Abstract
Hematopoietic cells and their progenitors play important roles in human cytomegalovirus latency and reactivation. Latent infection has been evaluated in defined populations of myeloid-lineage-committed progenitor cells coexpressing CD33 and CD15 or CD33 and CD14 along with the dendritic cell markers CD1a and CD10. These CD33+ cell populations were found to support latency and expression of viral latency-associated transcripts and to undergo reactivation of productive viral replication when differentiated in the presence of human fibroblasts. Reactivation was also observed when myeloid cells were carried in the presence of fibroblast-conditioned medium or medium supplemented with certain cytokines (interferon γ, tumor necrosis factor α, interleukin 4, or granulocyte–macrophage colony-simulating factor), suggesting that cell differentiation pathways act as determinants of reactivation. More primitive CD34+ hematopoietic cells were also found to be susceptible to viral infection and latency was maintained as these cells differentiated into CD33+-lineage-committed populations. Between 0.01% and 0.001% of CD33+ CD14+ or CD33+ CD15+ bone marrow mononuclear cells isolated from naturally infected individuals were found to express latent transcripts. Thus, cytomegalovirus is carried within a small percentage of myeloid and dendritic cell progenitors in the healthy seropositive host. Virus reactivation may be triggered by factors associated with the inflammatory response.
Human cytomegalovirus (CMV), a betaherpesvirus, is an important pathogen in immunocompromised patients (1) in whom virus reactivates from latency to cause disease. Previous work implicated monocytes, macrophages, and their progenitors in natural CMV infection (2–5), suggesting that macrophage-lineage cells may provide a long-lived site for CMV latency (6–9). Despite earlier suggestions to the contrary (10), productive viral gene expression is apparently shut down during latency in the naturally infected host (8, 9). Cultured CMV-DNA-positive monocytes from healthy seropositive individuals express productive-phase genes only when stimulated to differentiate with growth or proinflammatory cytokines (11) or after cultivation under conditions of allogeneic stimulation (8). These observations have been consistent with the view that CMV is carried by a primitive hematopoietic cell during latency and that reactivation occurs as macrophage- or dendritic-lineage cells differentiate. Permissiveness for CMV replication has long been associated with differentiation (9, 12) and the monocyte-derived macrophage differentiation state appears to dictate the level of permissiveness for CMV (2, 3, 8, 11, 13, 14). Studies carried out on experimentally infected granulocyte–macrophage progenitors (GM-Ps) have supported this view of latency and have identified specific CMV latency-associated transcripts (CLTs) encoded within the ie1/ie2 region of the viral genome. Although first identified in mixed populations of experimentally infected GM-Ps, latent transcripts are detected in bone marrow (BM) mononuclear cells from naturally infected CMV-seropositive adults (15).
On the one hand, primitive (CD34+) and lineage-committed (CD33+) cells may act as reservoirs for CMV in the naturally infected host (7, 15–17) and differentiated monocyte-derived macrophages with dendritic cell characteristics appear to support productive viral replication (8, 14). On the other hand, evidence that BM-derived myelomonocytic progenitors give rise to antigen-presenting dendritic cells in addition to granulocytes and macrophages is also accumulating (18, 19). Human-cord-blood-derived CD34+ progenitors differentiate into dendritic cells when grown in the presence of granulocyte–macrophage colony-simulating factor (GM-CSF) and tumor necrosis factor α (TNF-α) (19–22). Although hematopoietic-stem-cell-enriched populations by definition mature into a wide variety of cell lineages (18, 23, 24), CMV DNA has been associated only with myelomonocytic lineage cells in healthy latently infected individuals (6, 25), suggesting that the viral genome is preferentially maintained in this cell population. These results suggest that CMV may target a long-lived cell population that gives rise to monocyte-derived macrophages and dendritic cells as a site of latency.
Our focus has been to delineate the cell type distribution of latent transcripts in hematopoietic progenitor and peripheral blood (PB) cell types during CMV experimental infection and in the naturally infected host. To investigate sites of CMV latency and to determine whether cells related to dendritic progenitors have the capacity to carry latent virus, we separated progenitor and PB mononuclear cell types and evaluated these populations for latent transcripts with a highly sensitive reverse transcription-coupled PCR (RT-PCR) analysis. We present evidence that CD33+ progenitors of dendritic cells and monocyte–macrophages are important reservoirs of latent CMV and show that T cells, B cells, and CD33− mature granulocytes and macrophages are not. In an attempt to reveal the cellular determinants of latency and reactivation, we demonstrate that progenitor populations reactivate virus after coculture with human fibroblasts (HFs) and that reactivation is triggered by soluble HF cell factors or certain cytokines. Finally, we detect latency-associated transcripts in 0.01–0.001% of sorted CD33+ but not in CD33− cell populations within naturally infected individuals. Thus, natural latent infection is maintained in hematopoietic progenitors with viral DNA and latent gene expression accompanying the growth and differentiation of lineage-committed cells.
MATERIALS AND METHODS
Cell and Virus Culture.
Fetal liver cells (from 16- to 24-week gestation abortuses) were separated on Lymphoprep (GIBCO/BRL) and cultured as described (16) except that a high density (1 × 106 cells per ml) was maintained. These tissues were collected in accordance with human subjects guidelines at Stanford University. At day 4 of culture, cells were infected with RC256, a lacZ-positive derivative of human CMV Towne (26) or the low-passage strain Toledo (27) at a multiplicity of infection of 3 and passaged for 3–4 weeks.
Cocultivation and Transwell Chamber Assays.
Reactivation by cocultivation was as described (16) except that fluorescence-activated cell sorter (FACS)-sorted GM-Ps were seeded at 5,000 cells per well (six-well plate, Costar) in the presence of 70% confluent HFs and 0.1% pooled human gamma-globulin (Armour Pharmaceutical). Cultures were stained with 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-gal) (26) and the mAb 810 (1:500 dilution; Chemicon) followed by a fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (1:100 dilution; Vector Laboratories). For cytokine-mediated reactivation, 1 × 105 GM-Ps at 4 weeks after infection (p.i.) were seeded per well (96-well round-bottom plate, Costar) and cultured for 2 weeks in the presence of either interferon γ (IFN-γ; 100 units/ml; Boehringer), tumor necrosis factor α (TNF-α; 10 ng/ml; Boehringer), GM-CSF (50 units/ml; Boehringer), interleukin (IL) 4 (25 ng/ml; R & D Systems), IL-3 (50 units/ml; Boehringer), IL-6 (100 units/ml; Boehringer), transforming growth factor β (10 ng/ml; R & D Systems), IL-10 (10 ng/ml; R & D Systems), stem cell factor (50 ng/ml; R & D Systems), lymphocyte inhibitory factor (20 ng/ml; Boehringer), and one of the following chemokines at 50 ng/ml: regulated on activation of normal T cells, expressed and secreted macrophage inflammatory protein (MIP)-1α, MIP-1β, or macrophage chemotactic protein (MCP)-1 (all provided by Tom Schall, DNAX Research Institute). Cells were collected onto slides by using a cytocentrifuge and subjected to X-gal and IE1/IE2 antibody staining at the time of cytokine addition and at 6 and 10 weeks of culture.
cDNA Synthesis and RT-PCR.
Three hundred cells in 1 μl of PBS were sorted into 7 μl of RNA buffer containing 8% Triton X-100 (Sigma), BSA (Sigma) at 200 μg/ml, 1 mM spermidine (Sigma), 6 mM methylmercury hydroxide (Johnson Matthey, Karlsruhe, Germany, distributed by Alpha Chemical) (28), all four dNTPs (each at 2 mM; United States Biochemical), and random primers (GIBCO/BRL)at 300 ng per reaction. Seven microliters of a solution of 50 mM DTT (GIBCO/BRL), RNase inhibitor (40 units per reaction; Boehringer Mannheim), and Superscript II at 10 μl (200 units) per reaction (GIBCO/BRL; prepared in 2.5× reverse transcriptase buffer diluted from 5× buffer provided by the manufacturer) were added. cDNA synthesis (in a total volume of 25 μl) was for 90 min at 42°C followed by heating to 72°C for 10 min. Each subsequent PCR amplification used 3 μl of this mixture.
Nested PCR, electrophoresis in 3% agarose gels, immobilization, and hybridization with [γ-32P]ATP-end-labeled oligonucleotides IEP1M or IEP4AP were performed as described (15, 16). PCR with primers for cellular histidyl-tRNA synthetase transcripts (11) were used to assess the presence of amplifiable cDNA in all samples. All oligonucleotides were purchased from the Stanford University PAN Facility. Single transcript-positive cells were shown to be detectable in pools of up to 5 × 103 cells in a reconstruction experiments with 1 × 102, 3 × 102, 1 × 103, 5 × 103, 1 × 104, and 3 × 104 uninfected GM-Ps. cDNA was prepared as described above and amplified for the 206-bp product of sense CLTs. The confidence intervals of numbers of positive cells in pools of 5 × 103 cells was statistically evaluated by using a Poisson distribution. The 95% confidence interval for one positive cell in 5 × 103 cells in three or six pools of 5 × 103 cells was calculated to be 1.04 and 1.73 or 1.02 and 1.32, respectively.
Isolation and Culture of PB and BM Mononuclear Cell Populations.
Twenty milliliters of heparinized PB or 2 ml of BM mononuclear cells were layered over Lymphoprep (GIBCO/BRL) and sedimented by centrifugation for 15–20 min at 600–750 × g. Con A (10 μg/ml, final concentration; Sigma) stimulation of PB mononuclear cells was performed for 3 days before infection with CMV (multiplicity of infection of 3). Nonadherent cells were harvested at 3 days p.i. and subjected to flow cytometry. BM samples were cultured for 1 week under GM-P conditions before being subjected to flow cytometry.
Antibody Staining and Flow Cytometry.
Approximately 106 GM-Ps or BM cells were incubated with fluorochrome-conjugated antibodies for 30 min at 4°C in 500 μl of Hanks’ balanced saline solution (HBSS) containing 2% FBS and 1 μl of each undiluted antibody. After washing three times in HBSS, cells were suspended in HBSS supplemented with 5% FBS and subsequently sorted with a Flasher flow cytometer (Becton Dickinson) equipped with a 5-W argon argon laser (all channels) and 488-nm [FITC and phycoerythrin (PE) channel] and 604-nm (allophycocyanin channel) lasers or a FACStar flow cytometer (BD) equipped with a 5 W argon laser (all lines). The following band pass filters were used: FL1 (FITC) 530/30; FL2 (PE) 575/25; FL3 [propidium iodide and tricolor (TC)] 670/40, FL4 (allophycocyanin) 660/20 nm. Positive and negative staining with cell-surface-specific antibodies was defined as the emission of a level of fluorescence that exceeded or did not exceed levels obtained by 99% of the cells from the same starting population when these were stained with control fluorochrome-conjugated antibodies. The purity of each cell sort was typically greater than 97% as determined by reanalyses. Very bright fluorescent cells, with a fluorescence intensity above 160 on a linear scale, were referred to as positive; dull cells, with a staining intensity below 80, were referred to as negative; and cells that fell between 80 and 160 were referred to as intermediate. The following antibodies were used: CD34 (Becton Dickinson, BD), CD34-PE (Caltag, South San Francisco, CA), CD33-PE (BD), CD33-FITC (Caltag), CD38-TC (Caltag), CD3-FITC (BD), CD4-FITC (BD), CD19-FITC (BD), CD14-FITC (BD), CD15-FITC (BD), CD1a (BD), CD1a-PE (Caltag), CD10-TC (Caltag), CD11a (BD), CD11b (Dako), CD29 (BD), CD31 (BD), CD44 (BD), CD49d (BD), CD54 (BD), intercellular cell adhesion molecule 2 (BD), E-cadherin (BD), s-LE-x (BD), anti-murine IgG (heavy and light chains, H+L) TC (Fab fragment) (Caltag), anti-murine IgG (H+L) PE (Caltag), anti-murine IgG (H+L) FITC (Caltag), and anti-murine IgG (H+L) allophycocyanin (Caltag).
Methylcellulose Culture Assay.
Hematopoietic progenitor cells were FACS-sorted into CD34+ CD33+ and CD34+ CD33− cells, infected with either Towne (RC256) or Toledo at an multiplicity of infection of 3 and subsequently washed three times with HBSS. Methylcellulose assay (StemCell Technologies, Vancouver, BC, Canada) was performed according to the manufacturer’s instructions. The following cytokines were added: c-kit ligand (100 ng/ml; R & D Systems), erythropoietin (2 units/ml; R & D Systems), GM-CSF (20 ng/ml; Boehringer Mannheim), and IL-3 (20 ng/ml; R & D Systems) (23). Single CFU-GM colonies were subjected to cDNA preparation and RT-PCR analysis at 3 weeks p.i.
RESULTS
Detection of Latency-Associated Transcripts in Hematopoietic Progenitor Subpopulations.
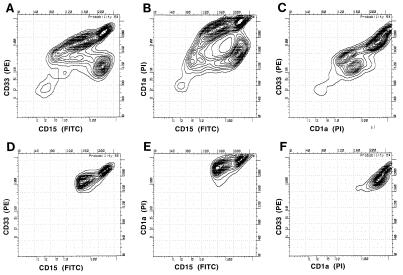
We had found that viral DNA was carried in a large percentage of CMV-infected fetal-liver-derived GM-Ps (16) and that transcription of the viral genome was restricted to latency-associated transcripts (15). GM-P cultures were infected with either RC256, a lacZ-tagged derivative of the Towne strain (26), or the low-passage Toledo strain (27). Three to 4 weeks p.i., we used a FACS in conjunction with antibody to cell surface markers to sort CD33+ lineage-committed cells carrying surface markers indicative of granulocyte (CD15), monocyte (CD14), or dendritic lineages (CD10 and CD1a) and subjected panels of 300 sorted cells to analysis for sense latency-associated transcripts. Fig. 1 shows one analysis of infected GM-P cultures for cells expressing CD33, CD1a, and CD15. CD33+ CD10+ CD15+ and CD33+ CD1a+ CD14+ populations were also analyzed (data not shown). This work confirmed the existence of a dendritic precursor coexpressing CD14 and CD1a (22) and revealed the existence of previously unrecognized cell populations expressing the markers CD33+ CD15+ CD10+ and CD33+ CD15+ CD1a+ that may represent dendritic cell progenitors in the myeloid lineage.
Figure 1.
Flow cytometry analysis of infected CD33+ CD15+ CD1a+ dendritic lineage cells. Three-color analysis of CMV-infected progenitors 3 weeks p.i. was performed with anti-CD33 (PE), anti-CD1a (TC, read on the propidium iodide channel), and anti-CD15 (FITC). A–C show the FACS profile of analyzed cells. D–F show the sorted populations.
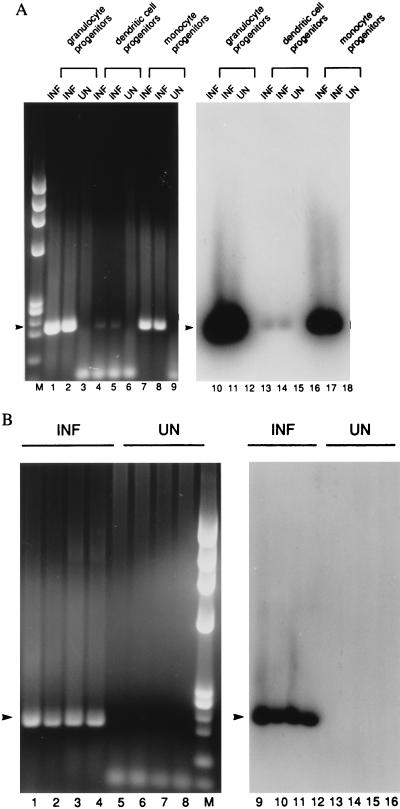
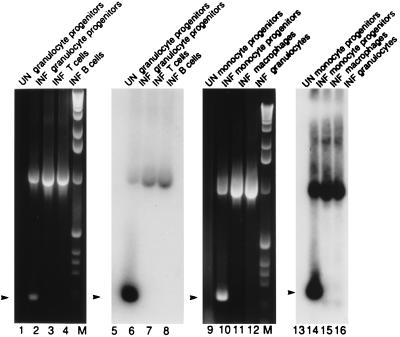
RNA from replicate aliquots of 300 FACS-sorted cells was reverse-transcribed and RT-PCR analysis was performed to assess the presence of latent viral transcripts. In the example shown in Fig. 2, sense transcripts were detected in progenitors of monocytes (CD33+ CD14+), granulocytes (CD33+ CD15+), and dendritic cells (CD33+ CD10+ CD15+, CD33+ CD1a+ CD15+, and CD33+ CD1a+ CD14+). In contrast to the progenitor populations, viral transcripts were not detected in differentiated CD33− cells (data not shown). In all, 83% of CD33+ CD15+ (10 of 12 cultures), 60% of CD33+ CD14+ (3 of 5 cultures), 83% of CD33+ CD1a+ CD15+ (5 of 6 cultures), 100% of CD33+ CD1a+ CD14+ (2 of 2 cultures), 67% of CD33+ CD10+ CD15+ (2 of 3 cultures), and neither of the more highly differentiated CD33− CD15+ populations (0 of 2 cultures) were sense-transcript-positive when the results from Towne- or Toledo-infected GM-P cultures were summarized. Thus, FACS-sorting suggested that all CD33+ cell populations, including a previously unrecognized population coexpressing the myeloid markers CD33 and CD15 along with the dendritic marker CD1a and the previously recognized CD33+ CD14+ CD1a+ predendritic population (22) were able to maintain latent CMV. These data implicate dendritic and myeloid progenitors as reservoirs of latent virus.
Figure 2.
RT-PCR analysis to detect latent transcripts in pools of 300 FACS-sorted monocyte-, granulocyte-, and dendritic-lineage cells 3 weeks p.i. RT-PCR for sense CLTs yields a 206-bp product (arrowhead). (A) The following cell types were analyzed. Lanes: 1–3 and 9–11, granulocyte progenitors (CD33+ CD15+); 4–6 and 12–15, dendritic cell progenitors (CD33+ CD10+ CD15+); 7–9 and 16–18, monocyte progenitors (CD33+ CD14+). (B) The following dendritic cell progenitors were analyzed. Lanes: 1, 2, 5, 6, 9, 10, 13, and 14, CD33+ CD1a+ CD15+; 3, 4, 7, 8, 11, 12, 15, and 16, CD33+ CD1a+ CD14+. (Left) Ethidium bromide-stained gel. (Right) DNA blot. INF, infected cells; UN, uninfected cells; M, molecular weight marker HaeIII-digested φX174.
To determine whether CMV infection induced any change in cell surface markers on GM-P populations, we evaluated a panel of lineage markers and adhesion molecules with mAbs and flow cytometry but found little or no difference in the levels of CD34, CD33, CD14, CD15, CD10, CD1a, CD11a, CD11b, CD29, CD31, CD44, CD49d, CD54, intercellular cell adhesion molecule 2, E-cadherin, or s-LE-x when CMV-infected cells were compared with uninfected cells (data not shown). Consistent with earlier analyses (16), the presence of latent virus did not appear to significantly alter the growth rate, cell surface phenotype, or differentiation state of hematopoietic progenitors.
Reactivation of CMV from CD33+ CD15+, CD33+ CD14+, and CD33+ CD15+ CD1a+ Populations After Coculture with Permissive Fibroblasts.
To determine whether latent-transcript-positive cell populations would reactivate after extended coculture with HFs (16), 5,000 virus-infected CD33+ CD15+, CD33+ CD14+, or CD33+ CD15+ CD1a+ sorted GM-Ps were cocultured in the presence of uninfected HFs. CMV (RC256)-infected GM-Ps did not exhibit any signs of productive infection (lack of detectable infectivity associated with cells or medium) and failed to express detectable IE1/IE2 antigen or β-galactosidase at the time coculture was initiated (data not shown). Reactivation of sorted populations of RC256-infected CD33+ cells was monitored by appearance of β-galactosidase- and virus-positive cells over a 28-day period (12) (Table 1). A majority of these cultures yielded reactivated virus within 21 days, indicating that CD33+ cell populations coexpressing CD14+, CD15+, or CD15+ and CD1a+ could support reactivation and productive replication when differentiated in the presence of HFs. The reactivation of virus after extended coculture demonstrates that various CD33+ cell populations had supported latent and not abortive infection.
Table 1.
Detection of virus reactivation in FACS-sorted cells
| Day | No. X-gal-positive samples/no. total samples
|
||
|---|---|---|---|
| CD33+15+ | CD33+14+ | CD33+15+1a+ | |
| 7 | 0/5 | 0/3 | 0/3 |
| 14 | 0/5 | 0/3 | 0/3 |
| 21 | 3/5 | 2/3 | 2/3 |
| 28 | 4/5 | 2/3 | 2/3 |
Day following the start of coculture with HFs is indicated.
Reactivation of CMV Induced by Cytokines.
To determine whether soluble factors produced by HFs were sufficient to trigger reactivation, latently infected GM-Ps (3 weeks p.i.) and uninfected HFs were placed on opposite sides of a 0.45-μm (pore size) membrane in a Transwell Chamber to separate these cell populations but allow the free diffusion of cytokines. In three individual experiments, RC256-infected GM-Ps that were negative for both β-galactosidase and IE1/IE2 expression before coculture became positive for productive infection within 4–6 weeks of additional coculture (data not shown). X-gal-positive cells were recovered in the top compartment and virus produced in the top compartment spread through the 0.45-μm membrane to form plaques on the HFs in the lower compartment (data not shown) over the course of several weeks. This result suggested that soluble factors might play a pivotal role in reactivation and led to an evaluation of proinflammatory and growth cytokines for ability to induce reactivation. GM-Ps at 3 weeks p.i. were either cocultured with HFs or cultured for a 2-week period (between 4 and 6 weeks p.i.) in the presence of IFN-γ, TNF-α, GM-CSF, IL-4, IL-3, IL-6, transforming growth factor β, IL-10, stem cell factor, lymphocyte inhibitory factor, RANTES, MIP-1α, MIP-1β, or macrophage chemotactic protein-1. All GM-Ps reactivated when cocultured with HFs. Viral antigen and β-galactosidase expression were monitored in all cytokine treated samples at 6 and 10 weeks p.i. As shown in Table 2, culture of cells in the presence of IFN-γ or TNF-α led to reactivation of CMV in all three cultures with the numbers of antigen- or X-gal-positive cells ranging from 5 to 30% at 10 weeks p.i. GM-CSF and IL-4 also induced reactivation but in a smaller percentage of cells. Repeated experiments with these cytokines continued to yield reactivation in approximately one-third of the attempts. Future experiments should investigate mixtures of these four cytokines. Our results are consistent with cytokines playing an important role as determinants of hematopoietic cell differentiation state.
Table 2.
Reactivation of latently infected GM-Ps in the presence of cytokines
| Factor | Expression | Reactivation
|
|||||
|---|---|---|---|---|---|---|---|
| Sample 1
|
Sample 2
|
Sample 3
|
|||||
| 6 wk | 10 wk | 6 wk | 11 wk | 6 wk | 11 wk | ||
| INF-γ | IE1-2/lacZ | −/− | +++/+++ | −/− | −/++ | −/− | ++/+++ |
| TNF-α | IE1-2/lacZ | −/− | ++/+++ | −/− | −/++ | −/− | −/+ |
| GM-CSF | IE1-2/lacZ | ND | ND | ND | ND | −/− | +/+ |
| IL-4 | IE1-2/lacZ | ND | ND | −/− | +/+ | −/− | −/− |
| None | IE1-2/lacZ | −/− | −/− | −/− | −/− | −/− | −/− |
Cytokine was present during weeks 4–6 p.i. Samples were taken 6, 10, and 11 weeks p.i. +++, 15–30%; ++, 5-15%, +, 0.1–5%, −, 0.1%; ND, not done.
Detection of Latent Transcripts in PB Mononuclear Cells.
To investigate the ability of different PB cell populations to express latent transcripts, PB mononuclear cells from three CMV seronegative individuals were isolated, Con A-stimulated for 3 days, and infected with CMV (RC256). At 3–5 days p.i. with RC256, these cells were sorted into monocyte-lineage and granulocyte-lineage populations and T and B cells (data not shown). Fig. 3 shows an example of this analysis of cells from one blood donor. Consistent with our findings on fetal-liver-derived GM-Ps, CD33+ cells of the GM lineage in the PB expressed latent transcripts, whereas T cells, B cells, and mature CD33− cells did not. PCR for cellular histidyl-tRNA synthetase showed that all samples contained amplifiable cDNA (data not shown). Viral DNA was detected by PCR in all samples (data not shown). T and B lymphocytes, mature granulocytes, and macrophages lacked latent transcripts; however, ie1 region RNA was detected in these cells with ie1 exon 3/4-specific primers (15), consistent with the establishment of an abortive infection (29) (data not shown). Thus, these data suggest that detection of sense CLTs is associated with the differentiation state of the host cell and the expression of the cell surface marker CD33 on a precursor of myelomonocytic cells.
Figure 3.
RT-PCR analysis to detect latent transcripts in 300 infected and sorted PB mononuclear cells. Cells with the following cell surface phenotypes were analyzed. Lanes: 1, 2, 5, and 6, granulocyte progenitors (CD33+ CD15+); 3 and 7, T cells (CD3+ CD4+); 4 and 8, B cells (CD19+); 9, 10, 13, and 14, monocyte progenitors (CD33int CD14+); 11 and 15, monocyte/macrophages (CD33− CD14+); 12 and 16, granulocytes (CD33− CD15+). Lanes 1–4 and 9–12 show ethidium bromide stained gels, and lanes 5–8 and 13–16 show the DNA blot. Conditions and abbreviations are as in Fig. 2. M, molecular weight marker 1-kb ladder. Arrowhead, sense CLTs (206 bp).
Maintenance of CMV in CFU-GM Originating from CD34+ CD33− or CD34+ CD33+ Fetal Liver Cell Populations.
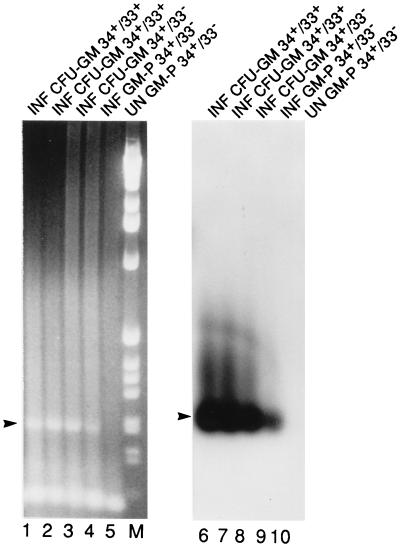
To determine whether primitive hematopoietic progenitors could be infected with CMV and whether latent virus could be maintained as these progenitors differentiated, we sorted mononuclear cells from two fetal liver samples into CD34+ CD33− or CD34+ CD33+ populations and, after infection, placed samples into GM-P culture or methylcellulose culture (23). After 3–4 weeks in GM-P culture, cells were collected and cDNA was prepared from pools of 300 FACS-sorted CD33+ CD15+ cells. Sense latency-associated transcripts were readily detected in CD33+ CD15+ populations originating either from CD34+ CD33− or CD34+ CD33+ populations (Fig. 4). Although both may contain progenitor cells, the CD34+ CD33− population has been considered more primitive because these cells retain the capacity to form colonies in long-term BM cultures (30). There were no significant differences in numbers of erythroid colony-forming units or CFU-GM in CMV (RC256)-infected as opposed to uninfected cultures (data not shown). After 3 weeks of culture, individual CFU-GM were picked and cDNA was prepared. Sense transcripts were detected in CFU-GM originating from both CD34+ CD33− and CD34+ CD33+ cell populations (Fig. 4). Nucleic acids were extracted from 12 colonies (CFU-GM) grown from CD34+ CD33+ cells, and 8 were sense-CLT-positive by RT-PCR. Ten colonies from CD34+ CD33− cells were analyzed and 5 were sense-CLT-positive. This work revealed the susceptibility of primitive hematopoietic progenitors and showed that latent transcripts were expressed as pluripotent progenitors divided into GM-lineage committed cells. In a preliminary analysis, we also cultured FACS-sorted CD34+ CD33− cells on the mouse stromal cell AC-6 (provided by Irving Weissman) (31) and were able to detect latency-associated transcripts in sorted CD34+ CD33+ cells but not in sorted CD34+ CD33− cells, consistent with the expression of this transcript in CD33+ but not CD33− cell populations.
Figure 4.
RT-PCR analysis to detect latent transcripts in CFU-GM colonies picked from methylcellulose culture. CFU-GM colonies originated from the following FACS-sorted and infected populations. Lanes: 1, 2, 6, and 7, CD34+ CD33+ cells; 3 and 8, CD34+ CD33− cells; 4, 5, 9, and 10, CD34+ CD33− cells grown in liquid culture for 3 weeks and then sorted into CD33+ CD15+ cells. (Left) Ethidium bromide-stained gel. (Right) DNA blot. Conditions and abbreviations are as in Fig. 2. M, molecular weight marker 1-kb ladder.
Detection of Latent Transcripts in the BM of Naturally Infected Healthy Donors.
To determine whether the results with cultured GM-Ps reflected the behavior of the virus in the naturally infected host, CD33+ and CD33− BM mononuclear populations cells from CMV-seropositive individuals were isolated and examined. We sought to extend the study by Kondo et al. (15) to include an analysis of cell types. BM contains a range of primitive hematopoietic progenitors, including stem cells (32), that might harbor latent CMV. Mononuclear cells from five CMV-seropositive and two CMV-seronegative BM donors were sorted into CD33+ or CD33− populations. Cells from four individuals (donors 1, 2, 4, and 6) were sorted into CD33+ CD15+ and CD33− CD15+ populations. As shown in Table 3, sense CLTs were detected in 1.5 × 104 (donor 1) and 3 × 104 (donor 2) CD33+ CD15+ cells when analyzed as pools of 5 × 103 cells. Sense CLTs were not detected in a total of 3 × 104 CD33− cells (donors 1–5). For donors 3 and 5, samples totaling 1.5 × 104 CD33+ CD14+ cells were found to be transcript-positive. Transcripts were not detected in any cell populations from CMV-seronegative individuals (donors 6 and 7, 1. 5 × 104 to 3.0 × 104 cells analyzed). A distribution frequency of 0.01–0.001% was estimated from this data, suggesting that seropositive individuals carry approximately one sense-CLT-positive cell in 1 × 104 to 1 × 105 CD33+ progenitors. Thus, the presence of latency-associated transcripts in CD33+ progenitors from healthy seropositive individuals corroborates the results derived from GM-P cultures and implicates these cells as an important reservoir of latent CMV.
Table 3.
Expression of sense CLTs in CD33+ progenitors during natural infection
| Donor | CMV serostatus | FACS-sorted cell type, % CLT-positive cells
|
|
|---|---|---|---|
| CD33− cells | CD33+ cells | ||
| 1 | + | <0.007 | <0.007 |
| 2 | + | <0.007 | 0.003 |
| 3 | + | <0.007 | <0.007 |
| 4 | + | <0.007 | <0.007 |
| 5 | + | <0.007 | 0.007 |
| 6 | − | <0.007 | <0.007 |
| 7 | − | <0.007 | <0.007 |
Donors: 1, 2, 4, and 6, CD33+CD15+, CD33− CD15+; donors 3, 5, and 7, CD33+CD14+, CD33−CD14+.
DISCUSSION
In this study we have established that transcripts previously determined to be latency-associated (15) are harbored in defined subpopulations of CD33+ progenitors that have characteristics in common with dendritic and myeloid lineages. Although a majority of the work herein was carried out with a lacZ-tagged derivative of the Towne strain, the same transcript and cell distribution pattern was observed when experiments were carried out using the Toledo strain, a low passage virus that is known to carry additional genes compared with Towne strain (33, 34). Once GM-Ps were infected, progenitors of monocytes, granulocytes, and dendritic cells expressed latent transcripts, whereas mature macrophages, granulocytes, T cells, and B cells lacked evidence of these transcripts. Viral gene expression was restricted to latency-associated transcripts in these progenitors. After coculture of infected CD33+-expressing dendritic and myeloid progenitors with permissive HFs, virus reactivated within 3–4 weeks in a pattern that was similar to that previously characterized for nonsorted GM-Ps (16). Latent infection of CD33+ cells was dependent on culture conditions and viral reactivation followed a period of coculture with uninfected HFs. Our FACS analyses corroborated the work of Caux et al. (22) and defined cell populations coexpressing the myeloid markers CD33 and CD15 or CD14 along with the dendritic markers CD1a and CD10, and the analysis of these cell populations for viral RNA suggested that progenitors of dendritic cells and progenitors of granulocytes and macrophages are responsible for maintaining CMV latency.
Our studies add significant evidence to the importance of the host cell as a determinant of CMV latency. Progenitors undergoing a nonproductive infections convert to productive viral replication when GM-Ps or sorted CD33+ GM-P populations are cultivated in the presence of HFs, HF soluble factors, or specific proinflammatory cytokines. Even in the absense of immunity that is important in controlling CMV replication in the natural host, the differentiation state of GM-Ps appears critical in dictating whether latency is maintained or is reactivated. Our demonstration that individual growth and proinflammatory cytokines can influence reactivation suggests a connection to the activation state of the host cell. This observation may provide an explanation previous reports suggesting that productive infection is the outcome in a proportion of BM-derived mononuclear cells (35). These earlier results may have been influenced by the level cytokines such as IFN-γ or TNF-α included in the culture medium and are consistent with a cell-differentiation-state dependence for CMV permissiveness (2, 3, 8, 11, 13, 14). Herein and previously (16) we have shown that GM-P culture conditions maintain a uniform level of latency without spontaneous or continuous differentiation-dependent generation of permissive cells. It will now be very important to understand the phenotype of different cell populations as well as the signals that lead to various differentiation states.
Previous evaluation for evidence of viral DNA has implicated CD34+ cells as a reservoir for CMV in the naturally infected host (7, 17). In our hands, experimental infection of sorted CD34+ cells resulted in a latent infection and the expression of CMV latent transcripts in CD33+ cell populations arising in either suspension or methylcellulose culture. Thus, experimental infection of pluripotent cells does not alter differentiation, suggesting that primitive (CD34+) hematopoietic progenitors may be a reservoir for CMV to retain a lifelong foothold in the host. We have also detected the CMV genome in cells expressing CD34 and Thy-1 cell surface markers (G.H., L. Murray, K. Lüns, and E.S.M., unpublished data) isolated from naturally infected individuals, which is consistent with this model.
From our evaluation of naturally infected myelomonocytic cells, it appears that a progenitor of dendritic and myeloid lineage cells harbors latent virus in a manner similar to that of cultured GM-Ps. Our study of BM reveals CD33+ progenitors in the BM of seropositive individuals as a source of latent CMV and suggests that the frequency of latent CMV in the CD33+ progenitor population can be greater than 0.001% in the naturally infected host. These findings extend a previous study by Kondo et al. (15, 16) who showed that sense CLTs (but not productive-phase transcripts) were present in mononuclear cells from seropositive but not seronegative healthy BM donors.
We propose a mechanism of latency in which a primitive hematopoietic cell population, possibly stem cells, acquire CMV during viremia that follows primary infection (1, 27). The viral genome may be retained with limited gene expression as primitive cells reside quiescently in the BM or self-renew. Upon differentiation into lineage-committed progenitors, latency-associated transcripts and proteins may be expressed in association with cell differentiation. Full differentiation to tissue dendritic cells or macrophages in a particular proinflammatory cytokine milieu may drive host cells into a permissive state that supports virus replication (2, 3, 8, 11, 13, 14).
Acknowledgments
We thank members of the Stanford University Hospital Bone Marrow Transplantation Program for donor samples, William Dworsky (Stanford University Hospital) for liver samples, Ellen Rozler (SyStemix) and Beth Hill (SyStemix) for sharing their expertise in methylcellulose culture, Hideto Kaneshima (SyStemix) for mAbs and many insights, Tim Knaak (Stanford) for advice in FACS analysis, Norbert Hain (Stanford) for helpful discussions, and Peter Parham (Stanford) and Mark Davis (Stanford) for comments on the manuscript. This work was supported by grants from the National Institute of Health (RO1 AI33852 and PO1 CA49605) and SmithKline Beecham. G.H. was supported by a fellowship from the Deutsche Forschungsgemeinschaft.
ABBREVIATIONS
- CMV
cytomegalovirus
- CLT
CMV latency-associated transcripts
- GM-P
granulocyte-macrophage progenitors
- FITC
fluorescein isothiocyanate
- PE
phycoerythrin
- X-gal
5-bromo-4-chloro-3-indolyl β-d-galactopyranoside
- BM
bone marrow
- PB
peripheral blood
- FACS
fluorescence-activated cell sorter
- IFN-γ
interferon γ
- TNF-α
tumor necrosis factor α, GM-CSF, granulocyte-macrophage colony-stimulating factor: IL, interleukin
- TC
tricolor
- RT-PCR
reverse transcription-coupled PCR
- HFs
human fibroblasts
- MIP
macrophage inflammatory protein
- p.i.
after infection
References
- 1.Alford C A, Britt W J. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. New York: Lippincott-Raven; 1996. pp. 2493–2534. [Google Scholar]
- 2.Ibanez C E, Schrier R, Ghazal P, Wiley C, Nelson J A. J Virol. 1991;65:6581–6588. doi: 10.1128/jvi.65.12.6581-6588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lathey J L, Spector S A. J Virol. 1991;65:6371–6375. doi: 10.1128/jvi.65.11.6371-6375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maciejewski J P, Bruening E E, Donahue R E, Sellers S E, Carter C, Young N S, St. Jeor S. Virology. 1993;195:327–336. doi: 10.1006/viro.1993.1383. [DOI] [PubMed] [Google Scholar]
- 5.Minton E J, Tysoe C, Sinclair J H, Sissons J G. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor-Wiedeman J, Sissons J G, Borysiewicz L K, Sinclair J H. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 7.Mendelson M, Monard S, Sissons P, Sinclair J. J Gen Virol. 1996;77:3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 8.Soderberg-Naucler C, Fish K N, Nelson J A. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 9.Sinclair J, Sissons P. Intervirology. 1997;39:293–301. doi: 10.1159/000150501. [DOI] [PubMed] [Google Scholar]
- 10.Schrier R D, Nelson J A, Oldstone M B. Science. 1985;230:1048–1051. doi: 10.1126/science.2997930. [DOI] [PubMed] [Google Scholar]
- 11.Taylor-Wiedeman J, Sissons P, Sinclair J. J Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson J A, Gnann J J, Ghazal P. Curr Top Microbiol Immunol. 1990;154:75–100. doi: 10.1007/978-3-642-74980-3_4. [DOI] [PubMed] [Google Scholar]
- 13.Sinclair J H, Baillie J, Bryant L A, A, T-W J, Sissons J G. J Gen Virol. 1992;73:433–435. doi: 10.1099/0022-1317-73-2-433. [DOI] [PubMed] [Google Scholar]
- 14.Fish K N, Britt W, Nelson J. J Virol. 1996;70:1855–1862. doi: 10.1128/jvi.70.3.1855-1862.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo K, Xu J, Mocarski E S. Proc Natl Acad Sci USA. 1996;93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo K, Kaneshima H, Mocarski E S. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Laer D, Meyer-Koenig U, Serr A, Finke J, Kanz L, Fauser A A, Neumann-Haefelin D, Brugger W, Hufert F T. Blood. 1995;86:4086–4090. [PubMed] [Google Scholar]
- 18.Inaba K, Inaba M, Deguchi M, Hagi K, Yasumizu R, Ikehara S, Muramatsu S, Steinman R M. Proc Natl Acad Sci USA. 1993;90:3038–3042. doi: 10.1073/pnas.90.7.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J. Nature (London) 1992;360:258–261. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- 20.Caux C, Massacrier C, Dezutter-Dambuyant C, Vanbervliet B, Jacquet C, Schmitt D, Banchereau J. J Immunol. 1995;155:5427–5435. [PubMed] [Google Scholar]
- 21.Young J W, Szabolcs P, Moore M A. J Exp Med. 1995;182:1111–1119. doi: 10.1084/jem.182.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caux C, Vanbervliet B, Massacrier C, Dezutter-Dambuyant C, de Saint-Vis B, Jacquet C, Yoneda K, Imamura S, Schmitt D, Banchereau J. J Exp Med. 1996;184:695–706. doi: 10.1084/jem.184.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galy A, Travis M, Cen D, Chen B. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 24.Steinman R M. Annu Rev Immunol. 1991;9:271–296. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 25.Stanier P, Taylor D L, Kitchen A D, Wales N, Tryhorn Y, Tyms A S. Br Med J. 1989;299:897–898. doi: 10.1136/bmj.299.6704.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spaete R R, Mocarski E S. Proc Natl Acad Sci USA. 1987;84:7213–7217. doi: 10.1073/pnas.84.20.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plotkin S A, Starr S E, Friedman H M, Gonczol E, Weibel R E. J Infect Dis. 1989;159:860–865. doi: 10.1093/infdis/159.5.860. [DOI] [PubMed] [Google Scholar]
- 28.Bailey J M, Davidson N. Anal Biochem. 1976;70:75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- 29.Rice G P, Schrier R D, Oldstone M B. Proc Natl Acad Sci USA. 1984;81:6134–6138. doi: 10.1073/pnas.81.19.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews R G, Singer J W, Bernstein I D. J Exp Med. 1989;169:1721–1731. doi: 10.1084/jem.169.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitlock C A, Tidmarsh G F, Muller-Sieburg C, Weissman I L. Cell. 1987;48:1009–1021. doi: 10.1016/0092-8674(87)90709-4. [DOI] [PubMed] [Google Scholar]
- 32.Baum C M, Weissman I L, Tsukamoto A S, Buckle A M, Peault B. Proc Natl Acad Sci USA. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown J M, Kaneshima H, Mocarski E S. J Infect Dis. 1995;171:1599–1603. doi: 10.1093/infdis/171.6.1599. [DOI] [PubMed] [Google Scholar]
- 34.Cha T A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maciejewski J P, Bruening E E, Donahue R E, Mocarski E S, Young N S, St. Jeor S C. Blood. 1992;80:170–178. [PubMed] [Google Scholar]