Abstract
Previous studies have shown that, in the Royal College of Surgeon rat, circadian rhythms in the retinal dopaminergic and melatonergic systems are still present after the photoreceptors have degenerated, thus demonstrating that circadian rhythmicity in the mammalian retina can be generated independently from the photoreceptors. The aim of the present study was to investigate the pattern of expression of the clock genes in the retina of the Royal College of Surgeons rat under different lighting conditions. Expression of clock genes was investigated in the retina of normal and dystrophic Royal College of Surgeons rats under 12 hours of light/12 hours of dark (LD), constant darkness (DD) and constant light (LL) using Real Time Quantitative RT-PCR. Our data indicate that, in control animals, Period1, Period2, Cryptochrome1, Cryptochrome2, Clock, Rora, Rev-Erbα and Npas2 mRNA levels showed a significant variation over the sampling period in LD cycles and in DD, whereas Bmal1 mRNA did not show any significant variation. In LL, the transcripts for Per1, Per2, Clock and Rev-Erbα showed significant the temporal variations. In the dystrophic retina, only Per1 and Per2 mRNA levels showed a temporal variation over the 20-hours period. Our work indicates that degeneration of the photoreceptor cells dramatically affected the expression levels and patterns of many clock genes. Finally, the present study suggests that investigating the expression pattern of clock genes using the whole retina or animals with photoreceptor degeneration may not provide any definitive answers about the working of the retinal circadian clock system.
Keywords: Retina; Circadian rhythm; Photoreceptor degeneration, Royal College Surgeon rat, Clock genes
1. Introduction
Circadian clocks generate endogenous circadian rhythms throughout autoregulatory transcriptional-translational feedback loops comprised of a well defined set of clock genes [Ko and Takahashi, 2006]. These feedback loops consist of positive and negative components. The positive components consist of the basic helix-loop-helix-PAS domain transcription factors, CLOCK and BMAL1. These transcription factors heterodimerize and bind to circadian E-box promoter elements that enhance the transcription of the genes encoding the negative components PERIOD 1, 2 and CRYPTOCHROME 1,2. The CRYPTOCHROME and PERIOD proteins' feedback inhibits the transcription of the Cryptochrome (Cry) and Period (Per) genes by blocking CLOCK/BMAL1-mediated transactivation. The second feedback loop involves the transactivation of the Rev-Erbα and Rora genes by CLOCK/BMAL1. The protein products of these genes compete for binding to Rev-erb/Ror element (RRE) in the Bmal1 promoter, driving a daily rhythm of Bmal1 transcription and closing the second feedback loop [Ko and Takahashi, 2006]. Rhythmic expression of these clock gene products produces circadian clock outputs by regulating transcription of clock-controlled genes (CCGs). At least some of these CCGs contain circadian E boxes, which have a core nucleotide sequence of CACGTG and are activated rhythmically by CLOCK/BMAL1 [Chen and Baler, 2000, Tosini and Fukuhara, 2002].
Several studies have demonstrated that the mammalian retina contains an intrinsic circadian clock that controls many of these rhythms [Iuvone et al., 2005]. Cultured mammalian retinas show a clear circadian rhythm in melatonin release [Tosini and Menaker, 1996; Sakamoto et al., 2004], and Arylalkylamine Nacetyltransferase (Aanat, a clock controlled gene) mRNA is rhythmic in animals in which the suprachiasmatic nuclei of the hypothalamus (SCN) have been lesioned [Sakamoto et al., 2000], thus indicating that the retinal clock can generate circadian rhythmicity independently from the master circadian clock located in the SCN. Further, we have recently shown that dopamine rhythmicity and the inner retinal neurons are not necessary for the generation of the circadian rhythm of Aanat mRNA in the photoreceptors (Sakamoto et al., 2006).
On the other hand, in the Royal College of Surgeons (RCS), circadian rhythms in the dopaminergic and melatonergic system are still present in animals with severe photoreceptor degeneration (Doyle et al., 2002, Sakamoto et al., 2004). Moreover, the circadian clock controlling the melatonergic system in the dystrophic retina appears to be somewhat different from the clock located in the photoreceptors, since kainic acid (KA) treatment abolished the circadian rhythm in the dystrophic retina (Sakamoto et al., 2004) but not in the intact retina (Sakamoto et al., 2006). All together, this evidence suggests that the mammalian retina is composed of a network of circadian clocks that may be located in different retinal layers.
The aim of the present work was to investigate the pattern of expression of the “clock genes” in the rat retina under different lighting conditions and after photoreceptor cells have degenerated.
2. Results
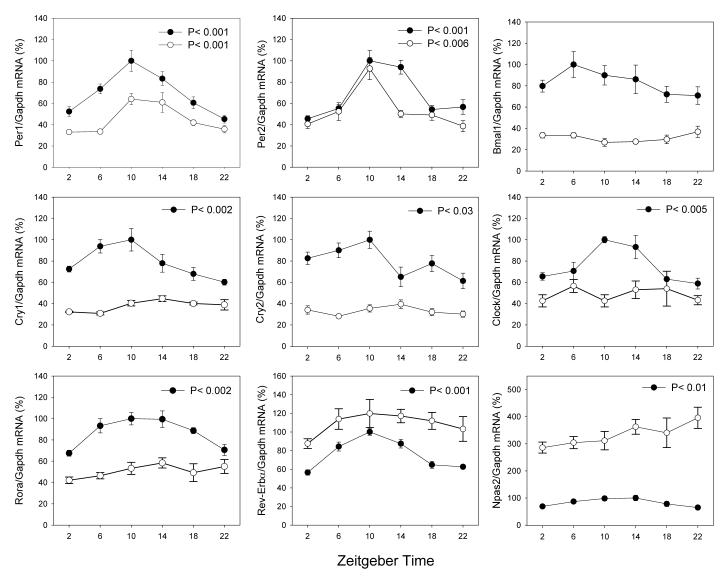
Figure 1 reports the normalized expression levels of the clock genes investigated under LD in the retina of RCS/N-rdy+ (−rdy+) rats homozygous for the normal rdy allele and congenic age-matched RCS/N-rdy (−rdy) rats. In the −rdy+ retina, only Bmal1 mRNA levels failed to show any significant variation over the 20-hour period (Figure 1, Kruskall-Wallis P > 0.1), whereas, in −rdy, only Period1 (Per1) and Period2 (Per2) showed a significant variation over the sampling period (Figure 1, Kruskall-Wallis, P < 0.01 in both cases). Per1 mRNA levels were slightly reduced in −rdy, whereas Per2 mRNA levels were almost unaffected by degeneration of photoreceptors. In both genotypes, Per1 mRNA and Per2 mRNA levels reached their maximum levels at zeitgeber time (ZT) 10. The mRNA levels for Bmal1, Cryptochrome1 (Cry1), Cryptochrome 2 (Cry2), Clock and Rora were reduced in −rdy, while the mRNA levels for Rev-erbα and Npas2 were significantly increased (Figure 1).
Figure 1.
Expression pattern of clock genes in the retina of −rdy+ (black circles) and −rdy (white circles) rats under 12 Light:12 Dark cycles. The presented values are normalized values. Comparison among treatment time points was carried out using non-parametric analysis of variance (Kruskall-Wallis tests, N = 4-6 for each time-point). In −rdy+, the peak-trough amplitude was: Per1 = 2.21; Per2 = 2.19; Cry1= 1.66; Cry2 = 1.63; Clock = 1.61; Rora = 1.48; Rev-Erbα = 1.77; Npas2 = 1.55. In −rdy, the peak-trough amplitude was 1.94 and 2.4, respectively, for Per1 and Per2. The statistical significance of the differences observed between the peak-trough levels was verified by Dunn's multiple comparisons test (P < 0.05).
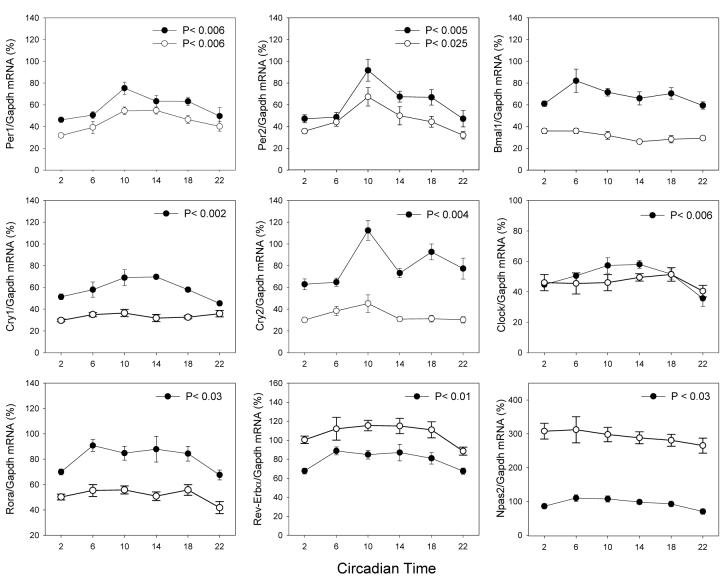
In −rdy+, exposure to DD did not change the number of genes that expressed a significant variation in their expression pattern (Figures 1-2). However, exposure to DD induced a 10-20% reduction in the overall expression level of many genes (Figures 1 and 2). In rdy, exposure to DD had little effect either on the pattern of expression or on the overall levels (Figure 2).
Figure 2.
Expression pattern of clock genes in the retina of −rdy+ (black circles) and −rdy (white circles) rats under constant darkness. The presented values are normalized values. Comparison among treatment time points was carried out using non-parametric analysis of variance (Kruskall-Wallis tests, N = 4-6 for each time-point). In −rdy+, the peak-trough amplitude was: Per1 = 1.63; Per2 = 1.94; Cry1= 1.54; Cry2 = 1.59; Clock = 1.63; Rora = 1.34; Rev-Erbα = 1.32; Npas2 = 1.57. In −rdy, the peak-trough amplitude was 1.82 and 2.11, respectively, for Per1 and Per2. The statistical significance of the differences observed between the peak-trough levels was verified by Dunn's multiple comparisons test (P < 0.05).
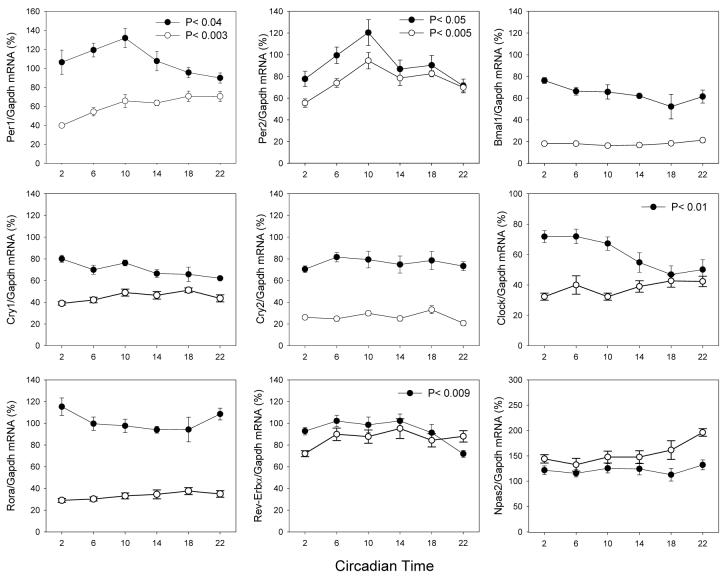
Exposure to LL induced significant changes in the expression level and pattern of several genes. In −rdy+, Per1 and Per2 mRNA levels showed a 10-20% increase (Figure 3). Clock mRNA levels showed a significant change in expression levels (Kruskall-Wallis, P < 0.01), but they reached their maximum level four to eight hours earlier from in LD or DD (ZT2-6 vs. ZT10, Figure 3). Reverb± mRNA levels showed a pattern (Kruskall-Wallis P < 0.009) not different from what we observed in LD and DD (Figures 1-3). Cry1, Cry2, Rora and Npas2 mRNA did not show any variation in their levels (Kruskall-Wallis, P > 0.1 in all cases, Figure 3). In the −rdy retina, Per2 mRNA levels showed an expression pattern similar to what was observed in LD and DD in −rdy+ and −rdy (Figures 1-3). Per1 mRNA levels showed a dramatic change in the expression pattern, (Figure 3). All the other genes failed to show any significant changes over the sampling period (Kruskall-Wallis, P > 0.1 in all cases), and their mRNA levels were significantly decreased (Figure 3).
Figure 3.
Expression pattern of clock genes in the retina of −rdy+ (black circles) and −rdy (white circles) rats under constant light. The presented values are normalized values. Comparison among treatment time points was carried out using non-parametric analysis of variance (Kruskall-Wallis tests, N = 4-6 for each time-point). In −rdy+, the peak-trough amplitude was: Per1 = 1.47; Per2 =1.69; Clock = 1.45; Rev-Erbα = 1.42. In −rdy, the peak-trough amplitude was 1.78 and 1.70, respectively, for Per1 and Per2. The statistical significance of the differences observed between the peak-trough levels was verified by Dunn's multiple comparisons test (P < 0.05).
3. Discussion
We, along with others, have previously reported that a circadian rhythm in melatonin and dopamine levels is still present in 60-day-old −rdy rats with severe photoreceptor degeneration (Doyle et al., 2002; Sakamoto et al., 2004). We have also shown that the circadian clock driving the melatonin rhythm in the dystrophic retina is sensitive to KA lesion, while the circadian clock driving Aanat rhythmicity in intact retina is insensitive to KA (Sakamoto et al., 2004; 2006). Such a result suggests that at least two different circadian-clock-generating mechanisms are present in the rat retina.
To gather more information about the functioning of the retinal circadian clock system, we decided to investigate the expression of clock genes in the rat retina under different lightning conditions and after photoreceptor degeneration. Our data demonstrate that many clock genes showed significant variation in the retina of −rdy+ under LD and DD, while under LL only Per1, Per2, Clock and RevErbα showed a significant variation over the sampling period (See Figures 1, 2 and 3.) In the dystrophic retina (−rdy), Per1 and Per2 showed a significant variation in LD, DD and LL, while the other clock genes did not any variation in their expression levels. Moreover, in constant light, most of the clock genes were down-regulated (Figures 1, 2 and 3), while Rev-erbα and Naps2 were up-regulated (Figures 1, 2 and 3).
A few studies have investigated the expression of circadian clock genes in the rat retina. Namihira et al. (1999, 2001) reported that Per2 and Clock mRNA showed a significant circadian rhythm, whereas Per1 and Bmal1 did not show any significant variation in their mRNA levels. More recently, Kamphuis et al. (2005) investigated the daily rhythms of many of the clock genes in the rat retina and reported that only Per2, Per3, Cry2 and Bmal1 were rhythmic. Finally, Numano et al. (2006) and Sakamoto et al. (2006) reported that Per1 and Per2 mRNA levels were rhythmic in the rat retina. Overall, our data in rdy+ agree with the data reported by Namihira et al. [1999; 2001], Numano et al. (2006) and Sakamoto et al. (2006), although some discrepancies are present with respect to the data presented by Kamphuis et al.(2005). Recent studies have also reported the expression pattern of several clock genes in the intact mouse retina and in the retina of mice (rd/rd) lacking photoreceptors [Ruan et al., 2006; Dinet et al., 2006]. In one study, the authors reported that Per1, Per2, Cry1, Cry 2 and Bmal1 showed a significant rhythm in LD and DD in the retina of C3H rd/rd mice (Ruan et al., 2006), while another investigation reported a circadian rhythm in Per1 mRNA in C57BL and C3H rd/rd, but not in Cry 2 mRNA (Dinet et al., 2007). Therefore, it is clear that there are significant differences among the results obtained in different species or even within the same species among different strains.
The distribution of clock genes in the rat and mouse retinas has been also investigated by some studies. In the rat, Per1 transcripts are present, but at low levels, in the photoreceptors, and are more abundantly present in the inner nuclear layer, whereas Per2 transcripts are present in the inner nuclear layer and the ganglion cell layer (Namihira et al., 1999), but not in the photoreceptor layer. In the mouse, Per1, Bmal1 and Clock transcripts are expressed in the photoreceptors (Gekakis et al., 1998, Yujnovsky et al., 2006; Dinet et al., 2007). Cry1 transcripts seem to be absent from the photoreceptors (Miyamoto and Sancar, 1997), whereas a low level of expression for Cry2 transcripts and immunoreactivity have been detected in the outer nuclear layer of C57BL mice (Miyamoto and Sancar, 1997: Dinet et al., 2007).
Therefore, we anticipated that degeneration of photoreceptors will only marginally affect the expression of Per1 and Cry2, and Per2 will not be affected. Indeed, Per1 RNA levels were slightly reduced by the degeneration of the photoreceptors (Figures 1-3), while Cry2 mRNA levels were dramatically affected, either in their pattern of expression or in their levels, by photoreceptor degeneration (Figures 1-3). Per2 mRNA levels and expression patterns were only slightly modified by the loss of the photoreceptors. This observation indicates that a re-arrangement of the clock gene expression within the different retinal layers must occur during the progression of the photoreceptor degeneration.
We have previously shown that, in −rdy rats, Aanat transcriptions are up-regulated in the inner nuclear layer where they showed a high-amplitude circadian rhythm (Sakamoto et al., 2004). As we already mentioned, Aanat transcription in the rat retina is under the direct control of the circadian clock via the action of CLOCK:BMAL1 on the E-box present on the promoter region of this gene (Chen and Baler, 2000; Tosini and Fukuhara, 2002). Therefore, we expected that the expression of clock genes in −rdy retinae would have shed some light on the working of this inner retinal clock and, thus, on the mechanisms that control Aanat transcription in the −rdy. Unfortunately, our data do not provide any useful insight on this mechanism.
In conclusion, our data indicated that a group of four clock genes is rhythmic in LD, DD and LL in −rdy+ retina, whereas only Per1 and Per2 are rhythmic in the retina of −rdy. The expression pattern of these clock genes was greatly affected by degeneration of the photoreceptor cells. Recent evidence suggests that the mammalian retina is composed of a network of circadian clocks that are located in different types of retinal cells or layers (Sakamoto et al., 2004; 2006; Ruan et al., 2006), where they can present a different phase and, thus, affect the overall levels of expression observed in the entire retina.
Therefore, our work suggests that investigating the expression pattern of clock genes using the whole retina or animals with photoreceptor degeneration may not provide any definitive answers about the working of the retinal circadian clock system.
4. Experimental Procedure
Tan-hooded RCS/N-rdy+ (−rdy+) rats homozygous for the normal rdy allele and congenic age-matched RCS/N-rdy (−rdy) homozygotes with retinal dystrophy were used in this study. Animals were raised from birth at Morehouse School of Medicine in a 12 hours light /12 hours dark (LD) cycle of illumination with light on from Zeitgeber Time (ZT) 0 – 12. Food and water were available ad libitum. All animals used for these experiments were 60 +/-2 days old at the time of killing. At this age, degeneration of the retina in −rdy has advanced to the point where photoreceptors are both histologically and functionally undetectable.
To investigate the expression of clock genes in the retina in LD cycles, animals were sacrificed at ZT 2, 6, 10, 14, 18 and 22. When animals were killed during the night, the procedure was carried out in dim red light (< 1 lux). To investigate the pattern of expression in constant conditions, rats were transferred into constant darkness (DD) or light (LL, 300 Lux) for two days before sacrifice. Samples were then collected on the third day at circadian time (CT) 2, 6, 10, 14, 18 and 22. Retinas were dissected, immediately frozen on dry ice and stored at −80°C.
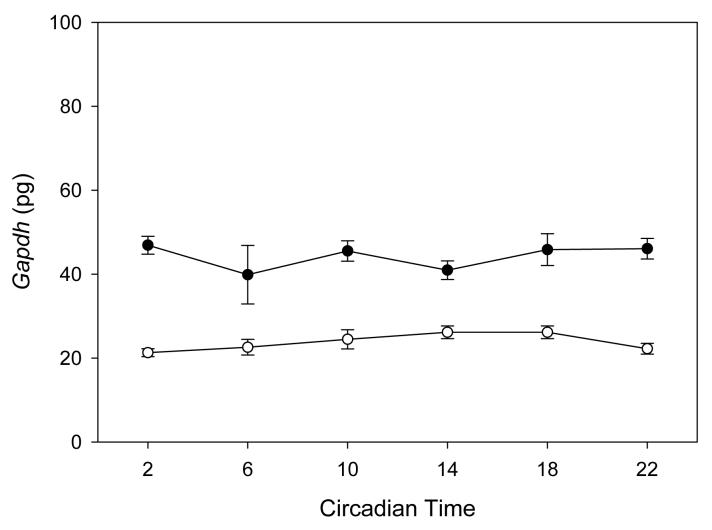
Total RNA was isolated form each retina using TRIZOL reagent (Life Technologies, Grand Island, NY) following sonication. RNA was treated with DNase I to remove any traces of genomic DNA. First-strand cDNA was synthesized from 1μg of each RNA sample using oligo (dT) and Omniscript reverse transcriptase (QIAGEN, Valencia, CA), according to the manufacturer's protocol. Each set of samples was simultaneously processed for RNA extraction, DNase I treatment, cDNA synthesis and PCR reaction. Real-time quantitative RT-PCR was performed with SYBR Green using an iCycler (BioRad, Hercules, CA), as described in (Sakamoto et al., 2006). Details about the primers used are reported in Table 1. Cycling parameters were 95°C for 8 minutes to activate Taq DNA polymerase, then 40 cycles of 95°C for 15 seconds, 60°C for 45 seconds and 72°C for 30 seconds. The fluorescence of the accumulating product was measured after each cycle at 4°C below the product melting temperature. To confirm the specificity of PCR products, melting curves were determined using iCycler software, and samples were run on an agarose gel. The amount of amplified product was calculated from plasmid standards. mRNA levels were normalized using glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA levels. For each of the clock genes investigated, the values were normalized with respect to the peak values measured in LD in rdy+. As previously reported, Gapdh mRNA levels did not show any circadian variation (Figure 4, but see also Sakamoto et al., 2006).
Table 1.
Real-time RT-PCR primers. Primers were designed using Primer3 on the indicated GenBank sequences. The length of the PCR product for each primer pair and the cDNA sequence number for each primer are indicated. Each primer pair yielded a single product, as determined by agarose gel electrophoresis and melt-curve analyses.
| Name | GenBank number |
Size (bp) |
Sequence (5′-3′) | Nucleodite number |
|---|---|---|---|---|
| Gapdh | X 02231 | 142 | F: AGACAGCCGCATCTTCTTGT | 24-43 |
| R: TGATGGCAACAATGTCCACT | 146-165 | |||
| Per1 | AB092976 | 164 | F: ACACCCAGAAGGAAGAGCAA | 2354-2373 |
| R: GCGAGAACGCTTTGCTTTAG | 2498-2517 | |||
| Per2 | NM031678 | 195 | F: GAGAGAGGAACAGGGCTTCC | 2285-2304 |
| R: TTGACACGCTTGGACTTCAG | 2460-2479 | |||
| Cry1 | NM198750 | 271 | F: TGCTCCTGGAGAGAATGTCC | 2084-2103 |
| R: TGACTCTCCCACCAACTTCA | 2335-2354 | |||
| Cry2 | NM133405 | 302 | F: CTGTGGCAGAGCCTGGTT | 1604-1621 |
| R: TCGCTCTGTCTGTTGGTGAC | 1886-1905 | |||
| Bmal1 | NM024362 | 215 | F: CCGATGACGAACTGAAACACCT | 1016-1037 |
| R: TGCAGTGTCCGAGGAAGATAGC | 1209-1230 | |||
| Clock | NM021856 | 110 | F: TCTCTTCCAAACCAGACGCC | 1939-1958 |
| R: TGCGGCATACTGGATGGAAT | 2029-2048 | |||
| Rev-Erba | NM145775 | 101 | F: ACAGCTGACACCACCCAGATC | 996-1016 |
| R: CATGGGCATAGGTGAAGATTTCT | 1074-1096 | |||
| Rora | M217192 | 89 | F: CCCGATGTCTTCAAATCCTTAGG | 1333-1355 |
| R: TCAGTCAGATGCATAGAACACAAACTC | 1395-1421 | |||
| Npas2 | XM244089 | 221 | F: CGGGACCAGTTCAATGTTCT | 612-631 |
| R: CCATCTAACGCCTCCAACAT | 813-832 |
Figure 4.
Gapdh mRNA levels in the retina of −rdy+ (black circles) and -rdy (white circles) rats under constant darkness. Comparison among treatment time points was carried out using non-parametric analysis of variance (Kruskall-Wallis tests, N = 4-6 for each time-point). No significant difference were observed (−rdy+ P > 0.57; −rdy P > 0.31). Similar results have been also obtained in LD and LL.
Results are presented as mean +/− the standard error of the mean (SEM). Comparisons among the different time points within the same group (i.e., LD, DD and LL) were carried out using non-parametric analysis of variance (Kruskall-Wallis test) followed, when appropriate, by Dunn's multiple comparisons test (Sigma Stat package version 9.0).
Acknowledgements
Supported by NIH grant (NS 43459) and the NASA Cooperative Agreement NCC 9-58 with the National Space Biomedical Research Institute to G.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature
- Chen W, Baler R. The rat arylalkylamine N- acetyltransferase E-box: different use in a master vs. slave oscillator. Mol Brain Res. 2000;81:43–50. doi: 10.1016/s0169-328x(00)00160-1. [DOI] [PubMed] [Google Scholar]
- Dinet V, Ansari N, Torres-Farfan C, Korf H-W. Clock gene expression in the retina of melatonin-proficient (C3H) and melatonin-deficient (C57BL) mice. J. Pineal Res. 2007;42:83–91. doi: 10.1111/j.1600-079X.2006.00387.x. [DOI] [PubMed] [Google Scholar]
- Doyle SE, McIvor WE, Menaker M. Circadian rhythmicity in dopamine content of mammalian retina: role of the photoreceptors. J. Neurochemistry. 2002;83:211–219. doi: 10.1046/j.1471-4159.2002.01149.x. [DOI] [PubMed] [Google Scholar]
- Iuvone PM, Tosini G, Haque R, Klein DC, Chaurasia SS. Circadian Clocks, Clock-Controlled Genes and Melatonin Biosynthesis in the Retina. Prog Retin Eye Res. 2005;24:433–456. doi: 10.1016/j.preteyeres.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Fukuhara C, Liu C, Ivanova TN, Chan GC-K, Storm DR, Iuvone PM, Tosini G. Gating of the cAMP signaling cascade and melatonin synthesis by the circadian clock in mammalian retina. J. Neuroscience. 2004;241:803–811. doi: 10.1523/JNEUROSCI.4988-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Kamphuis W, Cailotto C, Dijk F, Bergen A, Buijs RM. Circadian expression of clock genes and clock-controlled genes in the rat retina. Biochem. Biophys. Res. Commun. 2005;330:18–26. doi: 10.1016/j.bbrc.2005.02.118. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Human Molecular Genetics. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Sancar A. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as photoactive pigments for setting the circadian clock in mammals. Proc Natl Acad Sci U S A. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namihira M, Honma S, Abe A, Tanahashi Y, Ikeda M, Honma K-I. Circadian rhythms and light responsiveness of mammalian clock gene, Clock and BMAL1, transcripts in the rat retina. Neuroscience Lett. 1999;271:1–4. doi: 10.1016/s0304-3940(99)00407-3. [DOI] [PubMed] [Google Scholar]
- Namihira M, Honma S, Abe A, Masubuchi S, Ikeda M, Honma K-I. Circadian pattern, light responsiveness and localization of rPer1 and r Per2 gene expression in the rat retina. Neuroreport. 2001;12:471–475. doi: 10.1097/00001756-200103050-00010. [DOI] [PubMed] [Google Scholar]
- Numano R, Yamazaki S, Umeda N, Samura T, Sujino M, Takahashi R, Ueda M, Mori A, Yamada K, Sakaki Y, Inouye S-I, Menaker M, Tei H. Constitutive expression of the Period1 gene impairs behavioral and molecular circadian rhythms. Proc Natl Acad Sci U S A. 2006;103:3716–3721. doi: 10.1073/pnas.0600060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan G-X, Zhang D-Q, Zhou T, Yamazaki S, McMahon DG. Circadian organization of the mammalian retina. Proc Natl Acad Sci U S A. 2006;103:9703–9708. doi: 10.1073/pnas.0601940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Oishi K, Shiraishi M, Hamano S, Otsuka H, Miyake Y, Ishida N. Two circadian oscillatory mechanisms in the mammalian retina. Neuroreport. 2000;11:3995–3997. doi: 10.1097/00001756-200012180-00018. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Liu C, Tosini G. Circadian Rhythms in the retina of rats with photoreceptor degeneration. J. Neurochemistry. 2004;90:1019–1024. doi: 10.1111/j.1471-4159.2004.02571.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Liu C, Kasamatsu M, Iuvone PM, Tosini G. Intraocular injection of kainic acid does not abolish the circadian rhythm of Aanat mRNA in the rat photoreceptors. Molecular Vision. 2006;12:117–124. [PubMed] [Google Scholar]
- Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- Tosini G, Fukuhara C. The mammalian retina as a clock. Cell and Tissue Res. 2002;309:119–126. doi: 10.1007/s00441-002-0578-z. [DOI] [PubMed] [Google Scholar]
- Witkovsky P, Veseinberger E, LeSauter J, Yan L, Johnson M, Zhang DQ, McMahon DG, Silver R. Cellular location and circadian rhythm of expression of the biological clock gene Period 1 in the mouse retina. J. Neurosci. 2003;23:7670–7676. doi: 10.1523/JNEUROSCI.23-20-07670.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yujnovsky I, Hyrayama J, Doi M, Borrelli E, Sassoni-Corsi P. Signaling mediated by dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc Natl Acad Sci U S A. 2006;103:6386–6391. doi: 10.1073/pnas.0510691103. [DOI] [PMC free article] [PubMed] [Google Scholar]