Abstract
In addition to TATA-binding protein (TBP), a key factor for transcription initiation, the metazoan-specific TBP-like factor TLF/TRF2 and the vertebrate-specific factor TBP2/TRF3 are known to be required for transcription of specific subsets of genes. We have combined an antisense-knockdown approach with transcriptome profiling to determine the significance and biological role of TBP-independent transcription in early gastrula-stage Xenopus laevis embryos. Here, we report that, although each of the TBP family members is essential for embryonic development, relatively few genes depend on TBP in the embryo. Most of the transcripts that depend on TBP in the embryo are also expressed maternally and in adult stages, and show no functional specialization. In contrast, TLF is linked to preferential expression in embryos and shows functional specialization in catabolism. A requirement for TBP2 is linked to vertebrate-specific embryonic genes and ventral-specific expression. Therefore TBP paralogs are essential for the gene-regulatory repertoire that is directly linked to early embryogenesis.
Keywords: embryogenesis, TBP, transcription
Introduction
The general transcription machinery has been thought of as a mainly invariant biochemical entity that recruits RNA polymerase II. Metazoan species, however, encode different general transcription factor paralogs in their genomes (reviewed by Dantonel et al, 1999; Berk, 2000; Veenstra and Wolffe, 2001; Davidson, 2003; Hochheimer and Tjian, 2003), which could diversify the transcription machinery beyond the mechanisms of initiation observed in yeast. Among the initiation factor paralogs are proteins related to TATA-binding protein (TBP), a key factor known to play a central role in transcription mediated by all three eukaryotic RNA polymerases. TBP is essential for all transcription processes in yeast (Cormack and Struhl, 1992; Schultz et al, 1992; Hernandez, 1993; Roeder, 1996; Kim and Iyer, 2004). However, in higher eukaryotes, a number of TBP paralogs are present. In Drosophila, an insect-specific TBP-related factor (TRF1) was discovered (Crowley et al, 1993; Hochheimer and Tjian, 2003) that in vivo was found to stimulate one of the tudor promoters (Holmes and Tjian, 2000) and in vitro is able to stimulate basal transcription (Hansen et al, 1997).
Functional studies of TBP in embryos of different metazoans showed that a subset of genes is transcribed in a TBP-independent manner (Veenstra et al, 2000; Muller et al, 2001; Martianov et al, 2002a, 2002b). TBP-like factor (TLF, also known as TRF2, TRP or TBPL1) has been found in different metazoan species (Dantonel et al, 2000; Veenstra et al, 2000; Muller et al, 2001). TLF consists only of a core domain and shares 40% identity with TBP. It can interact with TFIIA and TFIIB, but does not bind to the TATA box (Moore et al, 1999; Rabenstein et al, 1999; Teichmann et al, 1999). There are reports about positive and negative roles of TLF in transcription: overexpression of human TLF was found to increase transcription from the NF1 promoter (Chong et al, 2005), but in an in vitro reconstituted system, TLF inhibited TBP-dependent basal transcription (Teichmann et al, 1999).
Knockdown studies of TLF in Caenorhabditis elegans (Dantonel et al, 2000; Kaltenbach et al, 2000), Xenopus (Veenstra et al, 2000), zebrafish (Muller et al, 2001) and Drosophila (Kopytova et al, 2006) showed that TLF is essential for embryonic development in these metazoan species and that it contributes to transcription in vivo. In contrast, TLF-knockout mice have defects in spermatogenesis, but not embryogenesis (Martianov et al, 2001, 2002a, 2002b; Zhang et al, 2001a, 2001b).
More recently, another TBP family member was discovered (Persengiev et al, 2003; Bartfai et al, 2004; Jallow et al, 2004). TBP2 (also referred to as TRF3 or TBPL2) is 95% identical to TBP in the core domain, features an N-terminal domain like TBP and is vertebrate-specific (Persengiev et al, 2003). Like TBP, TBP2 was found to interact with TFIIA and TFIIB and to be capable of binding to the TATA box and of stimulating transcription initiation in vitro (Bartfai et al, 2004; Jallow et al, 2004). TBP2 depletion by antisense (AS) approaches in zebrafish and Xenopus showed that TBP2 is essential for embryonic development. TBP2 is most abundant in the oocyte, but persists at low levels during early embryogenesis and could partly substitute for TBP in TBP-ablated embryos (Jallow et al, 2004).
Together, these studies have shown that TBP is not universally required for transcription in metazoans. What remains unknown however, is what the significance of TBP-independent transcription is. A general hypothesis posits that alternative mechanisms of transcription initiation may expand the gene-regulatory potential in metazoans in general and in vertebrates in particular. To uncover the specific connection between TBP-related factors and gene regulation, we performed expression profiling of TBP-, TLF- and TBP2-depleted gastrula-stage Xenopus embryos. We analyzed the extent to which embryonic transcription depends on either factor, the relation to published synexpression groups of genes (Baldessari et al, 2005) and the functional specialization of TBP family members in transcription of genes involved in specific cellular or molecular processes. On the basis of these results, we tested the relationship between TLF and catabolism and determined the requirement for TBP2 in dorso-ventral patterning. The recruitment of TBP and TBP2 to promoters was analyzed by chromatin immunoprecipitation (ChIP) experiments. We report a requirement for TLF and TBP2 in specific developmentally regulated processes, whereas TBP is functionally not specialized and linked to maternal-embryonic transcripts and to transcripts that are more abundant in adult stages.
Results
The majority of newly synthesized transcripts in the embryo depend on TLF or TBP2, but not TBP
To investigate the specific role of TBP, TLF and TBP2 during the early embryogenesis of Xenopus laevis, we depleted embryos of either of these factors (Figure 1) using chemically modified AS oligonucleotides that mediate RNaseH-dependent degradation of the target mRNA (Dagle et al, 2000; Dagle and Weeks, 2001; Lennox et al, 2006). The phenotypes of the embryos (Supplementary Figure S1) were as described previously (Veenstra et al, 2000; Jallow et al, 2004).
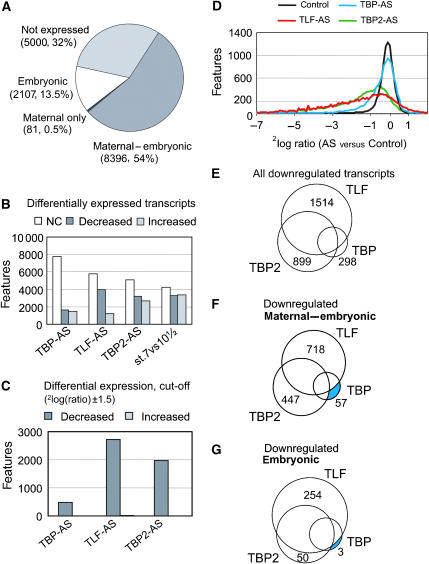
Figure 1.
Experimental design and AS knockdown. (A) TBP paralog requirements were determined for the early gastrula (stage 10½) transcriptome. To discriminate between maternal and embryonic transcripts, control blastula embryos (stage 7) were taken for hybridization as well. Indicated are post fertilization times at 23°C. AS: antisense ablation of the indicated factor. (B) AS knockdown of the TBP family members. AS-injected embryos were analyzed by Western blotting (upper panel) for the expression of TBP and α-tubulin and by qRT–PCR for the expression of TLF, TBP2 and GAPDH. GAPDH is a maternal transcript not affected by either of the knockdowns. The analysis shows that the expression of TBP, TLF and TBP2 is specifically reduced in embryos injected with AS oligonucleotide (cf. Veenstra et al, 2000; Jallow et al, 2004).
Before the midblastula transition (MBT, Nieuwkoop-Faber stage 8½) the embryonic genome is transcriptionally quiescent and the transcripts present during cleavage and blastula stages are of maternal origin. At the MBT, embryonic transcription starts. Gastrulation commences shortly after the MBT and requires new transcription (Newport and Kirschner, 1982; Sible et al, 1997). To minimize indirect effects of the AS knockdowns, control embryos, TBP-, TBP2- and TLF-depleted embryos were collected shortly after the MBT, at an early gastrula stage (stage 10½). To discriminate between persisting maternal and newly synthesized embryonic transcripts, control blastula embryos (stage 7) were collected as well. An overview of the experimental design is shown in Figure 1A. From two independent experiments, RNA was isolated from the embryos and hybridized to Affymetrix X. laevis GeneChips (14.4 k). The data were scaled using Affymetrix GeneChip Operating Software (Supplementary Figure S2A–C). Differentially expressed genes were defined using Affymetrix change calls that are based on a statistical test (Wilcoxon-rank-test) of the 16 probe-pairs for each transcript. To increase the stringency of the analysis, only genes that showed consistent change calls in the biological duplicates were considered in the subsequent analyses. The correlation between biological replicates varied between R2=0.64 and 0.89 (Supplementary Figure S3). The hybridization data were evaluated and validated using extensive quantitative RT–PCR (qRT–PCR, Supplementary Figure S2A–F). This analysis confirmed the strong decrease of transcripts that showed a similarly strong decrease by microarray analysis. Of the 15 610 features on the microarray (representing 14 400 ESTs), 10 930 features were found expressed at stage 10½, of these 9731 were already detected at stage 7, whereas approximately 30% of the represented features were neither expressed at stage 7 nor 10½. Most transcripts are maternal-embryonic, detected before and after the MBT without change in transcription level (8396, 54%, Figure 2A) in concordance with data reported by Graindorge et al (2006). Even though these transcripts have a strong maternal component of expression, many of them depend on embryonic transcription to maintain their transcription level. Fourteen percent of the features are embryonic transcripts; they are either exclusively expressed at stage 10½ or show a strong (>2.8-fold) increase between stage 7 and stage 10½. Only a few transcripts (81, 0.5%) are exclusively detected before the MBT (maternal transcripts, cf. Figure 2A).
Figure 2.
Contribution of TBP, TLF and TBP2 to the embryonic transcriptome. (A) Categories of transcripts detected on the microarray, grouped on the basis of absent and present calls in both biological replicas in stage 7 and stage 10½ control embryos. Embryonic transcripts are either exclusively present at stage 10½ or increased at least 2.8-fold in stage 10½ versus stage 7; maternal-embryonic transcripts are not increased between stage 7 and stage 10½. (B) Effect of TBP-, TLF- or TBP2-AS injection on transcription at stage 10½. Graph shows number of transcripts that are decreased, increased or not changed (NC). The ‘not changed' category includes ambiguous values: genes with inconsistent differential expression between replicates or a marginal present call. In (C), the cut-off of 2log(ratio) of ±1.5 (21.5=2.8-fold change) was applied to the genes shown in (B). (D) Distribution of 2log fold changes. Average 2log ratios of duplicate experiments are depicted as a curve showing, on the y-axis the number of genes with on the x-axis the corresponding 2log ratio (created by binning). Only genes present in the control sample of stage 10½ are represented, the control curve is calculated by the 2log ratio of experiment 1 versus average of experiment 1 and 2. (E–G) Venn diagrams show all decreased transcripts and their dependency on one or a combination of TBP family members. The relative and absolute numbers for all Venn compartments are given in Supplementary Table S1. (E) All downregulated genes (F) and (G) show maternal-embryonic and embryonic transcripts respectively.
Comparing the effect of TBP-AS, TLF-AS and TBP2-AS on the transcriptome shows that ablating the expression of TLF or TBP2 has a relatively strong impact on the transcriptome, both regarding the number of affected transcripts (respectively, 3974 and 3161 decreased transcripts) and the extent to which transcript levels are affected (Figure 2B–D). On the other hand, only 1647 transcripts are decreased in the absence of TBP and most of the expressed genes (at least 69%) are unaffected (Figure 2B–D).
For most of the analyses, an arbitrary cut-off of at least 2.8-fold change (2log(ratio) ±1.5) in transcript levels was applied. Figure 2D shows that the chosen cut-off is beyond the deviation of the control replicates. As we found relatively few transcripts that are strongly increased after the knockdown of either of the TBP family members (only 15 genes with TLF-AS and 1 with TBP2-AS, Figure 2C), we concentrated on decreased transcripts for further analyses.
K-means clustering revealed a complex organization of the embryonic transcriptome, with different subsets of transcripts affected differently by AS knockdown of TBP, TBP2 or TLF (Supplementary Figure S4). To analyze this in a more structural way, we grouped transcripts according to their transcriptional requirements as represented in a Venn diagram (Figure 2E). Transcripts that exclusively depend on TLF form the largest group (39%, see Supplementary Table S1 for absolute and relative numbers for all Venn categories). Another major group consists of transcripts that are dependent on TBP2 only (20%). A small number (2%) is exclusively dependent on TBP. Of these TBP-dependent transcripts, most (57 of 60) are maternal-embryonic (maternally derived but embryonic transcription contributes to steady-state levels in the embryo) rather than embryonic upregulated (3 of 60, Figure 2F–G and Supplementary Table S1). To uncover the characteristics of transcripts dependent on each of the TBP family members and to avoid complicating the analysis by indirect effects, transcripts that exclusively depend on one of the three TBP family members were analyzed in more detail.
Functional plasticity and promoter recruitment
The specificity of TBP, TLF and TBP2 knockdown has been established using control oligonucleotide injections, different AS oligonucleotides targeting the same factor, as well as ‘add back' rescue experiments (Veenstra et al, 2000; Jallow et al, 2004). We have also shown that overexpression of TBP2 can partially rescue the ablation of TBP in early embryos (Jallow et al, 2004). To explore in a more general way the regulatory specificity of TBP family members and the potential for crossregulation, we performed single factor knockdown rescue experiments as well as triple-knockdown rescue experiments. As shown previously, adding back the targeted transcription factor can restore transcription (Supplementary Figures S5 and S8). Surprisingly, when all three factors are ablated (triple-knockdown), adding back any of the three TBP family members frequently results in partial heterologous rescue of transcription, even though exquisite specificity is observed for the same transcripts in single-knockdown experiments (Supplementary Figure S6). One potential explanation is that overexpression of a single initiation factor poises a severely crippled transcription machinery for transcriptional rescue, regardless of the physiological roles of these factors. These physiological roles are uncovered in single-knockdown experiments. Nevertheless, these results raise the possibility of a context-dependent plasticity of TBP, TLF and TBP2 function.
To further explore this issue, the relationship between factor requirements and factor recruitment was analyzed using ChIP. We used X. tropicalis for ChIP because of the availability of genomic sequence. X. tropicalis is closely related to X. laevis as shown by viable interspecies hybrids and cross-species microarray experiments (Burki, 1985; Chalmers et al, 2005; Sartor et al, 2006).
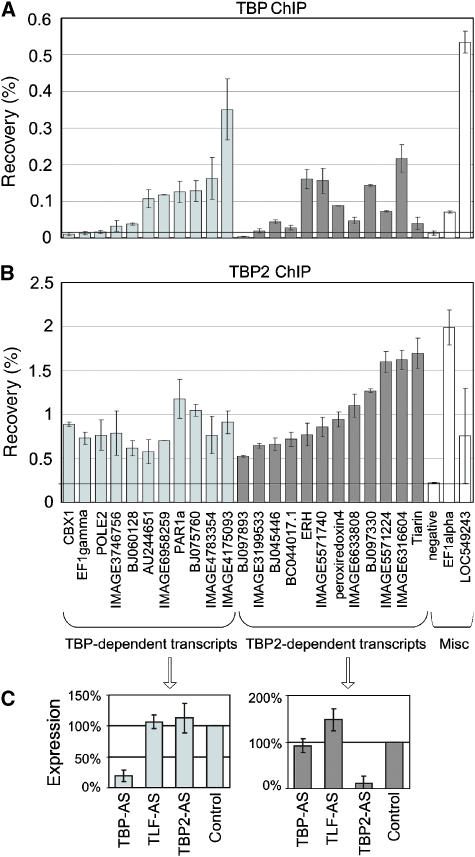
Promoters of TBP- and TBP2-dependent genes were randomly chosen for ChIP. No ChIP grade antibody is available for TLF. We detected TBP recruitment (>2.5-fold enrichment relative to genomic background) on 7 of 11 TBP-dependent promoters (Figure 3A, left panel), and enrichment of TBP2 on 11 of 12 TBP2-dependent promoters (Figure 3B, right panel). The relative recruitment varies significantly among promoters, which could be due to differences in expression level (Kim and Iyer, 2004).
Figure 3.
Recruitment of TBP and TBP2 to promoters of dependent genes. (A) TBP ChIP and (B) TBP2 ChIP on promoters of TBP- and TBP2-dependent transcripts. Promoters requiring TBP are shown on the left side of each panel and are sorted from low to high recovery of TBP. Promoters requiring TBP2 are shown on the right side of each panel and are sorted from low to high recovery of TBP2. Miscellaneous controls (Misc) in the far right of each panel include a genomic background control (negative), and the EF1alpha and LOC549243 loci, which represent examples of a relatively selective recruitment of TBP or TBP2. EF1alpha and LOC549243 show mixed requirements for TBP paralogs (i.e., overlap, Figure 2E). (C) Effect of AS injection on transcript levels as determined by qRT–PCR for transcripts identified as exclusively TBP-dependent (left panel) or TBP2-dependent (right panel) in the microarray analysis. Shown is the average of 14 (TBP) and 15 (TBP2) transcripts.
Gene-selective requirements for TBP family members could be explained by gene-selective factor recruitment. However, it is also possible that more than one factor is recruited to a promoter, but that one of these factors contributes more to productive transcription initiation and transcriptional output, due to, for example, protein–protein interactions. This latter possibility would reconcile both the highly selective functional requirements of the three factors, as well as the potential for crossregulation. Using ChIP, we find that the majority (16 of 23) of the promoters that functionally require either exclusively TBP or exclusively TBP2 are enriched for both factors (Figure 3A–C). Nevertheless, some promoters recruit TBP more efficiently than TBP2, or the other way around. Recruitment of TBP and TBP2 varies among promoters and TBP-dependent promoters tend to recruit TBP better than TBP2-dependent promoters do, and vice versa (Supplementary Figure S7). To investigate if recruitment of both factors is a general phenomenon in early embryos, an additional set of 48 promoters was randomly selected and tested by ChIP. Many promoters are substantially enriched for both factors (correlation 0.66, P<0.0001; data not shown). These results support the view that TBP and TBP2 recruitment is not exclusive for promoters requiring the respective factor for transcription, therefore factor binding is not necessarily sufficient for factor function under physiological conditions. Our data identify subsets of genes for which only one initiation factor contributes quantitatively to transcriptional output (Figure 3C).
TBP and TBP2 are linked to maternal-embryonic yeast orthologs and developmentally regulated vertebrate-specific genes, respectively
TBP is found in all eukaryotes, whereas TLF and TBP2 are restricted to the genomes of metazoans and vertebrates, respectively. We tested if this distribution among species is reflected in the genes dependent on either of the TBP family members. All ESTs represented on the Affymetrix X. laevis array were mapped to Ensembl X. tropicalis genes. The Ensembl ortholog predictions were used to analyze TBP family member requirements of yeast orthologs, metazoan-specific genes (ortholog in fly but not yeast) and vertebrate-specific genes (ortholog in human but not fly). The relative numbers of these three categories for the exclusively TBP-, TLF- and TBP2-dependent genes were calculated and presented as enrichment or underrepresentation compared to the complete Affymetrix data set (Figure 4A). Statistical significance was calculated using the hypergeometric distribution (Supplementary Table S2). The analysis shows that TBP-dependent genes are enriched for yeast orthologs (Figure 4B, 47% of TBP-dependent genes compared to 22% for all genes), whereas vertebrate-specific and metazoan-specific genes are underrepresented in this group. By contrast, TBP2- and TLF-dependent genes do not show a strong bias to specific gene orthology classes. We tested if the same orthology relationships were observed for embryonic transcripts and transcripts that have a significant maternal component of expression. The relationship between phylogenetic distribution and TBP family member requirements of all transcripts is very similar to that of maternal-embryonic transcripts (Figure 4C). For the embryonic transcripts (significantly upregulated during embryogenesis), a strikingly different pattern is observed (Figure 4D). Embryonic transcripts that depend exclusively on TBP2 are strongly enriched for vertebrate-specific genes (68% of TBP2-dependent genes compared to 43% for all genes) and have significantly fewer yeast orthologs (only 4.5% compared to 22% for all genes). Because there were no embryonic TBP-dependent Ensembl genes in our data set, the analysis could not be carried out for this category. A weak but significant enrichment of metazoan-specific orthologs was observed among TLF-dependent embryonic transcripts (Figure 4D).
Figure 4.
Gene orthology relationships. (A) Relative distribution of yeast orthologs, metazoan- and vertebrate-specific genes for the whole Affymetrix EST set. The relative values were used as a baseline to determine under- or overrepresentation of orthology groups among transcripts dependent on TBP, TLF or TBP2. The colors of the pie chart segments were continued to be used in (B–D). (B–D) Orthology relationships of transcripts dependent on TBP, TLF and TBP2. (B) Orthology relationships for all transcripts that are dependent on the TBP family members. For TBP-dependent genes, a significant enrichment of yeast orthologs was found. (C, D) Orthology relationships for maternal-embryonic genes and embryonic genes, respectively. Embryonic TBP2-dependent genes are enriched for vertebrate-specific genes. Indicated P-values are based on hypergeometric distribution; **indicates P-value<0.01, *indicates P-value<0.05.
Specialization of TBP family members in cellular functions and pathways
An important question to explore is how TBP, TLF and TBP2 contribute to pathways and cellular functions and to what degree these factors are functionally specialized. As a first step to address this question, we analyzed the effect of TBP, TLF or TBP2 ablation on different cellular pathways and processes using gene ontology (GO) annotation. Analysis of all the transcripts (including transcripts that depend on more than one family member) showed that major physiological cell processes appear to be regulated by TBP, TLF and TBP2 (data not shown). Transcripts annotated for basic processes, such as primary metabolism, transport, signal transduction and cell cycle are not exclusively TBP regulated, but depend on TBP2 and TLF to more or less the same extent (data not shown).
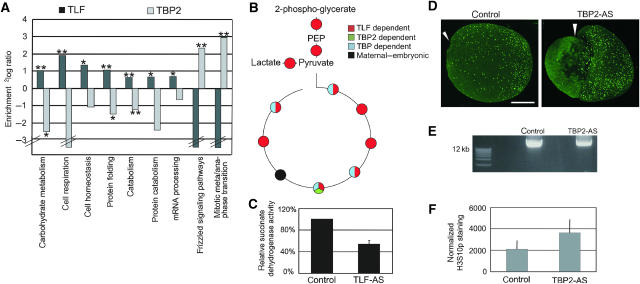
Transcripts exclusively dependent on one of the factors (excluding overlap groups, cf. Figure 2E) were chosen for the subsequent analysis (Figure 5A). The analysis was performed with and without application of the cut-off (2.8-fold change) for the extent to which transcripts were decreased. This revealed the same tendencies. Catabolism and some specific catabolism subcategories are strongly enriched among TLF-dependent transcripts (Figure 5A). TLF is especially linked to carbohydrate utilization. The transcripts that depend on TLF encode many enzymes of glycolysis and the Krebs cycle, including enolase, aldolase, lactate dehydrogenase and succinate dehydrogenase. To confirm the computational analysis, enzymes of the Krebs cycle (some not represented on the microarray) were analyzed by qRT–PCR. The results (Figures 5B and Supplementary Figure S8) showed that most transcripts (10/11) are TLF-dependent, six of which require only TLF but not TBP or TBP2. The decreased transcription upon TLF-AS injection of most of these genes can be increased by adding back TLF mRNA to the embryos showing that these enzymes are specific targets (Supplementary Figure S8). Additionally, we employed a biochemical assay of succinate dehydrogenase enzymatic activity. Comparison of control and TLF-knockdown embryos shows that in TLF-depleted embryos the succinate dehydrogenase activity is approximately twofold reduced (Figure 5C). As this gene is maternally expressed, the remaining enzyme activity may originate from the pool of maternal protein.
Figure 5.
Biological roles of TLF and TBP2: TLF and TBP2 are linked to catabolism and meta-anaphase transition, respectively. (A) Graph shows enrichment of TBP2- and TLF-dependent genes for different GO categories. Transcripts that depend exclusively on TLF or TBP2 were analyzed. Statistical significance was based on the hypergeometric distribution and is indicated; **indicates P-value<0.01, *indicates P-value<0.05. Broken bars indicate that no transcript was annotated for this category. For TBP, no significant correlation with functional groups was found. (B) Schematic overview of Krebs cycle. The dependency of the shown enzymes on TBP family members is indicated by color code. A detailed overview and RT–PCR data can be found in Supplementary Figure S8. (C) Succinate dehydrogenase activity in control embryos and TLF-AS injected embryos. (D) Immunofluorescence with anti-Histone H3 phospho-Serine 10 (H3S10p) in control and TBP2-AS-injected embryos. Lateral view of whole embryo projections, dorsal side is up. The triangle indicates the position of the blastopore in control embryos and of the dorsal blastopore lip in exogastrulating TBP2 embryos (cf. Jallow et al, 2004). The bar represents a size of 300 μm. (E) DNA content in control and TBP2-AS embryos. Agarose-ethidiumbromide gel showing three embryo equivalents of DNA isolated from control and AS-injected embryos. (F) Quantitative evaluation of the relative H3S10p signal after normalization for the DNA content (DNA content measured by spectrophotometer). Mean values and s.e.m. The number of embryos quantitated is 20 for the control, 22 for TBP2-AS injected embryos.
Annotation for the meta-anaphase transition of mitosis is strongly enriched among exclusively TBP2-dependent transcripts (3 out of 5 annotated transcripts, P=0.004, Figure 5A). One of the transcripts annotated for this function is the Xenopus homolog of Saccharomyces cerevisiae cdc23, also known as APC8, a subunit of the E3 ubiquitin ligase of the anaphase-promoting complex. APC8 is important for the progression through mitosis and mutants in yeast display an arrest in mitosis (Lamb et al, 1994; Irniger et al, 1995). As a part of the anaphase-promoting complex is TBP2-dependent in the early Xenopus embryo, we hypothesized that cells may either arrest or spend more time in mitosis in TBP2-depleted embryos compared to control embryos. To measure this biological effect, whole-mount immunofluorescence staining of early neurula (stage 14–15) embryos was performed using an anti-histone H3 phospho-Serine10 antibody as a marker for cells in mitosis (Mahadevan et al, 1991). The embryos were examined by confocal microscopy of serial optical sections covering whole embryos. In control embryos, mitotic cells are observed in all three germ layers with a high density of mitoses in the dorsal-anterior quadrant of the embryo, in particular in the brain and the eye anlagen (Figure 5D). The phospho-H3S10 signal was quantified on account of the pixel area occupied by phospho-H3S10 figures relative to the total embryo area in projections of serial sections. We found an increase of approximately 30% in mitosis figures between TBP2-AS-injected embryos and control embryos (Figure 5D). In addition, we isolated genomic DNA from control and TBP2-AS embryos. Knockdown of TBP2 causes a significant decrease in DNA content (Figure 5E), showing that the increase in mitotic cells in TBP2-AS embryos is associated with a decrease rather than an increase in cell proliferation. When normalized for DNA content (measured by spectrophotometer), TBP2-knockdown embryos contain almost twice the number of mitotic cells compared to control embryos (Figure 5F).
For TBP, no functional specialization was observed.
Preferential-embryonic and ventrally enriched expression
We asked how initiation by different TBP family members might relate to coexpressed groups of genes. Baldessari et al (2005) identified 25 synexpression groups on the basis of extensive microarray analysis of different developmental stages, tissues and experimental treatments of Xenopus. Genes in these synexpression groups are coexpressed and therefore may share common molecular mechanisms of regulation. The groups include gene clusters that are more highly expressed in somatic tissues than in embryos (preferential adult transcripts) or vice versa (preferential embryonic transcripts), furthermore, genes involved in anterior-posterior (A/P) or dorso-ventral (D/V) patterning. The ESTs of these 25 groups were matched to the Affymetrix data using TIGR TC identifiers and we investigated whether TBP-, TLF- or TBP2-dependent genes are enriched or underrepresented in any of the synexpression groups. For most of the groups, there was no enrichment for a requirement for any of the three TBP family members (data not shown). In contrast, transcripts that are expressed at higher levels in the adult organism than in the embryo, including transcripts highly expressed in epithelial and muscle cells, were strongly enriched for a requirement for TBP in the embryo, whereas TLF- and TBP2-dependent genes are underrepresented in these synexpression groups (Figure 6A). Conversely, genes that are primarily expressed in early gastrula embryos are enriched for TLF-dependent genes. TBP-dependent genes are found underrepresented in this group (Figure 6A). Analysis of TBP family member requirements of transcripts expressed differentially in anterior versus posterior halves or dorsal versus ventral halves of the early embryo, showed that these requirements are not linked to anterior-posterior patterning. However, a requirement for TBP2 is linked statistically to dorso-ventral differences, and more specifically to ventral-enriched expression (Figure 6A). Among the transcripts with ventrally enriched expression, 19 out of 30 require TBP2 for their expression, 11 of which exclusively depend on TBP2 rather than TBP or TLF (2.4-fold enrichment, P=0.003, Supplementary Figure S9). In zebrafish TBP2 has been reported to be localized at the ventral side in early (50% epiboly) embryos (Bartfai et al, 2004).
Figure 6.
Synexpression groups. (A) Graph shows enrichment and underrepresentation of TBP family requirements among transcripts that are preferentially expressed in adult or embryonic stages, or differentially expressed between dorsal and ventral (D/V) or anterior and posterior (A/P) sides of the embryo. Indicated P-values are based on hypergeometric distribution; **indicates P-value<0.01, *indicates P-value<0.05. (B) qRT–PCR analysis of control and TBP2-AS-injected embryos that were cut into ventral and dorsal halves. BJ060214.1, BC084093, BC086268.1, BC064290, BC084290.1, BC047969.1, BJ076691 and BJ049504 are ventral TBP2 targets, whereas BC099297.1 is a dorsal TBP2 target. PV.1 and XVent-1 are ventral marker genes, chordin and Xlim-1 are dorsal marker genes. Otx2 and FKH5 are dorsal genes whose expression is upregulated by TBP2 depletion. All values are relative to undissected control embryos.
Therefore, we tested how TBP2 might contribute to dorso-ventral patterning using dissected embryos. Control and TBP2-knockdown early gastrula embryos were cut into dorsal and ventral halves, and expression of a panel of putative targets as well as ventral (PV.1, XVent) and dorsal (chordin, Xlim-1, Otx2 and FKH5) marker genes was analyzed by qRT–PCR (Figure 6B). The dorsal and ventral marker genes showed the expected relative expression levels in dissected embryos. TBP2 is expressed at a higher level on the ventral side of the embryo. Most (seven of eight) ventral transcripts identified by microarray analysis are indeed more highly expressed on the ventral side and require TBP2. Five of these, as well as the two ventral marker genes (XVent-1 and PV.1) show a stronger effect of the AS injection on this side compared to the dorsal side. The dorsally enriched TBP2 target BC099297.1 is affected on both sides of the embryo in the same way. A number of dorsal marker genes (Xlim-1, FKH5, Otx2) do not require TBP2 and were used as markers for dorsal-ventral patterning. The increased expression of some dorsal markers, together with the decreased expression of a significant number of ventral transcripts, uncovers a significant contribution of TBP2 to dorsal-ventral polarity in gene expression.
Discussion
Until now, the extent and significance of TBP-independent transcription was not known. Our results highlight the role of TBP2 and TLF during embryogenesis, as a remarkably small proportion of transcripts depend on TBP. More genes appear to depend on TLF or TBP2 instead (Figure 2). Most of the transcripts that depend on TBP but not TBP2 or TLF in the early embryo are maternally derived transcripts that continue to be expressed in the embryo and that have true yeast orthologs (Figures 4 and 6). In contrast, yeast orthologs are strongly underrepresented in embryonically upregulated TBP2-dependent genes. Out of the genes in this group, 4.5% have orthologs in yeast, whereas among TBP-dependent genes 47% have true yeast orthologs (Figure 4). Among TBP2-dependent transcripts, vertebrate-specific orthologs are significantly overrepresented (68 versus 43% in total data set). Analysis of published data on coexpressed genes revealed links between TBP and genes that are more abundantly expressed in adult stages, whereas TLF is linked to preferential embryonic transcripts and TBP2 is linked to dorso-ventral differential expression (Figure 6). Analysis of glycolytic enzymes and dehydrogenase assays confirmed a role for TLF in the citric acid cycle (Figure 5 and Supplementary Figure S8). Immunostaining of histone H3 phospho-Ser10, and analysis of dissected embryos showed that TBP2-ablated embryos are enriched for mitotic cells (Figure 5) and show dorso-ventral polarity defects, in line with the ventral-enriched expression of TBP2 and a significant contribution to ventral-specific gene expression (Figure 6 and Supplementary Figure S9). Together, these data imply that TLF and TBP2 play specialized roles in the developmental regulation of transcription. Even though TBP also contributes to developmental gene regulation and is essential during embryogenesis (Veenstra et al, 2000), it plays a more generic and less specialized role.
We have shown before that in the oocytes there is abundant TBP2 but no TBP expression (Jallow et al, 2004). On the other hand, in embryos, TBP protein is abundant but TBP2 levels are low. Surprisingly, our analysis illustrates that most of the transcripts that are exclusively TBP-dependent in the embryo are maternally expressed as well (Figure 2F and G). This suggests that the maternal-embryonic transcripts that depend on TBP in the embryo are transcribed in a TBP2-dependent fashion in the oocyte. This can be explained by our earlier studies that demonstrate that a knockdown of TBP in embryos can be rescued partially by overexpression of TBP2 (Jallow et al, 2004). We have also shown developmental stage-specific recruitment of TBP2 (early development) and TBP (late development) to the EF1α promoter (Jallow et al, 2004, cf. Figure 3). Our current data indicate that all three factors show heterologous rescue in triple-knockdown embryos (Supplementary Figure S6). These data should be interpreted with care, because overexpression conditions may mask the physiological contribution of each individual factor. In addition, depleting all three factors may severely cripple the transcription machinery to the extent that even suboptimal initiation of transcription is detected as an increase in transcription relative to triple-knockdown conditions. Individual ablation of these factors is more suited to uncover the physiological role of each factor. Importantly, all three factors are essential during early development (Veenstra et al, 2000; Jallow et al, 2004). A striking observation is that factor recruitment by itself cannot predict the requirements of a promoter for transcription, although some relation is observed (Figure 3 and Supplementary Figure S7). This highlights the complex nature of transcriptional regulation, which involves the integration of many protein–DNA and protein–protein interactions rather than the recruitment of a single factor. It is known that TBP recruitment can lead to a ‘poised' transcriptionally inactive state, which only leads to activation after additional interactions are established. For example, TBP is recruited to the promoter but not the enhancer of the HNF-4alpha gene before it is activated during the course of differentiation. Transcriptional activation coincides with the crosslinking of TBP to both enhancer and promoter (Hatzis and Talianidis, 2002). This may be similar to multiple TBP paralogs being recruited to a promoter with only one of these contributing to promoter activity through productive interactions with other factors. Future work will address the mechanistic implications of our observations.
Despite the plasticity between TBP and TBP2, the phylogenetic distribution of TBP and TBP2 is reflected in over- and underrepresentation of gene orthology of dependent transcripts (Figure 4). The gene orthology relationships appear to be different in embryonic-maternal versus embryonic groups of transcripts. In part, this may be due to yeast orthologs serving developmental functions in metazoans, for example in catabolism for which TLF-dependent transcription is important (Figure 5). As catabolism is essential from yeast to man and in somatic tissues, this seems paradoxical. However, TLF is only required for embryogenesis in metazoan species that develop ex utero, like fish, fly, worm and frog (Dantonel et al, 2000; Veenstra et al, 2000; Muller et al, 2001; Kopytova et al, 2006). In mouse, TLF is not required for embryogenesis (Martianov et al, 2001, 2002a). This suggests that TLF is required in species in which developmental regulation of catabolism plays an important role. The dependency of catabolic enzymes on TLF suggests that some yeast orthologs serve developmental functions controlled by TBP paralogs rather than TBP.
We conclude that TBP plays a limited and generic role in transcription in the early gastrula embryo, which is linked to widespread expression across developmental stages and tissues. In contrast, TLF and TBP2 play a more dominant role that is linked to expression in embryonic stages and particular developmental functions. In addition, the phylogenetic distribution of TBP paralogs can be traced in the transcripts that depend on these factors. Therefore, the biological role of a diversified and non-universal basal transcription machinery is to expand the gene regulatory potential and facilitate embryonic development.
Materials and methods
Embryo manipulation and modified oligonucleotides
AS oligonucleotides modified with aminopropyl methyl piperazine (APMP) linkages (TBP) or N,N-diethyl-ethylenediamine linkages (TLF and TBP2) were injected into fertilized embryos as described before (Veenstra et al, 2000; Jallow et al, 2004). 0.8 ng of TBP-AS, 0.7 ng of TLF-AS or 1 ng of TBP2-AS was used. Embryos were staged according to Nieuwkoop and Faber and collected at the indicated stage. For dissecting embryos into dorsal and ventral halves the Gastromaster equipped with white tips was used. Western blotting was performed as described previously (Veenstra et al, 1999) using anti-TBP 58C9 (Santa Cruz Biotechnology, 1:100).
Quantitative RT–PCR
cDNA was synthesized with the Superscript II system (Invitrogen). For the PCR reactions, iQ SYBR Green Supermix (Biorad) was used and the PCRs were conducted on a Biorad real-time thermal cycler.
Labeling and hybridization of RNA samples
RNA was isolated with TRIzol and the RNeasy kit (Qiagen). 10 μg of total RNA was used in the Affymetrix one-cycle amplification kit. The standard Affymetrix protocol for labeling, hybridization, washing and staining of eukaryotic expression samples was used (Affymetrix GeneChip manual for eukaryotic sample and array processing).
Data analysis
Raw data were analyzed and scaled with the Affymetrix GeneChip Operating 1.2 software (GCOS) in a comparison analysis with the stage 10½ control sample as baseline (cf. legend Supplementary Figure S2). Data were transferred to Spotfire DecisionSite 7.3 for further analysis.
Chromatin immunoprecipitation
ChIP analysis was essentially performed as described previously (Jallow et al, 2004) on X. tropicalis chromatin except for two changes in the protocol: 12.5 μl of Prot A/G beads (Santa Cruz) were used and during reversal of the crosslinking proteinase K was omitted from the buffer.
Ortholog analysis
EST sequences represented on the microarray were mapped to the X. tropicalis genome using BLAT. The Ensembl genes intersecting with these positions were used to predict orthologs using a human-fly-yeast homolog table of the Ensembl database (http://www.ensembl.org). Statistical significance was calculated with the hypergeometric distribution. The hypergeometric distribution models the total number of successes drawn without replacement from a finite population.
Gene ontology analysis
The GO browser of the Affymetrix NetAffx center, which is based on the AmiGO database, was used. Genes decreased by depletion of TBP, TBP2 or TLF were compared to all annotated probe sets of the array and searched for enriched or underrepresented categories. The P-value of the hypergeometric distribution was calculated to determine statistical significance.
Succinate dehydrogenase assay
A total of 25 embryos were homogenized with a Dounce potter in homogenization buffer (0.25 M sucrose, 10 mM Tris, pH7.4, 1 mM EDTA and 2 mM PMSF), fractionated for 10 min at 1000 g and the supernatant was again fractionated 10 minutes at 4000 g. The obtained pellets and supernatant were used to perform a succinate dehydrogenase assay according to the protocol described in Centrifugation Techniques V (NYCOMED PHARMA, Oslo, Norway) using P-iodonitrotetrazolium violet (INT). The values of all embryo fractions were added for comparison.
Whole-mount immunofluorescence and confocal imaging
Whole-mount immunofluorescence procedure was performed as described in Veenstra et al (1995). The primary antibody anti-H3S10p (1:500) and the anti-rabbit ALEXA 647-nm conjugated secondary antibody (1:500, Molecular Probes) were incubated overnight at 4°C. After washing off the secondary antibody, the embryos were cleared with BABB. Fluorescence was imaged on a TCS SP2 AOBS system (Leica Microsystems) using a 633-nm HeNe laser for excitation and applying a 639–699-nm window for detection of the emission signal. Stacks of optical sections covering the entire volume of the embryo were flattened using the maximum projection function of the Leica software. The amount of fluorescence was evaluated by applying thresholds to these monochrome projections and measuring the total area (in pixels) occupied by bright regions of interest using the histogram function of the Adobe Photoshop program. Normalized values were obtained per embryo by correcting for the total DNA content per embryo determined by isolation of genomic DNA. Mean and standard deviation for normalized values were calculated from 20 control and 22 TBP2-AS-injected embryos analyzed over three independent experiments.
Analysis of synexpression groups
Lists of synexpression groups were taken from Baldessari et al (2005). Transcripts were matched to the Affymetrix data set using the TIGR contig (TC) numbers (The Gene Index Databases, Dana Farber Cancer Institute, Boston, MA 02115, http://www.danafarber.org/; Quackenbush et al, 2001). Numbers of transcripts exclusively depending on TBP, TLF or TBP2 were determined for groups of coexpressed genes and for the complete Baldessari data set. Statistical significance of enrichment or underrepresentation was determined with the hypergeometric distribution.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported financially by NWO-ALW (Netherlands Organization for Scientific Research—Research Council for Earth and Life Sciences) with grant numbers 864.03.002 and 813.08.008 to GJCV. The work of JMD is funded in part by Integrated DNA Technologies. We thank C Logie, M Lohrum and H Stunnenberg for critical reading of the manuscript, and E Jansen, N van Bakel and R Engels for help with frogs and animal care.
References
- Baldessari D, Shin Y, Krebs O, Konig R, Koide T, Vinayagam A, Fenger U, Mochii M, Terasaka C, Kitayama A, Peiffer D, Ueno N, Eils R, Cho KW, Niehrs C (2005) Global gene expression profiling and cluster analysis in Xenopus laevis. Mech Dev 122: 441–475 [DOI] [PubMed] [Google Scholar]
- Bartfai R, Balduf C, Hilton T, Rathmann Y, Hadzhiev Y, Tora L, Orban L, Muller F (2004) TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr Biol 14: 593–598 [DOI] [PubMed] [Google Scholar]
- Berk AJ (2000) TBP-like factors come into focus. Cell 103: 5–8 [DOI] [PubMed] [Google Scholar]
- Burki E (1985) The expression of creatine kinase isozymes in Xenopus tropicalis, Xenopus laevis laevis, and their viable hybrid. Biochem Genet 23: 73–88 [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Goldstone K, Smith JC, Gilchrist M, Amaya E, Papalopulu N (2005) A Xenopus tropicalis oligonucleotide microarray works across species using RNA from Xenopus laevis. Mech Dev 122: 355–363 [DOI] [PubMed] [Google Scholar]
- Chong JA, Moran MM, Teichmann M, Kaczmarek JS, Roeder R, Clapham DE (2005) TATA-binding protein (TBP)-like factor (TLF) is a functional regulator of transcription: reciprocal regulation of the neurofibromatosis type 1 and c-fos genes by TLF/TRF2 and TBP. Mol Cell Biol 25: 2632–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Struhl K (1992) The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69: 685–696 [DOI] [PubMed] [Google Scholar]
- Crowley TE, Hoey T, Liu JK, Jan YN, Jan LY, Tjian R (1993) A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila. Nature 361: 557–561 [DOI] [PubMed] [Google Scholar]
- Dagle JM, Littig JL, Sutherland LB, Weeks DL (2000) Targeted elimination of zygotic messages in Xenopus laevis embryos by modified oligonucleotides possessing terminal cationic linkages. Nucleic Acids Res 28: 2153–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagle JM, Weeks DL (2001) Oligonucleotide-based strategies to reduce gene expression. Differentiation 69: 75–82 [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Quintin S, Lakatos L, Labouesse M, Tora L (2000) TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol Cell 6: 715–722 [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Wurtz JM, Poch O, Moras D, Tora L (1999) The TBP-like factor: an alternative transcription factor in metazoa? Trends Biochem Sci 24: 335–339 [DOI] [PubMed] [Google Scholar]
- Davidson I (2003) The genetics of TBP and TBP-related factors. Trends Biochem Sci 28: 391–398 [DOI] [PubMed] [Google Scholar]
- Graindorge A, Thuret R, Pollet N, Osborne HB, Audic Y (2006) Identification of post-transcriptionally regulated Xenopus tropicalis maternal mRNAs by microarray. Nucleic Acids Res 34: 986–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SK, Takada S, Jacobson RH, Lis JT, Tjian R (1997) Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell 91: 71–83 [DOI] [PubMed] [Google Scholar]
- Hatzis P, Talianidis I (2002) Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol Cell 10: 1467–1477 [DOI] [PubMed] [Google Scholar]
- Hernandez N (1993) TBP, a universal eukaryotic transcription factor? Genes Dev 7: 1291–1308 [DOI] [PubMed] [Google Scholar]
- Hochheimer A, Tjian R (2003) Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev 17: 1309–1320 [DOI] [PubMed] [Google Scholar]
- Holmes MC, Tjian R (2000) Promoter-selective properties of the TBP-related factor TRF1. Science 288: 867–870 [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K (1995) Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 81: 269–278 [DOI] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ (2004) Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci USA 101: 13525–13530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach L, Horner MA, Rothman JH, Mango SE (2000) The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol Cell 6: 705–713 [DOI] [PubMed] [Google Scholar]
- Kim J, Iyer VR (2004) Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol Cell Biol 24: 8104–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopytova DV, Krasnov AN, Kopantceva MR, Nabirochkina EN, Nikolenko JV, Maksimenko O, Kurshakova MM, Lebedeva LA, Yerokhin MM, Simonova OB, Korochkin LI, Tora L, Georgiev PG, Georgieva SG (2006) Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol Cell Biol 26: 7492–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Michaud WA, Sikorski RS, Hieter PA (1994) Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J 13: 4321–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KA, Sabel JL, Johnson MJ, Moreira BG, Fletcher CA, Rose SD, Behlke MA, Laikhter AL, Walder JA, Dagle JM (2006) Characterization of modified antisense oligonucleotides in Xenopus laevis embryos. Oligonucleotides 16: 26–42 [DOI] [PubMed] [Google Scholar]
- Mahadevan LC, Willis AC, Barratt MJ (1991) Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 65: 775–783 [DOI] [PubMed] [Google Scholar]
- Martianov I, Brancorsini S, Gansmuller A, Parvinen M, Davidson I, Sassone-Corsi P (2002a) Distinct functions of TBP and TLF/TRF2 during spermatogenesis: requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development 129: 945–955 [DOI] [PubMed] [Google Scholar]
- Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, Davidson I (2001) Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell 7: 509–515 [DOI] [PubMed] [Google Scholar]
- Martianov I, Viville S, Davidson I (2002b) RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science 298: 1036–1039 [DOI] [PubMed] [Google Scholar]
- Moore PA, Ozer J, Salunek M, Jan G, Zerby D, Campbell S, Lieberman PM (1999) A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol Cell Biol 19: 7610–7620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Lakatos L, Dantonel J, Strahle U, Tora L (2001) TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr Biol 11: 282–287 [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M (1982) A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30: 675–686 [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Zhu X, Dixit BL, Maston GA, Kittler EL, Green MR (2003) TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc Natl Acad Sci USA 100: 14887–14891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenstein MD, Zhou S, Lis JT, Tjian R (1999) TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci USA 96: 4791–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 21: 327–335 [PubMed] [Google Scholar]
- Sartor MA, Zorn AM, Schwanekamp JA, Halbleib D, Karyala S, Howell ML, Dean GE, Medvedovic M, Tomlinson CR (2006) A new method to remove hybridization bias for interspecies comparison of global gene expression profiles uncovers an association between mRNA sequence divergence and differential gene expression in Xenopus. Nucleic Acids Res 34: 185–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MC, Reeder RH, Hahn S (1992) Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell 69: 697–702 [DOI] [PubMed] [Google Scholar]
- Sible JC, Anderson JA, Lewellyn AL, Maller JL (1997) Zygotic transcription is required to block a maternal program of apoptosis in Xenopus embryos. Dev Biol 189: 335–346 [DOI] [PubMed] [Google Scholar]
- Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, Chait BT, Roeder RG (1999) Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci USA 96: 13720–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Beumer TL, Peterson-Maduro J, Stegeman BI, Karg HA, van der Vliet PC, Destree OH (1995) Dynamic and differential Oct-1 expression during early Xenopus embryogenesis: persistence of Oct-1 protein following down-regulation of the RNA. Mech Dev 50: 103–117 [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Destree OH, Wolffe AP (1999) Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol Cell Biol 19: 7972–7982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Weeks DL, Wolffe AP (2000) Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science 290: 2312–2315 [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Wolffe AP (2001) Gene-selective developmental roles of general transcription factors. Trends Biochem Sci 26: 665–671 [DOI] [PubMed] [Google Scholar]
- Zhang D, Penttila TL, Morris PL, Roeder RG (2001a) Cell- and stage-specific high-level expression of TBP-related factor 2 (TRF2) during mouse spermatogenesis. Mech Dev 106: 203–205 [DOI] [PubMed] [Google Scholar]
- Zhang D, Penttila TL, Morris PL, Teichmann M, Roeder RG (2001b) Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292: 1153–1155 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information