Abstract
The Kir3.1 K+ channel participates in heart rate control and neuronal excitability through G-protein and lipid signaling pathways. Expression in Escherichia coli has been achieved by replacing three fourths of the transmembrane pore with the pore of a prokaryotic Kir channel, leaving the cytoplasmic pore and membrane interfacial regions of Kir3.1 origin. Two structures were determined at 2.2 Å. The selectivity filter is identical to the Streptomyces lividans K+ channel within error of measurement (r.m.s.d.<0.2 Å), suggesting that K+ selectivity requires extreme conservation of three-dimensional structure. Multiple K+ ions reside within the pore and help to explain voltage-dependent Mg2+ and polyamine blockade and strong rectification. Two constrictions, at the inner helix bundle and at the apex of the cytoplasmic pore, may function as gates: in one structure the apex is open and in the other, it is closed. Gating of the apex is mediated by rigid-body movements of the cytoplasmic pore subunits. Phosphatidylinositol 4,5-biphosphate-interacting residues suggest a possible mechanism by which the signaling lipid regulates the cytoplasmic pore.
Keywords: crystal structure, gating, G-protein-gated inward rectifier, potassium channel, selectivity filter
Introduction
Inward rectifier K+ channels (Kir channels) comprise one family of K+ selective ion channels. Kir1 channels are important for electrolyte flow across kidney epithelial cells (Hebert et al, 2005). Kir2 channels control the resting membrane potential in many different cells and are important for electrical activity in muscle and neurons (Stanfield et al, 2002). In cardiac atrial cells, Kir3 channels, also known as GIRK channels, are responsible for slowing the heart rate when the vagus nerve is stimulated and for membrane potential maintenance in certain neurons (Stanfield et al, 2002). Kir6 channels, which exist as a complex with a second membrane protein called the sulfonylurea receptor, are regulated by the intracellular concentration of ATP (Aguilar-Bryan et al, 1995). In pancreatic beta cells, this form of regulation allows Kir6 channels to mediate insulin secretion (Ashcroft, 2006). These cellular functions underscore the medical importance of Kir channels. Familial diseases characterized by abnormal electrolyte processing by the kidney, epilepsy, cardiac arrhythmia and diabetes mellitus have their basis in abnormal function of Kir channels (Simon et al, 1996; Plaster et al, 2001; Hattersley and Ashcroft, 2005). Thus, Kir channels are potentially important targets for pharmaceutical agents.
The name ‘inward rectifier' stems from the fact that under physiological conditions, Kir channels exhibit higher conductance for K+ flowing into the cell (Lu, 2004). This is because Mg2+ and polyamines abundant inside cells block the pore when the electrochemical gradient favors the outward flow of K+ (Matsuda et al, 1987; Vandenberg, 1987; Lopatin et al, 1994). Consequently Kir channels operate near the resting membrane potential and tend to be silenced when voltage-dependent Na+ or Ca2+ channels depolarize the membrane.
Kir channels, like other K+ channels, are tetramers of identical or similar subunits that surround a central ion conduction pathway. Each subunit contains two transmembrane α helices, a pore helix that crosses the membrane part way, and K+ channel signature sequence amino acids that form the selectivity filter (Doyle et al, 1998). The N- and C-termini, which are located on the intracellular side of the membrane, form a large ‘cytoplasmic pore' structure: this is one of the characteristic features of the Kir family (Nishida and MacKinnon, 2002). Electrophysiological studies show that the cytoplasmic pore is important for at least two purposes. First, it extends the effective length of the pore. The extra long pore contains some of the acidic residues essential for blockage of the channel by Mg2+ ions and polyamines from the intracellular side and give rise to inward rectification (Taglialatela et al, 1994; Yang et al, 1995; Guo and Lu, 2003). Second, the cytoplasmic pore is the structural component through which gating of Kir channels can be modulated by regulatory proteins, ATP, Na+ ions or pH (Stanfield et al, 2002; Hebert et al, 2005; Ashcroft, 2006). Opening of Kir3 channels, for example, is stimulated by beta–gamma G-protein subunits (Logothetis et al, 1987; Reuveny et al, 1994; Wickman et al, 1994).
Another prominent feature of Kir channels is their regulation by the signaling lipid phosphotidylinositol 4,5-bisphosphate (PIP2) (Hilgemann and Ball, 1996; Baukrowitz et al, 1998; Huang et al, 1998; Shyng and Nichols, 1998; Liou et al, 1999; Zhang et al, 1999; Lopes et al, 2002; Pegan et al, 2005). In Kir3 channels, there is a complex interplay between the effects on gating of beta–gamma G-protein subunits and PIP2: both appear to be important for channel opening through direct interactions with the ion channel (Huang et al, 1998).
Recently, three crystallographic studies have provided information on Kir structures. The cytoplasmic pores of Kir3.1 and Kir2.1 have been described at 1.8 and 2.6 Å, respectively (Nishida and MacKinnon, 2002; Pegan et al, 2005), and the complete structure of a prokaryotic Kir from Burkholderia pseudomallei (KirBac1.1) has been described at 3.65 Å (Kuo et al, 2003). The cytoplasmic pore structures correspond to the eukaryotic channels that have been studied extensively for many years using electrophysiological methods, but they are detached from the transmembrane pore. The prokaryotic channel structure, although complete, is only 19% identical in amino-acid sequence to Kir3.1. In combination, these structures have provided templates for homology models for interpreting functional data on eukaryotic Kir channels (Antcliff et al, 2005).
In this study, we have determined two structures at high resolution (2.2 Å) of a chimeric channel, whose transmembrane pore over the extracellular three-fourths of its length is of the prokaryotic Kir from Burkholderia xenovorans (KirBac1.3) (Durell and Guy, 2001) and the remainder, including the intracellular fourth of the transmembrane pore and the cytoplasmic pore, is from the mouse Kir3.1, which is 99% identical to human Kir3.1. The eukaryotic component encompasses the regulatory surfaces through which Kir3 channels are modulated by G-protein subunits and signaling lipids. Many disease-causing mutations map to these surfaces.
Results and discussion
Structure of the chimeric channel
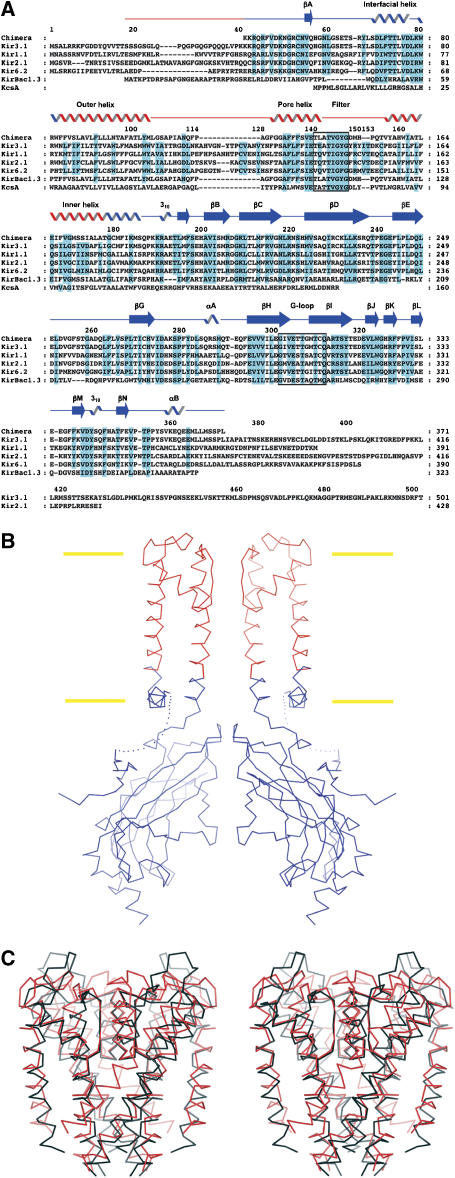
Figure 1A shows an amino-acid sequence alignment that includes four eukaryotic channels representing different subclasses of eukaryotic Kir channels, KirBac1.3 and a prokaryotic K+ channel from Streptomyces lividans (KcsA). We produced a chimeric channel by splicing Kir.3.1 and KirBac1.3 at the indicated junctions (Figure 1A and B). The N-terminus of KirBac1.3 was retained in order to achieve correct processing and membrane insertion using the Escherichia coli expression system. This N-terminus was then removed by thrombin cleavage before crystallization, whereas the N-terminal region of Kir3.1 was included in the final product from K41 in the Kir3.1 sequence. The transmembrane helix junction points were constructed sufficiently deep within the membrane to ensure that all of the cytoplasmic and membrane-surface exposed regions of the channel are of eukaryotic origin. In particular, the interfacial helix (lying parallel to the membrane plane, termed the slide helix in Kuo et al, 2003) and surrounding transmembrane helices are from Kir3.1. Numerous electrophysiological studies have shown that the eukaryotic components, colored blue in Figure 1B, are important for gating and regulation of Kir channels (Stanfield et al, 2002; Hebert et al, 2005; Ashcroft, 2006).
Figure 1.
(A) Sequence alignment of the chimera and K+ ion channels from mammals and prokaryotes. Abbreviations are as follows: Kir3.1: mouse Kir3.1, Kir1.1: human Kir1.1, Kir2.1: mouse Kir2.1, Kir6.2: mouse Kir6.2. Notation for secondary structures and numbering for the chimera above the sequences are from the published structure of the Kir3.1 cytoplasmic pore (Nishida and MacKinnon, 2002). The regions from Kir3.1 and KirBac1.3 in the chimeric channel are colored blue and red, respectively. Highly conserved residues are colored cyan. Signature sequence residues (140–147) and the G-loop residues (301–311) are enclosed in black boxes. (B) Overview of the chimeric channel. Only two subunits are shown for clarity. Cα atoms from Kir3.1 and KirBac1.3 are colored blue and red, respectively. The linker region between the N-terminus and the interfacial helix (dotted line) is disordered and not resolved. Boundaries of the lipid bilayer are drawn as yellow bars. (C) Stereo-view of the transmembrane pores of the chimera and KcsA. The chimera (red) and KcsA (black) are superposed. The interfacial helix of the chimera is omitted.
Construction of the chimeric channel that expressed as a stable membrane protein was not straightforward and required many attempts (Supplementary Figure S1). This fact suggests that there are strict structural requirements, such as achieving the proper phase of the helix attachments. Moreover, we were unable to construct a similar chimera using the pore of the KcsA K+ channel, which is not a member of the Kir family. The strict constraints on establishing a ‘correct' interface and the requirement of a Kir pore imply that the chimera probably replicates with good accuracy the basic structure of Kir3.1.
The channel was expressed and purified as a stable tetramer in the detergent nonylglucoside. Crystallization at pH 6.2 in 120 mM KCl, 200 mM potassium phosphate and polyethylene glycol 4000 yielded crystals that diffracted X-rays to 2.2 Å Bragg spacings (Table I). The tetragonal crystal contained two unique subunits per asymmetric unit, allowing the structure determination of two unique four-fold symmetric chimeric channels, each revealing a different conformation of the cytoplasmic pore. The transmembrane pores are essentially identical to each other. One of these transmembrane pores is compared in Figure 1C with the corresponding region of KcsA.
Table 1.
Data and refinement statistics
| Data statistics | |
| Space group | P4 |
| Lattice constants (Å) | a=b=98.42, c=92.62 |
| Source | Brookhaven X29 |
| Resolution (Å) | 100–2.2 |
| Wavelength (Å) | 1.1 |
| Total/unique observations | 163 094/43 991 |
| I/sigma (I)a | 17.0 (3.9) |
| Redundancyb | 3.7 (3.7) |
| Completeness (%) | 97.9 (99.7) |
| Rsym (%)c | 5.4 (30.9) |
| Refinement statistics | |
| Resolution (Å) | 29.5–2.2 |
| Number of reflections | 43 987 |
| Number of atoms | |
| Protein | 4662 |
| Nonylglucoside | 21 |
| K+ ion | 12 |
| Water | 226 |
| R-factors (%)d | |
| Rwork/Rfree | 23.1/25.6 (29.1/35.1) |
| R.m.s.ds of bonds | Length/angle 0.008 Å/1.28° |
| R.m.s.ds, root mean-square deviations. | |
| The number in parentheses is a statistic for the last resolution shell (2.3–2.2 Å). Same for redundancy, completeness, Rsym and R-factors. | |
| Redundancy=total observations/unique observations. | |
| Rsym=∑∣Ii–〈Ii〉∣/∑Ii, where 〈Ii〉 is the average intensity of symmetry equivalent reflections. | |
| R-factor=∑∣F (obs)–F (cal)∣/∑F (obs). A total of 5% of the total reflections were used for Rfree calculation. | |
Figure 2A shows one of the two channels in the crystal. A crystal soaked in RbCl was used to identify permeant ions in the conduction pathway (see Materials and methods). Four ion positions are observed in the selectivity filter, a fifth in the central cavity, and a sixth and seventh in between the transmembrane and cytoplasmic pores. Ion positions labeled 1 through 5 in Figure 2A correspond to ions observed in KcsA (Zhou et al, 2001). Ion positions labeled 6 and 7 are unique.
Figure 2.
(A) Conduction pore of the chimeric channel in the dilated conformation. A crystal was soaked in a RbCl-containing solution before data collection. The structure refinement and subsequent map calculation were carried out at 2.4 Å without waters and ions. Seven Rb+ ions (green spheres) were located on the Fo–Fc omit map contoured at 3.5 sigma (cyan meshes), on the crystallographic four-fold symmetry axis in the conduction pore. Two constriction sites along the pore (F181 side chain and residues 302–309 Cα atoms in the G-loop) are colored red. Two subunits have been removed from the tetramer for clarity. (B) Surface representation of the chimeric channel. The electrostatic potential of the surface was calculated using the program GRASP. The range of surface potentials is from −21.1 to +19.2 mV. Electropositive and electronegative surfaces are colored blue and red, respectively. Part of the channel has been removed to expose the cytoplasmic pore's inner surface to the viewer.
The channel contains two relatively constricted regions along its length (Figure 2A, red). One of these constrictions corresponds to the inner helix bundle ‘gate' (Doyle et al, 1998). Phenylalanine side chains completely occlude the pore at this point, suggesting that the inner helix bundle gate is closed. The closed gate is similar to that observed in KirBac1.1 (Kuo et al, 2003). The second constriction occurs at the apex of the cytoplasmic pore. It is formed by the ‘G-loop' (Figure 1A), which has been shown to influence gating of Kir channels (Pegan et al, 2005, 2006).
Figure 2B is colored according to the surface electrostatic potential. The cytoplasmic pore is electronegative inside, and therefore it should be attractive to cations such as K+, Mg2+ and polyamines. In contrast, the ‘top' or membrane-facing surface of the cytoplasmic pore and interfacial helices are electropositive. The abundant positively charged arginine and lysine amino acids on these surfaces are well positioned to interact with the negatively charged phospholipid head groups of the cell membrane's inner leaflet.
Structural conservation in the K+ selectivity filter
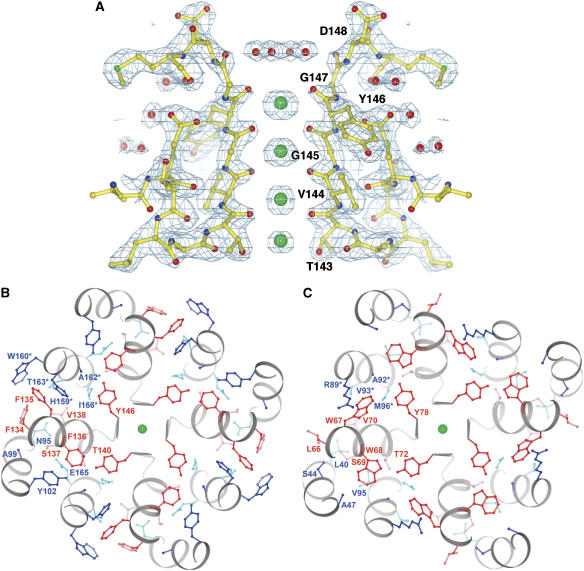
Among the several K+ channel structures determined, KcsA has yielded the most detailed description at a resolution better than 2.0 Å (Zhou et al, 2001; Zhou and MacKinnon, 2003). The chimeric channel presented in this study at 2.2 Å represents a second high-resolution K+ channel structure (Figure 3A). Given that Kir channels and KcsA are far removed from each other on the K+ channel family tree, it is interesting to ask how highly conserved is the three-dimensional structure of the selectivity filter. The signature sequence amino acids, which form the first layer of atoms surrounding the K+ ions in the selectivity filter, are highly conserved in all K+ channels (Figure 1A). But there is significant variation in nearby amino acids, particularly those forming the intricate protein core surrounding the selectivity filter (sequence labeled ‘pore helix' in Figure 1A, and structures shown in Figure 3B and C).
Figure 3.
(A) 2Fo–Fc map of the selectivity filter of the chimeric channel. K+ ions (green) and waters (red) are shown as spheres. Residues 143–148, K+ ions and water molecules were omitted during simulated-annealing refinement against the native data set and subsequent map calculation at 2.2 Å. The map is contoured at 1.3 sigma (cyan meshes). Only two subunits are shown for clarity. Numbering in panels A and B corresponds to the line above the sequences in Figure 1A. (B) Cross section through the selectivity filter of the chimeric channel. The selectivity filter in panel A is rotated by 90° about a horizontal axis in the plane and viewed from the extracellular side. Outer helix, pore helix and inner helix are colored gray. Side chains from the pore helix (red) and from other regions (blue) form a well-packed protein core around the selectivity filter. Residues labeled with an asterisk are from a neighboring subunit. (C) Cross section through the selectivity filter of KcsA. Same view as panel B.
Figure 4A shows a superposition of the selectivity filter atoms from KcsA (the conductive structure) (Zhou et al, 2001) and the chimera. The deviations between these two structures are quantified in a graph of the absolute distance separating corresponding KcsA and chimera main chain atoms (Figure 4B). The blue dashed line in Figure 4B marks the average deviation (0.12 Å) for selectivity filter atoms observed in separate refinements of independent KcsA crystal structures, one with K+ and the other with Tl+ (Zhou et al, 2001; Zhou and MacKinnon, 2003). This value should serve as a reasonable estimate of the uncertainty with which selectivity filter atom positions are defined by data around 2.0-Å resolution. The comparison of KcsA and the chimera therefore seems to indicate that the three-dimensional structure of the selectivity filter is conserved to the limit of our ability to detect differences (0.1–0.2 Å).
Figure 4.
(A) Superposition of the chimera (red) and KcsA (black) selectivity filters. K+ ions in the selectivity filter are shown as spheres (green). (B) Atomic deviations between main chain atoms around the selectivity filter of the chimera and KcsA K+ channels. Deviations between main chain atoms (N, Cα, C, O) for amino acids corresponding to W67-Y82 in KcsA are shown (black). Rad(K)−rad(Na) is the difference in radius between K+ and Na+ ions (0.38 Å, red). Uncertainty is the average deviation for all atoms in from E71 to D80 observed in two KcsA crystal structures (0.12 Å, blue) (Zhou and MacKinnon, 2003).
The precision with which selectivity filter atoms from the chimera and KcsA coincide is not a consequence of bias imposed by chemical terms in the refinement program. By refining the selectivity filter in the absence of Van der Waals radii restraint, significant effects on the mean atomic positions (i.e., greater than 0.2 Å) are not observed. Given the data resolution and quality, the atomic positions determined in this study are dominated by the experimentally measured X-ray intensities.
Quantitative analysis of amino-acid sequence variation in K+ channels shows that the protein core surrounding the selectivity filter forms a highly coupled network of amino acids (Figure 3B and C) (Lockless et al, 2007). In other words, variation at one position is associated with a compensatory variation at other positions in the region. It seems probable that such constrained variation stems at least in part from the fact that the selectivity filter must adopt a very specific structure in order to carry out its catalytic function. Figure 4 shows that deviations of mean atomic positions in the selectivity filter are actually smaller than the amplitudes of thermal fluctuations expected to occur under normal operating temperatures. The deviations are also smaller than the difference in radius between K+ and Na+ ions (0.38 Å, red dashed line in Figure 4B). The fact that the KcsA and chimera crystals were both studied at liquid nitrogen temperature does not diminish the significance of the structural identity of the two selectivity filters: we expect temperature to affect the atomic fluctuations around the mean but not the mean positions of the atoms. In all likelihood, the striking structural conservation (of the mean atomic positions) is a requirement for selectivity filter function.
Two conformations of the cytoplasmic pore
Figure 5A shows a superposition of the two channels present in the crystal. The transmembrane pores are essentially identical, but the cytoplasmic pores adopt different conformations through rigid-body displacements of the subunits with respect to one another. In one conformation, the apex of the cytoplasmic pore is dilated, whereas in the other it is constricted (Figure 5B and C). The dilated conformation is sufficiently wide and exposes oxygen atoms (T306 and G307) on its surface, so that a mostly hydrated K+ should be able to pass through the pore at this point (Figure 5B). The constricted conformation is lined by methionine sulfur atoms (M308) and is too narrow to permit the passage of even a dehydrated K+ ion (Figure 5C). The G-loop (Figure 1A) (Pegan et al, 2006) encompasses the region of the cytoplasmic pore at which the dilation and contraction occurs (I301–T309).
Figure 5.
(A) Superposition of two chimeric channels within the same crystal. The cytoplasmic pore in one channel (blue) is dilated and the other (red) is constricted. Residues 302–309 in the G-loop are highlighted. (B) The apex of the dilated cytoplasmic pore. The cytoplasmic pore is viewed from the transmembrane side. Residues 306–309 in the G-loop are shown as a CPK model. Color code is blue for nitrogen, green for carbon, red for oxygen and yellow for sulfur. The regions of the cytoplasmic pore surrounding the apex are shown with a wire model. The scale bar is 10 Å. (C) Same view as in panel B for the cytoplasmic pore in the constricted conformation.
Ultimately the biological significance of conformational differences observed in crystal structures has to be assessed through correlation with functional studies. Several published studies are relevant to the two conformations observed in the crystal. In Kir6.2 the mutation I296L results in a severe form of DEND syndrome, which is characterized by neonatal diabetes, developmental abnormalities, epilepsy and muscle weakness (Hattersley and Ashcroft, 2005). Electrophysiological studies showed that this mutation alters gating by increasing the open probability and reducing the ability of ATP to close the channel (Proks et al, 2005). The amino acid I296 in Kir6.2 corresponds to M308 in Kir3.1, which is the methionine residue that occludes the cytoplasmic pore when the G-loop constricts (Figure 5B and C). This location supports the conclusion of the Kir6.2 study, that the I296L mutation implicates a new region of Kir channels involved in gating.
In Kir2.1 several mutations within the G-loop cause Andersen's syndrome, which is characterized by periodic paralysis, abnormal electrocardiogram and cardiac arrhythmias (G300V, V302M and E303K) (Lopes et al, 2002; Tristani-Firouzi et al, 2002; Bendahhou et al, 2003; Pegan et al, 2005). Structural studies of the Kir2.1 cytoplasmic pore, combined with electrophysiological studies of Kir2.1 led the authors to conclude that the G-loop may function as a gate (Pegan et al, 2005; Ma et al, 2007).
In Kir3 channels, three independent electrophysiological studies identified the amino acid corresponding to L333 as being important for channel activation by beta–gamma G-protein subunits (He et al, 1999; Ivanina et al, 2003; Finley et al, 2004). In the crystal structure, L333 is located on the outer surface of the cytoplasmic pore at the interface between the subunits (Figure 6A, cyan sphere). This region undergoes a large conformational change when the channel switches between the two conformations observed in the crystal. If the binding of G-protein subunits was to be associated with such a conformational switch, then mutation of L333 would likely affect G-protein activation.
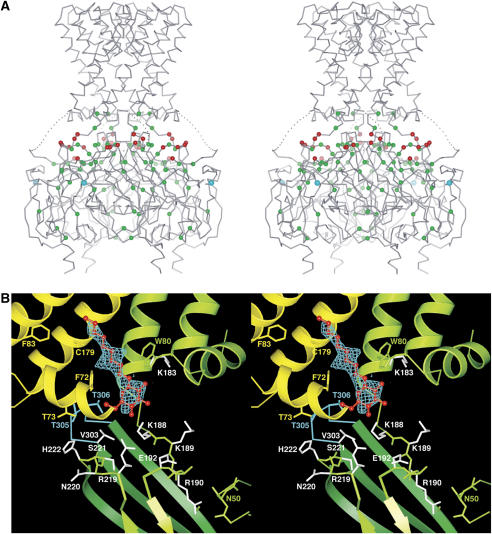
Figure 6.
(A) Stereo-view of the chimeric channel in the dilated conformation with PIP2-related mutations. All mutations that impair channel activation by PIP2 in published studies on the eukaryotic Kir family are mapped onto the chimera structure (see text). Green spheres mark residues buried beneath the protein surface, or that otherwise make close contact with residues on the protein surface. Red spheres show exposed residues on the protein surface and constitute four positively charged amino-acid clusters at the interfacial region, which are related to each other by the crystallographic four-fold symmetry. Cyan spheres mark L333 of Kir3.1, which is important for activation by G-protein subunits. (B) A positively charged amino-acid cluster and a bound detergent molecule. The figure is a close-up view of the interfacial region in panel A. Nonylglucoside (red) and its Fo–Fc omit map contoured at 2.5 sigma (cyan mesh) are shown. The map was calculated at 2.2 Å after the simulated-annealing refinement, with the detergent molecule omitted. Channel subunits and selected side chains near the detergent molecule are colored green and yellow. Residues 303–308 in the G-loop are colored cyan. Side chains colored white are known to alter activation by PIP2 when mutated (colored red or green in panel A). Among them, side chains of K188, K189, E192 and R219 were disordered and poorly defined in the electron-density map. Residues 67 and 68 on the N-terminal side of the interfacial helix (yellow) are omitted for clarity. Numbering corresponds to the line above the sequences in Figure 1A.
In summary, the currently available functional data suggest that conformations similar to those described in this study may occur in the living cell. The two structures imply that gating of the G-loop is mediated by rigid-body movements of the cytoplasmic pore subunits. The inner helix bundle is closed in both structures. Further data will be necessary to understand whether gating of the G-loop is coupled to conformational changes at the inner helix bundle (Zhang et al, 2006).
Amino acids mediating PIP2 activation
Inward rectifier K+ channels also deserve the name lipid-regulated K+ channels. Signaling lipids, especially PIP2, produce activation of all members of this ion channel family in which lipid regulation has been studied, including the Kir1.1, Kir2.1, Kir4.1/5.1, Kir6.2 and Kir3 channels (Hilgemann et al, 2001). For channels in which an intracellular ligand regulates gating, for example beta–gamma G-protein subunits in Kir3 channels and ATP in Kir6.2, complex dual control by the cytoplasmic ligand and membrane lipid is observed (Baukrowitz et al, 1998; Huang et al, 1998; Shyng and Nichols, 1998).
It has been difficult to quantify the absolute affinity and specificity of lipid interactions, because of the uncertainty involved in determining local concentrations of lipid molecules in the membrane. Nevertheless, the following facts have been established through electrophysiological studies. Depletion of PIP2 that occurs upon membrane patch excision results in loss of channel activity (Huang et al, 1998; Zeng et al, 2002; Schulze et al, 2003). Activity loss is accelerated by perfusion with anti-PIP2 antibodies and activity can be recovered by patch perfusion with PIP2-containing lipid vesicles (Huang et al, 1998; Zhang et al, 1999; Shyng et al, 2000; Zeng et al, 2002). Anionic lipids other than PIP2 also activate Kir6.2, and the effectiveness of a lipid tends to be proportional to the magnitude of negative charge on the head group (i.e., its valence) (Fan and Makielski, 1997; Krauter et al, 2001). Polycationic agents such as polylysine and neomycin diminish the ability of PIP2 to activate, presumably through electrostatic screening or direct interference, and mutation of specific lysine and arginine residues on the channel reduce PIP2 activation (Fan and Makielski, 1997; Krauter et al, 2001; Lopes et al, 2002). These findings suggest that the interaction of PIP2 with Kir channels is mediated in part through electrostatic interactions between the negatively charged head group of PIP2 and positively charged protein surfaces.
Many mutations are known to affect PIP2 activation of Kir channels in electrophysiological experiments. Some of these mutations underlie diseases such as Andersen's and Bartter's syndromes (Simon et al, 1996; Plaster et al, 2001). Mutated sites from all Kir channels that impair activation by PIP2 have been mapped onto the crystal structure in Figure 6A (Fan and Makielski, 1997; Huang et al, 1998; Shyng and Nichols, 1998; Liou et al, 1999; Zhang et al, 1999; Shyng et al, 2000; Lopes et al, 2002; Zeng et al, 2002; Garneau et al, 2003; Schulze et al, 2003; Du et al, 2004; Koike-Tani et al, 2005; Pegan et al, 2005; Ribalet et al, 2006; Ma et al, 2007). The structure helps in the interpretation of how a mutation might influence channel gating. Positions colored green are buried beneath the protein surface or otherwise make close contact with the other residues at the protein surface (R190, E304, R313). These residues could potentially destabilize the protein structure or influence motions of the domains associated with PIP2-mediated gating; they are distributed over the entire cytoplasmic pore and most likely have an indirect effect on lipid activation. Positions colored red are exposed on the protein surface and therefore should not disrupt the protein structure. Most of the red-colored positions are lysine or arginine residues located on the membrane-facing surface of the cytoplasmic pore; they tend to occur in a cluster (Figures 2B, 6A and B). Accordingly, these surface amino acids are possible candidates for interacting with the head group of PIP2.
Figure 6B shows a close-up view of a cationic residue cluster on the membrane-facing surface of the cytoplasmic pore. This region was largely disordered or missing in the crystal structure of the cytoplasmic pore alone (Nishida and MacKinnon, 2002), and amino-acid sequences of eukaryotic Kir channels of this region are significantly different from prokaryotic Kir channels. For instance, a linker sequence between the inner helix and the cytoplasmic pore, which is essential for PIP2 binding, is longer and adopts a 310 helix conformation in the chimera structure (Figure 1A). Electron density not accounted for by the protein is observed near the cluster. Although the crystal was grown in the presence of a short-chain analog of PIP2, the shape of the electron density is most compatible with a molecule of nonylglucoside, the detergent used in the purification and crystallization. It is possible that the detergent, present in large excess compared with the short-chain analog of PIP2, has occupied the site at which PIP2 would normally bind. High concentrations of phosphate included in the crystallization solution might also prevent the PIP2 analogue from binding to the protein. This location would place the PIP2 head group near the arginine and lysine residues that are known to affect PIP2 activation when mutated (shown in white, Figure 6B), and not far from the G-loop (shown in cyan). The tail of a lipid molecule at this location would lie very near residues F83 and C179, which when mutated, have dramatic effects on the gating of Kir1.1, Kir6.2 and Kir3 channels (Trapp et al, 1998; Sadja et al, 2001; Yi et al, 2001; Cui et al, 2003; Rapedius et al, 2006; Ribalet et al, 2006). It is perhaps significant that in the constricted conformation, the binding site for the detergent (residues 187–192 in Figure 6B) is disordered (Figure 5A) and the detergent is not observed in the electron-density map. The close contact of the G-loop (T305) with the interfacial helix (F72 and T73) is also not observed in the constricted conformation. Therefore, the structural integrity of this region of the channel seems to depend on lipid/detergent binding and protein conformation. Additional experiments on eukaryotic Kir channels will be required to establish whether this location is indeed a functional binding site for PIP2.
One notable feature of anionic lipid activation of Kir channels is the absence of strong molecular specificity. PIP2 may be the most effective lipid regulator, but other anionic lipids influence gating of Kir6.2 (Fan and Makielski, 1997; Krauter et al, 2001). These observations imply that PIP2 can affect function of Kir channels through interactions that are not highly specific, and that perhaps anionic lipid molecules on the inner leaflet of the cell membrane could influence gating, even if their alkyl chains are not directly bound to the channel. Such a hypothesis has been put forth for Kir6.2 (Fan and Makielski, 1997).
Superposition of the two structures in Figure 5A shows that the open cytoplasmic pore (blue) is closer to the transmembrane pore than the closed cytoplasmic pore (red) by approximately 5 Å. This relationship between the conformation of the cytoplasmic pore and its position relative to the transmembrane pore may be a coincidental consequence of crystal packing. But it may also suggest a possible mechanism if the open conformation of the cytoplasmic pore is indeed mechanically favored when its position is near the transmembrane pore. Through electrostatic attraction of the positive surface on the cytoplasmic pore, addition of PIP2 or other anionic lipids to the membrane might affect the cytoplasmic pore's position along with its conformation, and thus its open probability. Such a mechanism in which the cytoplasmic pore conformation is controlled by lipid surface charge would explain why an anionic lipid's potency tends to be proportional to its valence, and why polycationic molecules such as polylysine and neomycin interfere with PIP2 activation (Fan and Makielski, 1997; Krauter et al, 2001; Lopes et al, 2002).
Conclusion
By producing a chimera between prokaryotic and eukaryotic Kir channels, we have determined the structure of a channel in which the cytoplasmic regulatory surfaces from a eukaryotic G-protein-gated channel Kir3.1 have been preserved. The chimeric channel provides an atomic model with which to interpret electrophysiological data and design future experiments on eukaryotic Kir channels. We emphasize four ideas supported by the structure.
First, at 2.2-Å resolution, the chimera structure represents the second high-resolution K+ channel structure, following KcsA. The three-dimensional structures of the chimera and KcsA selectivity filters superimpose to within the limit of our ability to detect variation: a conservative estimate of this limit is 0.2 Å. The extreme conservation of atomic positions lining the selectivity filter presumably reflects the catalytic requirement of the filter to conduct K+ rapidly and selectively.
Second, K+ ions are present not only inside the selectivity filter but also at non-filter locations within the long pore. Further, the electronegative interior of the cytoplasmic pore may permit delocalized K+ ions that are not observed in the crystal structure. The detection of multiple ions within and outside the filter supports the hypothesis that the high electrical valence of voltage-dependent blockers depends on the coupled movement of permanent ions (Shin and Lu, 2005). The high valence gives rise to a sharply rectifying current–voltage relationship (Lu, 2004).
Third, two structures with different conformations existing in a single crystal form suggest that rigid-body motions of the cytoplasmic pore subunits are coupled to the position and conformation of the G-loop, which may function as a gate in the cytoplasmic pore. This finding supports numerous other studies identifying the G-loop as a potential gate (Lopes et al, 2002; Tristani-Firouzi et al, 2002; Bendahhou et al, 2003; Pegan et al, 2005; Proks et al, 2005).
Fourth, PIP2 might affect gating by binding to specific sites on the channel and/or by inserting into the inner membrane leaflet in the vicinity of the channel. These possibilities are not mutually exclusive. Several amino acids on the surface of the cytoplasmic pore are well positioned to interact with PIP2 and thereby influence the cytoplasmic pore.
One deficiency of this study is the absence of functional analysis of the chimera. No channel activity is observed in planar lipid membranes consisting of 1-palmitoyl-2-oleoyl-glycero-3-phosphoethanolamine and 1-palmitoyl-2-oleoyl-glycero-3-phospho-1-glycerol in a 3:1 ratio. There are several potential reasons for the lack of function: there may be an unmet lipid requirement, Kir3.1 is normally functional as a heteromultimer with other members of the Kir3 family, whereas the chimera is a homomultimer (Krapivinsky et al, 1995), the chimera might lack the proper coupling between the cytoplasmic and transmembrane pores, and finally, single channel activity has yet to be demonstrated for any of the prokaryotic Kir channels. Even in the absence of function, we believe that these structural data provide a valuable model to advance the understanding of eukaryotic Kir channels.
Materials and methods
Construction of the chimeric channel
Genomic DNA was isolated from Bacterium B. xenovorans (formerly known as Burkholderia fungorum) strain LB400 and a region encoding a prokaryotic Kir was amplified by the standard polymerase chain reaction (Stratagene). A chimeric channel gene encoding the amino-acid sequence shown in Figure 1A was constructed by sequential polymerase chain reactions: amino acids 41–82 and 178–371 correspond to mouse Kir3.1 with a mutation M180A. The DNA was also modified to encode amino acids LV in between positions 23 and 24 (KirBac1.3 sequence), and to encode the sequence VPRGSGGLEHHHHHH at the C-terminus. The gene was inserted between NcoI and XhoI restriction sites of the pET-28b(+) expression vector (Novagen).
Channel expression and purification
E. coli BL21(DE3) cells were transformed and used to inoculate 16 l of LB medium in the presence of 50 μg/ml kanamycin. After growing cells at 37°C in shaker flasks to an optical density of 2.0 (wavelength 600 nm) the temperature was lowered to 23°C and 300 μM of isopropyl thiogalactopyranoside was added to induce protein expression. Cells were harvested 12–15 h following induction and suspended in 400 ml of 50 mM Tris–HCl pH 7.5, 150 mM KCl, 100 μM phenylmethanesulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin and 1000-fold dilution of aprotinin from bovine lung (Sigma). Cells were disrupted using a french pressure cell press at 15 000 pounds per square inch and 4°C. Channel protein was extracted from the membrane by adding dodecylmaltopyranoside (Anatrace) to 20 mM and shaking the mixture gently for 2 h at room temperature. Cell debris and insoluble material were removed from the extract by centrifugation at 40 000 g for 20 min. The supernatant was incubated with 6 ml of cobalt resin (Clontech) for 1 h. The resin was added to a column and washed using five column volumes of 20 mM Tris–HCl pH 7.5, 150 mM KCl, 20 mM imidazole and 5 mM dodecylmaltopyranoside. The channel was eluted using the same solution, with imidazole increased to 100 mM. After addition of 2 mM dithiothreitol, the elution fraction was concentrated and treated with 8 U of thrombin at room temperature for 2 h to remove both the N- and C-termini. The tetrameric channel was purified on a Superdex200 gel-filtration column (GE Healthcare) pre-equilibrated with 8 mM Bis–Tris pH 6.5, 120 mM KCl, 3 mM dithiothreitol and 16 mM nonylglucoside. Following elution the channel was concentrated to 10–20 mg/ml and dialyzed overnight with a membrane of molecular weight cut-off 100 kDa (Spectrum Laboratories), at room temperature in the equilibration buffer in order to reduce the concentration of the detergent. Final yield of the channel was approximately 8 mg from 16 l of the culture.
Crystallization and structure determination
Tris (2-carboxyethyl) phosphine hydrochloride at 20 mM and the short-chain PIP2 analogue 1,2-dihexanoyl phosphatidylinositol 4,5-biphosphate (Cayman Chemical) at 2.6 mM were added to the sample solution before crystallization. P4 crystals were grown using sitting drop vapor diffusion of a 1:1 protein:reservoir solution against a reservoir containing 10–15% (w/v) PEG4000, 50 mM sodium citrate and 200 mM potassium phosphate (pH 6.2) at 20°C. Rectangular crystals grew over 2–4 weeks to 400 microns in the longest dimension. These were cryoprotected in a solution containing 400 mM KCl and 32% (w/v) glycerol and frozen in liquid nitrogen. A data set was collected from a single crystal at beamline X29 at Brookhaven National Laboratory, and processed with the programs Denzo and Scalepack (Otwinowski and Minor, 1997) (Table I). Initial phases were determined by molecular replacement using as search models the Kir3.1 cytoplasmic pore and KirBac1.1 transmembrane pore structures with the program Molrep (Collaborative Computational Project, Number 4, 1994). Model building and refinement were carried out using the programs O and CNS, respectively (Brunger et al, 1998). The reflections between 29.5 to 2.2 Å were used without a sigma cut-off for the refinement and free R-factor calculation, during which bulk solvent and anisotropic temperature factor corrections were applied. Two-fold symmetry restraints between two subunits in an asymmetric unit were applied throughout the refinement to residues 44–53, 70–184 and the structured components of the C-terminal cytoplasmic region independently. The following regions corresponding to the Kir3.1 sequence were disordered and therefore not modeled: residues 60–66 and 365–371 in the dilated pore and residues 41–42, 56–69 and 187–192 in the constricted pore. In the Ramachandran plot, 473 residues among non-glycine or non-proline residues were included in the most favored regions, 45 residues were in the additional allowed regions and one residue was in the generously allowed region (Laskowski et al, 1993). Water molecules were incorporated in the model using the program CNS, with manual inspection. Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the ID code 2QKS. In order to identify the K+ ion-binding sites in the conduction pore, K+ ions in a native crystal were exchanged for Rb+ ions by soaking in a solution containing 400 mM RbCl for several hours before cryoprotection and data collection. The Fo–Fo omit map between RbCl-soaked and native crystals was not interpretable due to non-isomorphism between individual crystals, and therefore Fo–Fc Fourier coefficients were used (Figure 2A).
Supplementary Material
Supplementary Figure S1
Acknowledgments
We thank Dr Lily Y Jan for the mouse Kir3.1 gene, Dr Tamara V Tsoi for B. xenovorans strain LB400, Dr Wang Qingjun for mass spectrometry, Alice Lee for helpful discussions on the manuscript, and the staff at stations X25 and X29 of the Brookhaven National Laboratory Light Source. Supported by NIH GM43949 to RM and grant RR00862 to BTC. RM is an investigator in the Howard Hughes Medical Institute.
References
- Aguilar-Bryan L, Nichols CG, Wechsler SW, Clement JP IV, Boyd AE III, Gonzalez G, Herrera-Sosa H, Nguy K, Bryan J, Nelson DA (1995) Cloning of the beta cell high-affinity sulfonylurea receptor: a regulator of insulin secretion. Science 268: 423–426 [DOI] [PubMed] [Google Scholar]
- Antcliff JF, Haider S, Proks P, Sansom MS, Ashcroft FM (2005) Functional analysis of a structural model of the ATP-binding site of the KATP channel Kir6.2 subunit. EMBO J 24: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft FM (2006) KATP channels and insulin secretion: a key role in health and disease. Biochem Soc Trans 34: 243–246 [DOI] [PubMed] [Google Scholar]
- Baukrowitz T, Schulte U, Oliver D, Herlitze S, Krauter T, Tucker SJ, Ruppersberg JP, Fakler B (1998) PIP2 and PIP as determinants for ATP inhibition of KATP channels. Science 282: 1141–1144 [DOI] [PubMed] [Google Scholar]
- Bendahhou S, Donaldson MR, Plaster NM, Tristani-Firouzi M, Fu YH, Ptacek LJ (2003) Defective potassium channel Kir2.1 trafficking underlies Andersen–Tawil syndrome. J Biol Chem 278: 51779–51785 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kuntsleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 Suite: programs for X-ray crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cui N, Wu J, Xu H, Wang R, Rojas A, Piao H, Mao J, Abdulkadir L, Li L, Jiang C (2003) A threonine residue (Thr71) at the intracellular end of the M1 helix plays a critical role in the gating of Kir6.2 channels by intracellular ATP and protons. J Membr Biol 192: 111–122 [DOI] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral JH, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280: 69–77 [DOI] [PubMed] [Google Scholar]
- Du X, Zhang H, Lopes C, Mirshahi T, Rohacs T, Logothetis DE (2004) Characteristic interactions with phosphatidylinositol 4,5-bisphosphate determine regulation of Kir channels by diverse modulators. J Biol Chem 279: 37271–37281 [DOI] [PubMed] [Google Scholar]
- Durell SR, Guy HR (2001) A family of putative Kir potassium channels in prokaryotes. BMC Evol Biol 1: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Makielski JC (1997) Anionic phospholipids activate ATP-sensitive potassium channels. J Biol Chem 272: 5388–5395 [DOI] [PubMed] [Google Scholar]
- Finley M, Arrabit C, Fowler C, Suen KF, Slesinger PA (2004) βL–βM loop in the C-terminal domain of G protein-activated inwardly rectifying K+ channels is important for Gβγ subunit activation. J Physiol 555: 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau L, Klein H, Parent L, Sauve R (2003) Contribution of cytosolic cysteine residues to the gating properties of the Kir2.1 inward rectifier. Biophys J 84: 3717–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Lu Z (2003) Interaction mechanisms between polyamines and IRK1 inward rectifier K+ channels. J Gen Physiol 122: 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley AT, Ashcroft FM (2005) Activating mutations in Kir6.2 and neonatal diabetes: new clinical syndromes, new sientific insights, and new therapy. Diabetes 54: 2503–2513 [DOI] [PubMed] [Google Scholar]
- He C, Zhang H, Mirshahi T, Logothetis DE (1999) Identification of a potassium channel site that interacts with G protein βγ subunits to mediate agonist-induced signaling. J Biol Chem 274: 12517–12524 [DOI] [PubMed] [Google Scholar]
- Hebert SC, Desir G, Giebisch G, Wang W (2005) Molecular diversity and regulation of renal potassium channels. Physiol Rev 85: 319–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgemann DW, Ball R (1996) Regulation of cardiac Na+, Ca2+ exchange and KATP potassium channels by PIP2. Science 273: 956–959 [DOI] [PubMed] [Google Scholar]
- Hilgemann DW, Feng S, Nasuhoglu C (2001) The complex and intriguing lives of PIP2 with ion channels and transporters. Sci STKE 2001: 1–8 [DOI] [PubMed] [Google Scholar]
- Huang C-L, Feng S, Hilgemann DW (1998) Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gβγ. Nature 391: 803–806 [DOI] [PubMed] [Google Scholar]
- Ivanina T, Rishal I, Varon D, Mullner C, Frohnwieser-Steinecke B, Schreibmayer W, Dessauer CW, Dascal N (2003) Mapping the Gβγ-binding sites in GIRK1 and GIRK2 subunits of the G protein-activated K+ channel. J Biol Chem 278: 29174–29183 [DOI] [PubMed] [Google Scholar]
- Koike-Tani M, Collins JM, Kawano T, Zhao P, Zhao Q, Kozasa T, Nakajima S, Nakajima Y (2005) Signal transduction pathway for the substance P-induced inhibition of rat Kir3 (GIRK) channel. J Physiol 564: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Gordon EA, Wickman K, Velimirovic B, Krapivinsky L, Clapham DE (1995) The G-protein-gated atrial K+ channel IKACh is a heteromultimer of two inwardly rectifying K+-channel proteins. Nature 374: 135–141 [DOI] [PubMed] [Google Scholar]
- Krauter T, Ruppersberg JP, Baukrowitz T (2001) Phospholipids as modulators of KATP channels: distinct mechanisms for control of sensitivity to sulphonylureas, K+ channel openers, and ATP. Mol Pharmacol 59: 1086–1093 [PubMed] [Google Scholar]
- Kuo A, Gulbis JM, Antcliff JF, Rahman T, Lowe ED, Zimmer J, Cuthbertson J, Ashcroft FM, Ezaki T, Doyle DA (2003) Crystal structure of the potassium channel KirBac1.1 in the closed state. Science 300: 1922–1926 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Liou H-H, Zhou S-S, Huang C-L (1999) Regulation of ROMK1 channel by protein kinase A via a phosphatidylinositol 4,5-bisphosphate-dependent mechanism. Proc Natl Acad Sci USA 96: 5820–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless SW, Zhou M, MacKinnon R (2007) Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol 5: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis DE, Kurachi Y, Galper J, Neer EJ, Clapham DE (1987) The subunit βγ subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature 325: 321–326 [DOI] [PubMed] [Google Scholar]
- Lopatin AN, Makhina EN, Nichols CG (1994) Potassium channel block by cytoplasmic polyamines as the mechanism of intrinsic rectification. Nature 372: 366–369 [DOI] [PubMed] [Google Scholar]
- Lopes CMB, Zhang H, Rohacs T, Jin T, Yang J, Logothetis DE (2002) Alterations in conserved Kir channel–PIP2 interactions underlie channelopathies. Neuron 34: 933–944 [DOI] [PubMed] [Google Scholar]
- Lu Z (2004) Mechanism of rectification in inward-rectifier K+ channels. Annu Rev Physiol 66: 103–129 [DOI] [PubMed] [Google Scholar]
- Ma D, Tang XD, Rogers TB, Welling PA (2007) An Andersen–Tawil syndrome mutation in Kir2.1 (V302M) alters the G-loop cytoplasmic K+ conduction pathway. J Biol Chem 282: 5781–5789 [DOI] [PubMed] [Google Scholar]
- Matsuda H, Saigusa A, Irisawa H (1987) Ohmic conductance through the inwardly rectifying K channel and blocking by internal Mg2+. Nature 325: 156–159 [DOI] [PubMed] [Google Scholar]
- Nishida M, MacKinnon R (2002) Structural basis of inward rectification: cytoplasmic pore of the G protein gated inward rectifier GIRK1 at 1.8 Å resolution. Cell 111: 957–965 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pegan S, Arrabit C, Slesinger PA, Choe S (2006) Andersen's syndrome mutation effects on the structure and assembly of the cytoplasmic domains of Kir2.1. Biochemistry 45: 8599–8606 [DOI] [PubMed] [Google Scholar]
- Pegan S, Arrabit C, Zhou W, Kwiatkowski W, Collins A, Slesinger PA, Choe S (2005) Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat Neurosci 8: 279–287 [DOI] [PubMed] [Google Scholar]
- Plaster NM, Tawil R, Tristani-Firouzi M, Canun S, Bendahhou S, Tsunoda A, Donaldson MR, Iannaccone ST, Brunt E, Barohn R, Clark J, Deymeer F, George AL Jr, Fish FA, Hahn A, Nitu A, Ozdemir C, Serdaroglu P, Subramony SH, Wolfe G et al. (2001) Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen's syndrome. Cell 105: 511–519 [DOI] [PubMed] [Google Scholar]
- Proks P, Girard C, Haider S, Gloyn AL, Hattersley AT, Sansom MS, Ashcroft FM (2005) A gating mutation at the internal mouth of the Kir6.2 pore is associated with DEND syndrome. EMBO Rep 6: 470–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapedius M, Haider S, Browne KF, Shang L, Sansom MS, Baukrowitz T, Tucker SJ (2006) Structural and functional analysis of the putative pH sensor in the Kir1.1 (ROMK) potassium channel. EMBO Rep 7: 611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuveny E, Slesinger PA, Inglese J, Morales JM, Iniguez-Lluhi JA, Lefkowitz RJ, Bourne HR, Jan YN, Jan LY (1994) Activation of the cloned muscarinic potassium channel by G protein βγ subunits. Nature 370: 143–146 [DOI] [PubMed] [Google Scholar]
- Ribalet B, John SA, Xie LH, Weiss JN (2006) ATP-sensitive K+ channels: regulation of bursting by the sulphonylurea receptor, PIP2 and regions of Kir6.2. J Physiol 571: 303–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadja R, Smadja K, Alagem N, Reuveny E (2001) Coupling Gβγ-dependent activation to channel opening via pore elements in inwardly rectifying potassium channels. Neuron 29: 669–680 [DOI] [PubMed] [Google Scholar]
- Schulze D, Krauter T, Fritzenschaft H, Soom M, Baukrowitz T (2003) Phosphatidylinositol 4,5-bisphosphate (PIP2) modulation of ATP and pH sensitivity in Kir channels. A tale of an active and a silent PIP2 site in the N terminus. J Biol Chem 278: 10500–10505 [DOI] [PubMed] [Google Scholar]
- Shin H-G, Lu Z (2005) Mechanism of the voltage sensitivity of IRK1 inward-rectifier K+ channel block by the polyamine spermine. J Gen Physiol 125: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyng S-L, Cukras CA, Harwood J, Nichols CG (2000) Structural determinants of PIP2 regulation of inward rectifier KATP channels. J Gen Physiol 116: 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyng S-L, Nichols CG (1998) Membrane phospholipid control of nucleotide sensitivity of KATP channels. Science 282: 1138–1141 [DOI] [PubMed] [Google Scholar]
- Simon DB, Karet FE, Rodoriguez-Soriano J, Hamdan JH, DiPietro A, Trachtman H, Sanjad SA, Lifton RP (1996) Genetic heterogeneity of Barter's syndrome revealed by mutation in the K+ channel, ROMK. Nat Genet 14: 152–156 [DOI] [PubMed] [Google Scholar]
- Stanfield PR, Nakajima S, Nakajima Y (2002) Constitutively active and G-protein coupled inward rectifier K+ channels: Kir2.0 and Kir3.0. Rev Physiol Biochem Pharmacol 145: 47–179 [DOI] [PubMed] [Google Scholar]
- Taglialatela M, Wible BA, Caporaso R, Brown AM (1994) Specification of pore properties by the carboxyl terminus of inwardly rectifying K+ channels. Science 264: 844–847 [DOI] [PubMed] [Google Scholar]
- Trapp S, Proks P, Tucker SJ, Ashcroft FM (1998) Molecular analysis of ATP-sensitive K channel gating and implications for channel inhibition by ATP. J Gen Physiol 112: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristani-Firouzi M, Jensen JL, Donaldson MR, Sansone V, Meola G, Hahn A, Bendahhou S, Kwiecinski H, Fidzianska A, Plaster N, Fu YH, Ptacek LJ, Tawail R (2002) Functional and clinical characterization of KCNJ2 mutations associated with LQT7 (Andersen syndrome). J Clin Invest 110: 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg CA (1987) Inward rectification of a potassium channel in cardiac ventricular cells depends on internal magnesium ions. Proc Natl Acad Sci USA 84: 2560–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickman KD, Iniguez-Lluhl JA, Davenport PA, Taussig R, Krapivinsky GB, Linder ME, Gilman AG, Clapham DE (1994) Recombinant G-protein βγ-subunits activate the muscarinic-gated atrial potassium channel. Nature 368: 255–257 [DOI] [PubMed] [Google Scholar]
- Yang J, Jan YN, Jan LY (1995) Control of rectification and permeation by residues in two distinct domains in an inward rectifier K+ channel. Neuron 14: 1047–1054 [DOI] [PubMed] [Google Scholar]
- Yi BA, Lin YF, Jan YN, Jan LY (2001) Yeast screen for constitutively active mutant G protein-activated potassium channels. Neuron 29: 657–667 [DOI] [PubMed] [Google Scholar]
- Zeng W-Z, Liou H-H, Krishna UM, Falck JR, Huang C-L (2002) Structural determinants and specificities for ROMK1–phosphoinositide interaction. Am J Physiol Renal Physiol 282: F826–F834 [DOI] [PubMed] [Google Scholar]
- Zhang H, He C, Yan X, Mirshahi T, Logothetis DE (1999) Activation of inwardly rectifying K+ channels by distinct PtdIns(4,5)P2 interactions. Nat Cell Biol 1: 183–188 [DOI] [PubMed] [Google Scholar]
- Zhang Y-Y, Sackin H, Palmer LG (2006) Localization of the pH gate in Kir1.1 channels. Biophys J 91: 2901–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, MacKinnon R (2003) The occupancy of ions in the K+ selectivity filter: charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates. J Mol Biol 333: 965–975 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R (2001) Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 Å resolution. Nature 414: 43–48 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1