Abstract
The circadian phosphorylation cycle of the cyanobacterial clock protein KaiC has been reconstituted in vitro. The phosphorylation profiles of two phosphorylation sites in KaiC, serine 431 (S431) and threonine 432 (T432), revealed that the phosphorylation cycle contained four steps: (i) T432 phosphorylation; (ii) S431 phosphorylation to generate the double-phosphorylated form of KaiC; (iii) T432 dephosphorylation; and (iv) S431 dephosphorylation. We then examined the effects of mutations introduced at one KaiC phosphorylation site on the intact phosphorylation site. We found that the product of each step in the phosphorylation cycle regulated the reaction in the next step, and that double phosphorylation converted KaiC from an autokinase to an autophosphatase, whereas complete dephosphorylation had the opposite effect. These mechanisms serve as the basis for cyanobacterial circadian rhythm generation. We also found that associations among KaiA, KaiB, and KaiC result from S431 phosphorylation, and these interactions would maintain the amplitude of the rhythm.
Keywords: circadian rhythm, cyanobacteria, in vitro, KaiC, phosphorylation
Introduction
Circadian rhythms, biological oscillations with a period of ∼24 h that are found ubiquitously among living cells, allow organisms to match their metabolism to the earth's day/night cycle (Pittendrigh, 1993). Cyanobacteria are the simplest organisms known to exhibit circadian rhythms. In the unicellular cyanobacterium Synechococcus elongatus PCC 7942, the kaiABC gene cluster has been shown to encode three essential circadian clock proteins: KaiA, KaiB, and KaiC (Ishiura et al, 1998).
KaiC has two sets of Walker A and B motifs, which are conserved in ATPases (Ishiura et al, 1998), suggesting that ATP-utilizing reactions play key roles in the cyanobacterial circadian system. One such candidate reaction is phosphorylation of KaiC; KaiC exhibits a circadian rhythm in its phosphorylation level in vivo (Iwasaki et al, 2002), and has been demonstrated to have both autokinase and autophosphatase activities in vitro (Nishiwaki et al, 2000, Xu et al, 2003).
KaiA and KaiB affect the phosphorylation/dephosphorylation of KaiC both in vivo and in vitro (Iwasaki et al, 2002, Williams et al, 2002, Kitayama et al, 2003). When KaiC is incubated in vitro with ATP and MgCl2, it is gradually dephosphorylated (Xu et al, 2003). On the other hand, KaiC phosphorylation is promoted in the presence of KaiA (Iwasaki et al, 2002, Williams et al, 2002). Although KaiB alone has little effect on these reactions, when all three proteins are incubated together, KaiB negates the KaiA activity and promotes KaiC dephosphorylation (Kitayama et al, 2003).
We succeeded in reconstituting a robust circadian oscillation of KaiC phosphorylation in vitro by mixing KaiA, KaiB, KaiC, and ATP (Nakajima et al, 2005). Reconstitution of this circadian rhythm enabled the in vitro elucidation of the mechanism underlying the generation of the cyanobacterial circadian rhythm. Using this in vitro reconstituted system, we examined the associations between the clock proteins and found that a high-molecular-weight protein complex that included KaiA, KaiB, and KaiC formed during the dephosphorylation phase. In addition, high levels of KaiC phosphorylation coincided with the formation of the Kai protein complex (Kageyama et al, 2006).
Previously, we used mass spectrometry to identify two phosphorylation sites in KaiC at serine 431 (S431) and threonine 432 (T432), both of which are required for circadian rhythm generation in vivo (Nishiwaki et al, 2004). In our recent studies using the in vitro reconstituted system (Nakajima et al, 2005, Kageyama et al, 2006), however, we only detected two species of KaiC, phosphorylated and nonphosphorylated, because of technical limitations. Therefore, it has not yet been elucidated how both S431 and T432 function to generate the circadian rhythm, and how these sites regulate the Kai protein interactions.
In the present study, we employed nanoflow liquid chromatography/electrospray ionization-mass spectrometry (nano-LC/ESI-MS) and improved SDS–PAGE to analyze these sites individually, which allowed us to examine the roles of S431 and T432 in the reconstituted system. We found that the phosphorylation states of S431 and T432 oppositely regulate the phosphorylation/dephosphorylation reactions at the adjacent residue. Furthermore, we clarified that interactions among KaiA, KaiB, and KaiC occur as a result of S431 phosphorylation, which maintains the amplitude of the rhythm.
Results
Circadian profiles of three phosphorylated forms of KaiC
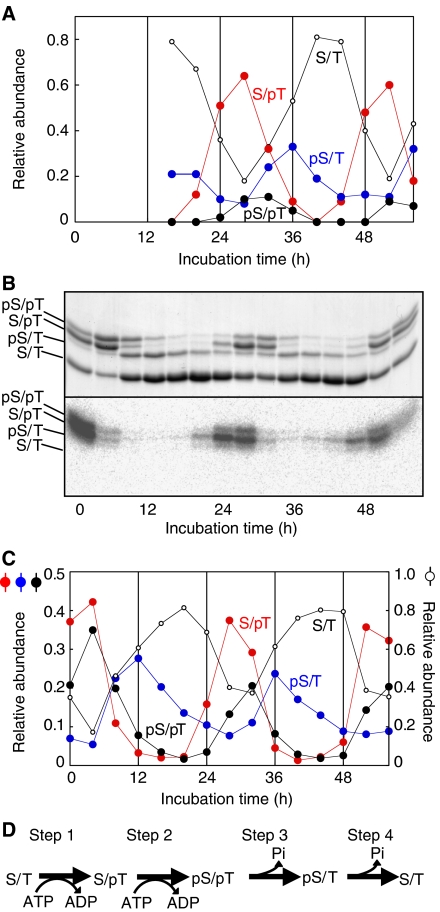
We incubated 3.5 μM KaiC with 1.2 μM KaiA, 3.5 μM KaiB, 5 mM MgCl2, and 1 mM ATP at 30°C (the standard conditions) to reconstitute the KaiC phosphorylation cycle in vitro. Then, we analyzed the circadian profiles of phosphorylation of the two sites separately using nano-LC/ESI-MS. Based on the observed masses and integrated ion peak areas of synthetic peptides (D427SHIpSTIT, D427SHISpTIT, D427SHIpSpTIT, and D427SHISTIT), which had been identified using nano-LC/ESI-MS/MS in a separate experiment (Nishiwaki et al, 2004), we examined the accumulation profiles every 4-h intervals of the nonphosphorylated and the three phosphorylated forms of KaiC (Figure 1A). The level of the S/pT (T432-phosphorylated) form peaked at 28 h, which was followed by the peaks of the pS/pT (S431- and T432-phosphorylated) form and the pS/T (S431-phosphorylated) form after 4-h intervals.
Figure 1.
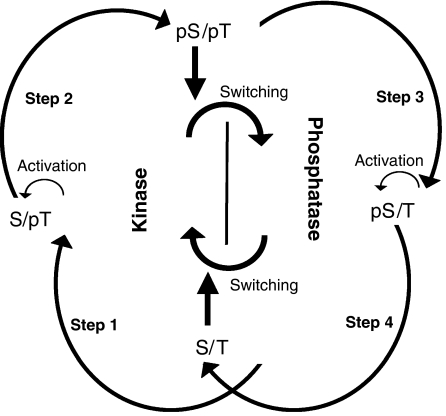
Semiquantitative analysis of the phosphorylation forms of KaiC. (A) KaiA, KaiB, and KaiC were incubated under the standard conditions. Aliquots of the reaction mixture were collected every 4 h, and subjected to SDS–PAGE and in-gel protease digestion followed by nano-LC/ESI-MS. The levels of the phosphorylated (pS/T: D427SHIpSTIT; S/pT: DSHISpTIP; and pS/pT: DSHIpSpTIT) and nonphosphorylated (S/T: DSHISTIT) peptides were measured. The ratio of the amount of each peptide to the total amount of all four peptides is plotted against the incubation time. (B) Aliquots of the reaction mixtures were collected at 4-h intervals, mixed with [γ-32P] ATP, and incubated for an additional 30 min. Samples were subjected to SDS–PAGE and autoradiography. After CBB staining, four bands with different mobilities were detected (upper panel). The circadian profile of phosphate incorporation was detected using autoradiography (lower panel). Phosphate was incorporated mainly into the two upper bands. (C) Densitometry of the bands shown in the upper panel of (B). The ratio of the amount of each form of KaiC to the total amount of KaiC is plotted against the incubation time. By comparing the time profiles of the bands in (B), we identified three of the KaiC bands as the pS/T, S/pT, and pS/pT forms of KaiC. (D) A schematic diagram of the KaiC phosphorylation cycle. The KaiC phosphorylation cycle can be considered to be a sequence of four reactions.
Because the ionization efficiencies of these peptides in nano-LC/ESI-MS could have been slightly modified by the phosphorylation state, the estimates of the amounts relative to each peptide may have deviated from the actual values. Thus, we tuned the composition of a polyacrylamide gel and performed long-distance electrophoresis to obtain maximum separation of the phosphorylation forms. As shown in Figure 1B, four forms of KaiC were sufficiently separated for quantitative assessment. Then, the abundance of each form in the mixture was assessed from the optical densities of the bands (Figure 1C). Because the upper three bands disappeared after λ-protein phosphatase digestion (data not shown), the lowest band was identified as the nonphosphorylated S/T form of KaiC, the relative amount of which was obtained about twice as much as that of corresponding nonphosphorylated peptide observed in ESI-MS (Figure 1A). However, by comparing the phase angle of accumulation profiles of each peptide obtained by mass spectrometry with those of each band obtained by SDS–PAGE, we identified the three phosphorylated KaiC bands from most to least mobile as the pS/T, S/pT, and pS/pT forms of KaiC. As shown in Figure 1C, we observed more S/pT form than the pS/pT and pS/T forms. The amount of the rhythmically accumulated pS/T form was almost equal to that of the pS/pT form. Unlike S/pT and pS/pT forms, a small amount of pS/T form remained even at the troughs (28 and 52 h).
The KaiC phosphorylation cycle is a sequence of four steps
To obtain insight into the nature of the phase differences in the accumulation of the three phosphorylated forms, we performed a radioactive phosphate uptake assay and examined the profile of phosphate incorporation into each form. We incubated the Kai proteins under the standard conditions and an aliquot of the reaction mixture taken every 4 h was incubated for 30 min with [γ-32P] ATP, followed by SDS–PAGE. As shown in the autoradiogram in Figure 1B, phosphate was mainly incorporated into KaiC to form the S/pT and pS/pT forms between hour 24 and 28. These observations indicated that the KaiC phosphorylation cycle proceeds following the scheme shown in Figure 1D. Phosphate was first incorporated onto T432 of the S/T form (step 1), followed by phosphorylation of the adjacent S431 residue, resulting in the formation of the pS/pT form (step 2). Subsequently, the pS/T form was generated by dephosphorylation of T432 of the pS/pT form (step 3), according to the following rationale: (i) little radioactivity was detected in the pS/T bands (Figure 1B, autoradiogram); (ii) the accumulation of the pS/T form coincided with a decrease in the level of the pS/pT form (Figure 1C); and (iii) the amount of the rhythmically accumulated pS/T form was almost equal to that of the pS/pT form (Figure 1C). Finally, the pS/T form was dephosphorylated to return to the S/T form (step 4). Some of the S/pT form may be dephosphorylated to the S/T form, because we observed more of the S/pT form than the pS/pT form.
Mutant analyses to investigate the mechanism underlying steps 1 and 2
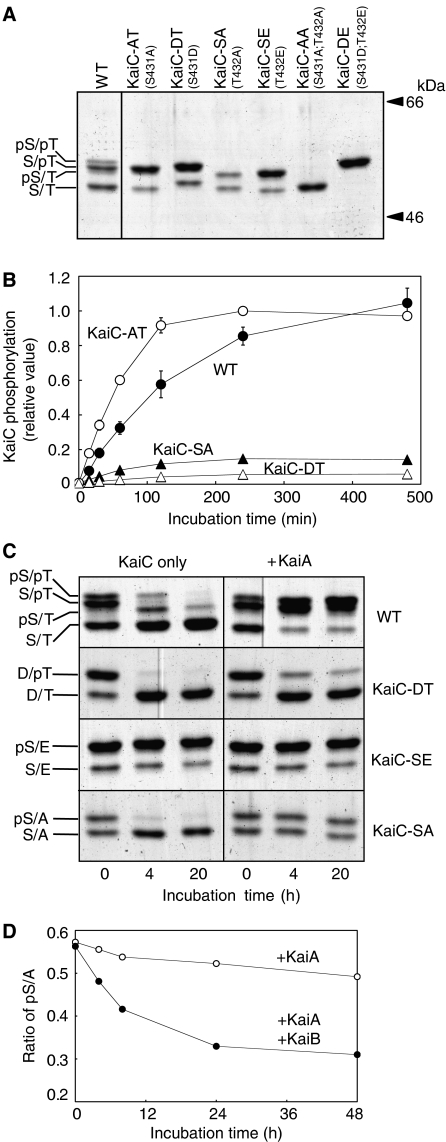
To elucidate the mechanism underlying the sequence of reactions, we prepared several mutant KaiC proteins. The phosphorylation sites were substituted for alanine (A) residues, which mimic a constitutively dephosphorylated state, or for negatively charged aspartic acid (D) or glutamic acid (E) residues to mimic the constitutively phosphorylated state. Figure 2A shows the results of SDS–PAGE of wild-type KaiC (WT-KaiC) and the mutant KaiC protein preparations. KaiC-AA and KaiC-DE yielded single bands, whereas the four mutant proteins with single amino-acid substitutions yielded two bands. The upper and lower bands for each of the single mutants corresponded to the phosphorylated and nonphosphorylated forms, respectively. We then incubated these mutant KaiC proteins under the standard conditions, and confirmed that the phosphorylation/dephosphorylation of all the mutant proteins did not show rhythmic profiles (Supplementary Figure 1).
Figure 2.
Regulation between the two KaiC phosphorylation sites. (A) WT and mutant KaiC protein preparations separated by SDS–PAGE. The mutations carried by the individual proteins are indicated in parentheses. We assigned the bands of WT-KaiC from most to least mobile as the S/T, pS/T, S/pT, and pS/pT forms. The upper and the lower bands of KaiC-AT, KaiC-DT, KaiC-SA, and KaiC-SE are corresponding to the phosphorylated and the dephosphorylated species, respectively. (B) Effect of the phosphorylation state of the adjoining site on phosphate uptake. Before the assay, WT or mutant KaiC was completely dephosphorylated by incubating the protein overnight. Dephosphorylated KaiC was incubated with KaiA and [γ-32P] ATP for the periods of time indicated on the abscissa, followed by SDS–PAGE and autoradiography. During the initial 2 h, phosphate was incorporated into each protein linearly. The maximum signal obtained with KaiC-AT was set to 1. Data were represented as mean±s.d. (n=3). (C) WT, KaiC-SE, KaiC-DT, and KaiC-SA preparations were incubated in the presence (right) or absence (left) of KaiA. Aliquots of the reaction mixtures were collected at 0, 4, and 20 h, and separated by SDS–PAGE. In the presence of KaiA, the intensities of the bands of phosphorylated WT-KaiC increased and remained elevated during the 20-h incubation. WT-KaiC was gradually dephosphorylated when it was incubated without KaiA. The band of phosphorylated KaiC-SE (pS/E) never decreased in intensity even in the absence of KaiA, whereas the amount of D/pT decreased rapidly even in the presence of KaiA. The pS/A band of KaiC-SA was maintained or disappeared in the presence or absence of KaiA, respectively. (D) Time course of pS/A dephosphorylation in the presence or absence of KaiA and KaiB. KaiB inhibited KaiA, resulting in a gradual decrease in the intensity of the pS/A band.
We examined phosphate uptake onto the intact residues (S431 or T432) of the single mutants in the presence of KaiA and [γ-32P] ATP (Figure 2B). Before the assay, WT-KaiC and mutant KaiCs were completely dephosphorylated by incubating the protein overnight in the absence of KaiA. Because the intensity of the pS/E band of KaiC-SE did not decrease even in the absence of KaiA (Figure 2C), we assayed the other three mutants. During the initial 2 h, phosphate was incorporated linearly into each protein (Figure 2B), and the initial incorporation rate into KaiC-AT was approximately eightfold faster than that into KaiC-SA. This result explains the selective phosphorylation of T432 when both sites were dephosphorylated. On the other hand, the initial rate of phosphorylation for KaiC-DT was ∼4% of that for Kai-AT, indicating that T432 is rarely phosphorylated when S431 is phosphorylated (Figure 2B). The biochemical properties of the phosphorylation of these two sites indicated that the doubly phosphorylated pS/pT form would be only generated efficiently by phosphorylation of S431 after phosphorylation of T432.
Mutant analyses of the mechanism underlying steps 3 and 4
We examined the effects of the phosphorylation state of one residue on the dephosphorylation of the other (Figure 2C). We incubated WT-KaiC and the three single mutants in the presence or absence of KaiA, and aliquots taken at 0 h, the first peak (4 h), and first trough (20 h) were subjected to SDS–PAGE, followed by Coomassie Brilliant Blue (CBB) staining. The phosphorylated forms of WT-KaiC disappeared in the absence of KaiA, whereas the presence of KaiA increased and maintained the levels of the phosphorylated forms. Each mutant protein, however, exhibited different behaviors from that of WT-KaiC. Phosphorylated T432 of KaiC-DT was dephosphorylated immediately regardless of the presence or absence of KaiA, whereas more than 95% of KaiC-SE remained in the pS/E form even in the absence of KaiA. KaiB had little effect on these processes (data not shown). These findings (Figure 2C, KaiC-DT and KaiC-SE) account for the observation that dephosphorylation of T432 occurs before that of S431 after the protein is phosphorylated at both residues (step 3).
Next, we examined the basis of step 4. The purified pS/A form of KaiC-SA was dephosphorylated rapidly in the absence of KaiA, whereas the amount of the pS/E form of KaiC-SE showed no decreases (Figure 2C). Thus, when T432 is not phosphorylated, phosphorylated S431 can be dephosphorylated, resulting in the nonphosphorylated S/T protein. On the other hand, the pS/A level was constant in the presence of KaiA (Figure 2C). As KaiB negates the activity of KaiA, thereby promoting dephosphorylation of KaiC (Kitayama et al, 2003), KaiB could inactivate any KaiA during step 4 in the reconstituted system in the presence of KaiA and KaiB. Therefore, we examined the inhibitory effects of KaiB against the ability of KaiA to dephosphorylate S431 (Figure 2D). The levels of the pS/A form decreased gradually when both KaiA and KaiB were incubated with KaiC-SA. As shown in Figure 1C, a small amount of pS/T remained even at the troughs, which may have been due to this moderate effect of KaiB on dephosphorylation of S431 in the presence of KaiA.
Taken together, the data in Figure 2 indicate that the phosphorylation states of S431 and T432 have opposite effects on the reactions at the adjacent residue. Dephosphorylated T432 allows the dephosphorylation of S431 (Figure 2C). Similarly, S431 phosphorylation can proceed efficiently after T432 phosphorylation (Figure 2B). In contrast, phosphorylated S431 allows the dephosphorylation of T432 (Figure 2C), and dephosphorylated S431 increases the rate of T432 phosphorylation (Figure 2B).
Associations among the Kai proteins in the phosphorylation cycle
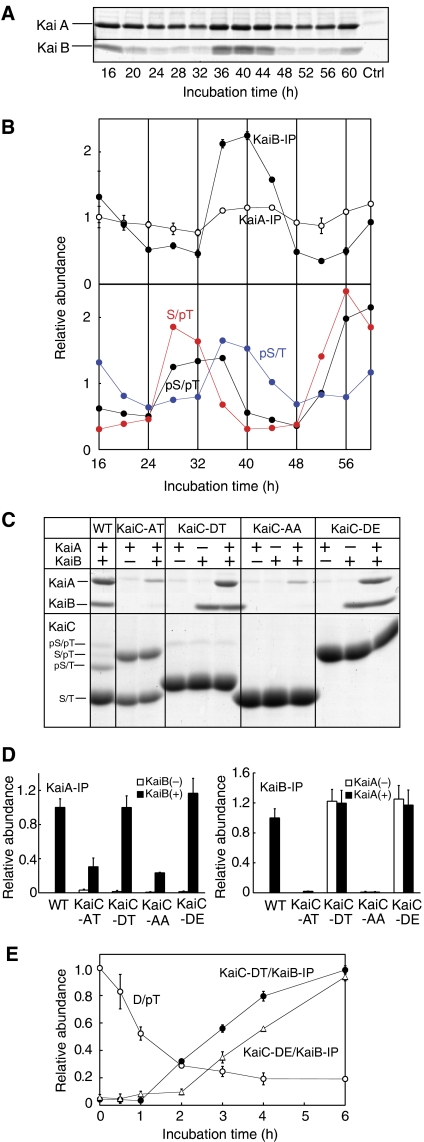
KaiA and KaiB had little effect on dephosphorylation of KaiC-DT, whereas dephosphorylation of KaiC-SA was affected by KaiA and KaiB (Figure 2C and D). These results suggested that the phosphorylation state of KaiC allowed or inhibited KaiA and KaiB to function. Therefore, we examined the relationship between associations among the Kai proteins and the phosphorylation state of KaiC. We previously demonstrated the circadian formation of a Kai protein complex and its correlation with high levels of KaiC phosphorylation (Kageyama et al, 2006). It, however, was not clear whether the phosphorylation state of KaiC regulates the formation of the Kai protein complex or vice versa. To address this issue, we examined the association profiles of KaiC with KaiA and KaiB by immunoprecipitation using KaiC bearing a FLAG tag and simultaneous monitoring of the phosphorylation state of each KaiC protein. As shown in Figure 3A, the KaiB–KaiC association showed a robust rhythm, which closely coincided with the rhythm observed for the accumulation of the pS/T form of KaiC. A weak rhythm was observed for KaiA–KaiC association, the phase of which was almost identical to the KaiB–KaiC interaction. These results raise the following possibilities: (i) the phosphorylated S431 residue in KaiC is crucial for KaiB binding; and (ii) KaiB–KaiC binding modulates the affinity of KaiC to KaiA.
Figure 3.
Relationships between the physical interactions among the KaiA, KaiB, and KaiC and KaiC phosphorylation. (A) KaiA, KaiB, and FLAG-KaiC were incubated under the standard conditions. Aliquots of the reaction mixture were collected every 4 h and subjected to immunoprecipitation using anti-FLAG M2-conjugated beads. KaiA and KaiB in the precipitates were separated by SDS–PAGE. A sample of KaiA and KaiB incubated without FLAG-KaiC was used as a control. (B) The relative amounts of KaiA, KaiB (upper panel), and each phosphorylated form of KaiC (lower panel) in the precipitates are plotted against the incubation time. The average level of each trace was normalized to 1. Data were represented as mean±s.d. (n=3 for KaiA and KaiB). (C) Binding of KaiA and KaiB to the KaiC mutants. WT and mutant FLAG-KaiC proteins were incubated in the presence or absence of KaiA and KaiB for 40 h, the time at which the maximum amounts of KaiA and KaiB bound to WT-KaiC. The samples were then subjected to immunoprecipitation. KaiA and KaiB in the precipitates were resolved by SDS–PAGE. The phosphorylation states of the KaiC proteins were examined simultaneously. (D) The relative amounts of KaiA and KaiB copurified with the FLAG-KaiC variants in the presence or absence of KaiB and KaiA, respectively. The amounts of KaiA (left) and KaiB (right) bound to WT-KaiC after a 40-h incubation under the standard conditions were assigned a value of 1. Data were represented as mean±s.d. (n=3). (E) Changes in the relative amounts of the D/pT form of KaiC-DT and the coprecipitated KaiB with respect to time. After KaiC-DT was incubated with KaiA and KaiB under the standard conditions, aliquots of the reaction were subjected to immunoprecipitation at the time points indicated on the abscissa, followed by SDS–PAGE. The time course of the interaction of KaiC-DE with KaiB was also examined. KaiB similarly associated with KaiC-DE and KaiC-DT. The relative amounts of the D/pT form of KaiC-DT at time 0 and the KaiB that coprecipitated with KaiC-DT or KaiC-DE after 40 h of incubation (shown in (C)) were assigned a value of 1. Data were represented as mean±s.d. (n=3).
S431 phosphorylation plays a key role in the interactions between the Kai proteins
To examine the role of phosphorylated S431 in the interactions among the Kai proteins and to clarify the relationship between KaiA–KaiC and KaiB–KaiC associations, we performed immunoprecipitation analysis using the phosphorylation mutants of KaiC. WT or mutant KaiC proteins bearing a FLAG tag were incubated under the standard conditions, and the KaiC proteins were precipitated with anti-FLAG antibodies at 40h, when the maximum amounts of KaiA and KaiB bound to WT-KaiC. The results confirmed that KaiB bound to KaiC-DT and KaiC-DE, but did not bind to KaiC-AT or KaiC-AA (Figure 3C and D, left panel), indicating that S431 must be phosphorylated for the KaiB–KaiC association. KaiA had no effect on KaiB–KaiC binding (Figure 3C and D, left panel).
In contrast, KaiA bound to all the KaiC mutants only in the presence of KaiB. KaiB was more efficient in promoting KaiA binding to KaiC-DT and KaiC-DE than to KaiC-AT or KaiC-AA. Taken together, these results suggest that KaiB induced the association between KaiA and KaiC, and this effect was enhanced when KaiB was bound to KaiC (Figure 3C and D, right panel). These results account for the observation in Figure 3A that KaiA bound to KaiC throughout the circadian cycle, and this binding was enhanced when KaiB interacted with KaiC, resulting in weak rhythm of KaiA–KaiC binding which is in phase with KaiB–KaiC binding.
KaiB did not associate with the pS/pT form of WT-KaiC (Figure 3B), whereas KaiB bound strongly to both KaiC-DT and KaiC-DE (Figure 3C). To explain this discrepancy, we simultaneously monitored the time courses of the association of KaiB with KaiC-DT and the dephosphorylation of the D/pT form of KaiC. T432 in the D/pT form was rapidly dephosphorylated within 3 h, whereas the amount of KaiB that was associated with KaiC-DT continued to increase gradually for 6 h (Figure 3E). These results explain why KaiB–KaiC binding coincided with the accumulation of only pS/T form. When the pS/pT form is generated, dephosphorylation of T432 occurs regardless of the presence of KaiB (Figure 2C and data not shown). KaiB–KaiC interaction occurs more slowly than dephosphorylation of T432 in the pS/pT form; conversion of the pS/pT to the pS/T form will have already been completed by the time KaiB–KaiC binding reaches the maximum level. Thus, KaiB appears to bind only with the pS/T form.
In contrast to S431, T432 phosphorylation appeared to have no effect on the Kai protein interactions, because KaiC-AA (a nonphosphorylatable mutant) and KaiC-AT (∼50% was phosphorylated after 40 h of incubation) produced similar results in the immunoprecipitation assays (Figure 3C). As seen in Figure 3E, the association between KaiB and KaiC-DE produced a binding profile that was similar to that of the association between KaiB and KaiC-DT, which suggests that S431 phosphorylation is crucial for the KaiB–KaiC association, whereas T432 phosphorylation has little effect. Based on the results shown in Figure 3, we concluded that S431-phosphorylated KaiC recruits KaiB and resulting KaiB–KaiC complex then binds KaiA.
Phosphate uptake occurred in the ∼440-kDa KaiC homohexamer fraction
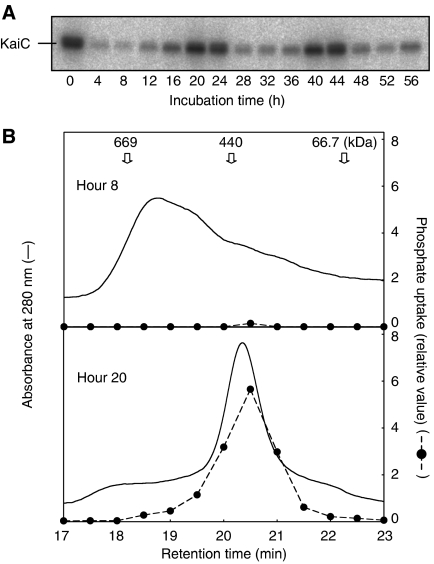
What is the role of the interactions among the Kai proteins that are induced by S431 phosphorylation? Because KaiA promotes KaiC phosphorylation and KaiB negates this KaiA activity (Kitayama et al, 2003), the interactions among the Kai proteins are likely to regulate phosphate incorporation into KaiC. Based on the molar ratio of the Kai proteins both in vivo and in vitro, we previously presented models in which KaiA enhances phosphorylation of KaiC by repeated weak association with KaiC. On the other hand, KaiB–KaiC complex binds strongly to KaiA to prevent the incorporation of phosphate (Kitayama et al, 2003, Kageyama et al, 2006). To test this model experimentally, we examined the size of the Kai protein complex into which phosphate was incorporated. We incubated KaiA, KaiB, and KaiC with ATP, and aliquots of the reaction mixtures were incubated for 30 min with [γ-32P] ATP every 4 h. To confirm the rhythmic phosphorylation of KaiC, the samples were subjected to SDS–PAGE followed by autoradiography. As shown in the autoradiogram in Figure 4A, we observed robust phosphate uptake rhythm.
Figure 4.
Phosphate was incorporated into the ∼440 kDa fraction, which contained the KaiC homohexamer. (A) KaiA, KaiB, and KaiC were incubated at pH 7.5. Aliquots of the reaction mixture were collected every 4 h, [γ-32P] ATP was added, and incubation was continued for an additional 30 min. Samples were subjected to SDS–PAGE followed by autoradiography. Under these conditions, the period of the phosphorylation cycle was ∼20 h. (B) The samples were separated by gel filtration chromatography at 8 and 20 h, which corresponded to first trough and second peak, respectively. Proteins were monitored by absorbance at 280 nm. Fractions were collected and subjected to SDS–PAGE, followed by autoradiography to determine the size of the protein complex in which the autophosphorylation reaction took place. Thyroglobulin (669 kDa), Ferritin (440 kDa), and albumin (67 kDa) were used as standards. After 20 h of incubation, a single ∼440-kDa (homohexamer) peak was predominant. At 8 h, the proteins were eluted as >440-kDa complex. Radioactivity was eluted only with the 440-kDa fraction. After 20 h of incubation, high levels of 32P were detected in the fractions corresponding to the ∼440-kDa peak, whereas little phosphate was incorporated after 8 h of incubation. As we used a silica-based column in this experiment, which is unstable above pH 8.0, reactions and subsequent separations at pH 7.5.
Samples were analyzed using gel filtration chromatography and each fraction was further subjected to SDS–PAGE and autoradiography. The results obtained at 8 and 20 h, corresponding to the first trough and the second peak in the circadian phosphate incorporation profile, respectively (Figure 4A), were then compared. At 20 h, a single peak corresponding to a ∼440 kDa KaiC homohexamer (Nishiwaki et al, 2004) was predominant and high levels of radioactivity were detected in this peak, suggesting that KaiA–KaiC binding was too weak to be detected by gel filtration chromatography. At 8 h, the major peak corresponded to a larger protein complex (>440 kDa), and no radioactivity was detected in this peak. A trace amount of radioactivity was detected in the fraction corresponding to ∼440 kDa. These results support our previous models based on the molar ratio of KaiA, KaiB, and KaiC (Kitayama et al, 2003, Kageyama et al, 2006). KaiA enhanced the autokinase activity of KaiC by repeated weak association and dissociation with KaiC without forming stable complex. In the dephosphorylation phase, the large protein complex trapped KaiA and prevented repeated transient KaiA–KaiC interactions, resulting in inhibition of phosphate incorporation into KaiC.
Discussion
Phosphorylation state regulates autokinase and autophosphatase activities of KaiC
In this study, we succeeded in separately analyzing the profiles of S431 and T432 phosphorylation in KaiC (Figure 1A–C), and found that the KaiC phosphorylation cycle was a sequence of four steps. To clarify the mechanisms underlying these sequential reactions, we mutated KaiC at the phosphorylation sites. Using these mutants, we revealed that the phosphorylation state of each of these two residues regulates the phosphorylation/dephosphorylation of the other residue.
As illustrated in Figure 5, it was suggested that the product of step 1 (phosphorylated T432) allowed the same type of reaction in step 2 (phosphorylation of S431), as S431 phosphorylation can proceed efficiently after T432 phosphorylation (Figure 2B). Similarly, in steps 3 and 4, dephosphorylated T432 enabled dephosphorylation of S431, resulting in the completely dephosphorylated S/T form (Figure 2C). Thus, the phosphorylation state of T432 is crucial for the formation of the double-phosphorylated and double-dephosphorylated (nonphosphorylated) forms during the reaction cycle.
Figure 5.
A model for the basis of the KaiC phosphorylation cycle. The KaiC phosphorylation cycle consists of four sequential reactions. During steps 1 and 2, KaiC has an autokinase activity and during steps 3 and 4, KaiC serves as an autophosphatase. Phosphorylation of S431 to create the double-phosphorylated form (pS/pT) switches KaiC from an autokinase to an autophosphatase, whereas dephosphorylation of S431 to form the completely dephosphorylated form (S/T), switches KaiC from a phosphatase to a kinase. Phosphorylation of T432 activates phosphorylation of S431. Likewise, dephosphorylation of S431 activates dephosphorylation of T432.
Importantly, changes in the phosphorylation state of S431 resulted in switching the KaiC activity between an autokinase activity and an autophosphatase activity (Figure 5). The switching from the autokinase activity to the autophosphatase activity occurred between steps 2 and 3; the product of the former step (phosphorylated S431) promoted the opposite reaction in the latter step (dephosphorylation of T432). In the same way, the switching from the autophosphatase activity to the autokinase activity took place between steps 4 and 1. This explains why both S431 and T432 are necessary for the in vitro oscillation of KaiC phosphorylation. When S431 is not phosphorylated, KaiC can serve as kinase, whereas, when S431 is phosphorylated, KaiC functions as autophosphatase.
Effect of the phosphorylation states of S431 and T432 on the structure of KaiC
How does the phosphorylation state of one residue regulate the phosphorylation/dephosphorylation of the other residue? Pattanayek et al (2004) determined the crystal structure of KaiC and found that ATP molecules bound at the interfaces between the monomer subunits. The phosphorylation sites on the adjacent monomer were within 10 Å of the ATP γ-phosphate (Xu et al, 2004). These facts indicate that the autophosphorylation reaction takes place at the subunit interfaces. Thus, it is possible that phosphorylation of S431 and T432 affects the structure at the subunit interfaces in the KaiC hexamer to regulate the activity of KaiC.
Xu et al (2004) reported that phosphorylation of T432 results in additional contacts at the subunit interfaces; the phosphate group on T432 forms a hydrogen bond with the catalytic carboxylate (E318) from the adjacent subunit. Catalytic carboxylates in ATPases are known to activate water molecules that then attack the ATP γ-phosphate (Story et al, 1992). Therefore, it is expected that E318 activates hydroxyl groups at the phosphorylation sites of KaiC, resulting in the transfer of ATP γ-phosphates to these residues. Here, we demonstrated that phosphorylated T432 allowed the phosphorylation of S431 (step 2). Newly established hydrogen bonding between phosphorylated T432 and E318 might affect the orientation of E318 to facilitate S431 phosphorylation. Phosphorylation of T432 also results in intersubunit contacts with other residues (summarized in Xu et al, 2004). Although the functions of these residues are not clear, phosphorylated T432 might regulate the phosphorylation of S431 by tightening the intersubunit associations within the KaiC hexamer.
In contrast to T432, the contributions of phosphorylated S431 to the intersubunit contacts are relatively small (Xu et al, 2004), although the phosphate group of S431 forms hydrogen bonds with H429 and T426 in the same subunit (Xu et al, 2004). Because the phosphorylation state of S431 determines whether KaiC functions as an autokinase or an autophosphatase (Figure 5), it is possible that these additional intramolecular bonds switch KaiC to the autophosphatase activity.
KaiA and KaiB interact with KaiC to maintain the high-amplitude rhythm of the phosphorylation cycle
Last year, we reported the time-dependent protein complex formation including KaiA, KaiB, and KaiC in vitro (Kageyama et al, 2006). More recently, Mori et al (2007) provided structural evidence for this observation using electron microscopy. However, whether the phosphorylation cycle drives the protein associations, or vice versa was unclear. In the present study, we showed that interactions among the Kai proteins occur as a result of phosphorylation of S431 (Figure 3).
We found that KaiA and KaiB had little effect on dephosphorylation of KaiC-DT or KaiC-SE (Figure 2C and data not shown), suggesting that switching of KaiC from an autokinase to an autophosphatase is predominantly regulated by the phosphorylation state of KaiC (step 3). Furthermore, our previous radioactive phosphate uptake assay showed that a small amount of phosphate was incorporated into KaiC even when KaiA was not included in the reaction (Iwasaki et al, 2002, Kitayama et al, 2003). We also confirmed that the ratio of phosphorylated serine and threonine was not altered by the presence of KaiA, whereas KaiA increased the total amount of phosphorylation of both residues (Iwasaki et al, 2002). These results imply that steps 1 and 2 can proceed without KaiA. In addition, dephosphorylation of S431 (step 4) proceeded efficiently in the absence of KaiA and KaiB (Figure 2C and D). Taken together, these observations indicate that KaiC has the potential to progress through all the steps of the phosphorylation cycle without KaiA and KaiB.
However, robust circadian oscillations were not observed when KaiC was incubated in the absence of KaiA and KaiB (Kageyama et al, 2006). It is possible that in the absence of KaiA only a small fraction of the KaiC subunits in the system start the phosphorylation cycle, which may be difficult to detect with CBB staining. KaiA may be responsible for generating the high-amplitude rhythm by increasing the number of KaiC subunits into which phosphate groups are incorporated by weak interaction with KaiC.
If each KaiC molecule is phosphorylated and starts the phosphorylation cycle ad libitum, the overall rhythmicity would disappear. This, however, is not the case. In fact, phosphate uptake to KaiC was rhythmic and limited to time-windows of 20–32 or 44–56 h (Figure 1B). We assume that KaiB could be responsible for synchronizing the phosphate incorporation into the KaiC subunits. As shown in Figures 3 and 4, association between KaiB and S431-phosphorylated KaiC could inhibit aberrant phosphorylation by trapping KaiA to decrease the concentration of active KaiA in the system. Furthermore, the phosphorylatable forms coexisting with the S431-phosphorylated KaiC subunit in the same hexamer should be integrated into the high-molecular-weight Kai protein complex that could not incorporate phosphate (see Figure 4). As the dephosphorylation of S431 proceeds, KaiA and the KaiC hexamer are released from the Kai protein complex and the next cycle of phosphate uptake proceeds.
Multisite phosphorylation may play important roles in circadian rhythm generation
In this study, we found that the phosphorylation states of S431 and T432 regulate the phosphorylation/dephosphorylation reactions at the adjacent residue, which is the basis for the phosphorylation cycle. Recent computational studies have shown that a multisite phosphorylation mechanism, such as the one found in the MAPK cascade, can theoretically generate oscillations (Chickarmane et al, 2007). Several eukaryotic clock proteins are also phosphorylated at multiple sites (Gallego and Virshup, 2006). In eukaryotic circadian systems, transcription/translation-derived oscillatory processes based on the negative feedback regulation of clock genes have been proposed as the core generators of the oscillations (Dunlap et al, 2004). Multisite phosphorylation of eukaryotic clock proteins, however, may also be involved in the generation of circadian rhythms. In mouse cryptochrome 2, phosphorylation of S557 allows subsequent phosphorylation at S553, resulting in degradation by ubiquitin–proteasome pathway (Harada et al, 2005). Twenty-one phosphorylated residues of mouse period 2 (mPER2) have been identified (Vanselow et al, 2006). Phosphorylation at S659 and downstream serine and threonine residues results in stabilization of mPER2, whereas phosphorylation at other sites leads to mPER2 degradation (Vanselow et al, 2006). Thus, phosphorylation cycles of eukaryotic clock proteins would be generated by alternating phosphorylation and degradation processes. The latter process may serve as an alternative process to the dephosphorylation of KaiC, because it could decrease the amount of the phosphorylated species.
Materials and methods
Bacterial strains
We used Escherichia coli DH5α and BL21 cells as hosts for plasmid construction and expression of glutathione S-transferase (GST) fusion proteins, respectively.
Site-directed mutagenesis
The kaiC ORF cloned into the BamHI and NotI sites of pGEX6P-1 (GE Healthcare) (Iwasaki et al, 2002) was mutagenized using the overlap extension method and PCRs as described previously (Nishiwaki et al, 2000) to obtain constructs for expression of GST–KaiC fusion proteins containing the following mutations: Ser431 to aspartic acid (S431D); Thr432 to glutamic acid (T432E); and Ser431 to aspartic acid and Thr432 to glutamic acid (S431D;T432E).
The primers used to introduce the S431D, T432E, and S431D;T432E mutations were 5′-ATTACTGACTCCCATATCGATACAATT-3′ and 5′-AATTGTATCGATATGGGAGTCAGTAAT-3′; 5′-ATTACTGACTCCCATATCTCAGAAATT-3′ and 5′-AATTTCTGAGATATGGGAGTCAGTAAT-3′; and 5′-ATTACTGACTCCCATATCGATGAAATT-3′ and 5′-AATTTCATCGATATGGGAGTCAGTAAT-3′, respectively. The primers used to introduce the S431A, T432A, and S431A;T432A mutations were described by Nishiwaki et al (2004). We named the proteins containing the S431A, S431D, T432A, T432E, S431A;T432A, and S431D;T432E mutations as KaiC-SA, KaiC-DT, KaiC-AT, KaiC-SE, KaiC-AA, and KaiC-DE, respectively.
The expression construct for KaiC tagged with the FLAG epitope at its C terminus (Kageyama et al, 2006) was subjected to site-directed mutagenesis using a QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. Primers used in the PCRs were the same as those described above.
Bacterial expression and purification of the Kai proteins
Recombinant KaiA protein was produced in E. coli and purified as described previously (Nishiwaki et al, 2004). Recombinant KaiB protein was expressed in E coli as described previously (Kitayama et al, 2003). Cells expressing KaiB were collected, resuspended in buffer Z (20 mM Tris at pH 8.0, 10 mM NaCl, and 0.5 mM EDTA), and homogenized by sonication. The homogenate was centrifuged at 24 000 × g, and the supernatant was applied to a glutathione Sepharose 4B column (GE Healthcare). After washing the column with 20 column volumes of buffer Z, the GST–KaiB fusion protein was eluted with three column volumes of glutathione solution (50 mM Tris at pH 8.0, 10 mM NaCl, and 20 mM reduced glutathione). After removing glutathione using an Econo-Pac 10DG column (BioRad), GST–KaiB was subjected to PreScission protease digestion (GE Healthcare) at a concentration of 20 U/mg of protein in the presence of 1 mM DTT overnight at 4°C. The digestion reaction was applied to a Resource Q column (GE Healthcare). KaiB was eluted from the column with a 0–300 mM NaCl gradient. After the NaCl concentration of the fractions containing KaiB was adjusted to 300 mM, KaiB was further purified using a Superdex 75 column (GE Healthcare). Recombinant KaiC protein was expressed and purified as described previously (Nishiwaki et al, 2004) with some modifications. The eluate from the glutathione Sepharose 4B column was first applied to a HiPrep Sephacryl S300 column (GE Healthcare), and the fractions containing KaiC were collected and applied to a Resource Q column. KaiC was then eluted with a 90–450 mM NaCl gradient. Protein concentrations were determined by the Bradford method using the Quick Start Bradford dye reagent (BioRad). The purity of the proteins was >95% as determined by SDS–PAGE.
In vitro assay of KaiC phosphorylation
The reconstitution of the KaiC phosphorylation cycle was performed as described previously (Kageyama et al, 2006). Briefly, KaiA, KaiB, and KaiC were incubated in buffer A (20 mM Tris at pH 8.0, 150 mM NaCl, 5 mM MgCl2, and 1 mM ATP) at 30°C. The final concentrations of KaiA, KaiB, and KaiC were 1.2, 3.5, and 3.5 μM, respectively. The conditions described here are referred to as the ‘standard conditions' in this study.
Nanoflow liquid chromatography/electrospray ionization mass spectrometry
The digested protein samples were separated using the Ultimate nano-LC system (Dionex) running at a column flow rate of 200 nl/min on a 75-μm i.d. −150-mm column (C18-PepMap; Dionex). The peptides were eluted using a linear gradient (solution A: 0.05% formic acid/water; solution B: 0.05% formic acid/acetonitrile), which started at 5% solution B and rose to 20% solution B over 80 min. The column effluent was introduced continuously into the nanoflow electrospray ion source of a quadrupole/time-of-flight mass spectrometer (Micromass). The phosphorylated (D427SHIpSTIT, DSHISpTIT, and DSHIpSpTIT) and nonphosphorylated (DSHISTIT) peptides were identified in the tandem mass spectrometry mode as described previously (Nishiwaki et al, 2004). The relative amounts of the peptides were roughly estimated in the positive-ion mode of ESI-MS from the integrated peak areas of the corresponding doubly protonated ions (Figure 1A), which gave approximately the same ionization efficiencies, irrespective of the sites or degree of phosphorylation (Satomi et al, 2005).
Separation of KaiA, KaiB, and each phosphorylated form of KaiC by SDS–PAGE
Kai proteins were incubated under the standard conditions. At each time point, an aliquot of the reaction mixture was mixed with an equal volume of 2 × SDS sample buffer (125 mM Tris at pH 6.8, 4% SDS, and 20% glycerol). To separate the phosphorylated forms of KaiC, samples were subjected to SDS–PAGE on a gel of 11% T with 0.67% C using an EPL156DA (Advantec) or a NA-1012L (Nihon Eido) electrophoresis apparatus, followed by CBB staining using Quick-CBB PLUS (Wako). To resolve KaiA and KaiB, we used a gel of 13% T with 2.7% C. For densitometric analysis, we used a GS-800 calibrated densitometer (BioRad) and ImageJ 1.33u software (National Institutes of Health, USA).
Phosphate uptake analysis
Kai proteins were incubated under the standard conditions. At each time point, an aliquot of the mixture was mixed with [γ-32P] ATP (12.3 kBq/μl, 12.3 GBq/mmol) and incubated for an additional 30 min at 30°C. The reaction was stopped by adding 2 × SDS sample buffer. To examine the circadian profile of phosphate uptake, the samples were separated by SDS–PAGE (11% T with 0.67% C), stained with CBB, and subjected to autoradiography using a BAS2000 image analyzer (Fuji). To compare the phosphate incorporation into WT and mutant KaiC, KaiC proteins were incubated without KaiA and KaiB for ∼16 h to completely dephosphorylate the proteins before the start of the assays. KaiA, KaiB, and [γ-32P] ATP were then added to the reaction mixtures at time 0. The samples were separated by SDS–PAGE, transferred onto Immobilon-P membranes (Millipore), and detected using autoradiography.
Immunoprecipitation
KaiA, KaiB, and KaiC with the FLAG epitope tag (KaiC-FLAG) were incubated under the standard conditions in the presence of 2 mg/ml BSA. At each time point, a 135-μl aliquot of the reaction mixture was mixed with 540 μl of Buffer B (20 mM Tris at pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM ATP, and 2 mg/ml BSA) and a 30-μl bed volume of prewashed anti-FLAG M2 Affinity Gel freezer-safe beads (Sigma), followed by incubation for 30 min at room temperature with gentle agitation. Then, the beads were washed five times with 1 ml of Buffer B. Trapped proteins were eluted twice by the addition of 35 μl of 2 × SDS sample buffer in the absence of 2-mercaptoethanol. For detection of KaiA and KaiB, samples were separated by SDS–PAGE (13% T with 2.7% C).
High-performance gel filtration chromatography
KaiA (4.8. μM), KaiB (14 μM), and KaiC (14 μM) were incubated in buffer C (20 mM Tris at pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 1 mM ATP) at 30°C. At each time point, a 20-μl aliquot of the reaction was mixed with [γ-32P] ATP (12.3 kBq/μl, 12.3 GBq/mmol) and incubated for an additional 30 min at 30°C. Then, the proteins were size-fractionated at 30°C using a Lachrom D-7000 system (Hitachi) and a TSKgel G4000SWXL (Tosoh Bioscience) equilibrated with buffer C. Proteins were monitored by absorbance at 280 nm. Thyroblobulin (669 kDa), ferritin (440 kDa) and albumin (67 kDa) were used as standards. The fractionated proteins were precipitated with acetone, dissolved in SDS sample buffer, and subjected to SDS–PAGE, followed by autoradiography. As we used a silica-based column in this experiment, which is unstable above pH 8.0, we performed reactions and subsequent separations at pH 7.5.
Supplementary Material
Supplementary Figure 1
Acknowledgments
We thank H Kondo and M Tamura for excellent technical assistance, and Dr T Oyama for helpful discussion. This research was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (15GS0308 to TK, 15GS0320 to TT). YK was supported by a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (16010326). Analysis of DNA sequences was conducted in conjunction with the Life Research Support Center at Akita Prefectural University.
References
- Chickarmane V, Kholodenko BN, Sauro HM (2007) Oscillatory dynamics arising from competitive inhibition and multisite phosphorylation. J Theor Biol 244: 68–76 [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, DeCoursey PJ (2004) Chronobiology: Biological Timekeeping. Sunderland, MA, USA: Sinauer Associates Inc [Google Scholar]
- Gallego M, Virshup PM (2006) Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8: 139–148 [DOI] [PubMed] [Google Scholar]
- Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y (2005) Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3β. J Biol Chem 280: 31714–31721 [DOI] [PubMed] [Google Scholar]
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T (1998) Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281: 1519–1523 [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T (2002) KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacterial. Proc Natl Acad Sci USA 99: 15788–15793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama H, Nishiwaki T, Nakajima M, Iwasaki H, Oyama T, Kondo T (2006) Cyanobacterial circadian pacemaker: Kai protein complex dynamix in the KaiC phosphorylation cycle in vitro. Mol Cell 23: 161–171 [DOI] [PubMed] [Google Scholar]
- Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T (2003) KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J 22: 2127–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Williams DR, Byrne MO, Qin X, Egli M, McHaourab HS, Stewart PL, Johnson CH (2007) Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol 5: e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308: 414–415 [DOI] [PubMed] [Google Scholar]
- Nishiwaki T, Iwasaki H, Ishiura M, Kondo T (2000) Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci USA 97: 495–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Nakajima M, Lee C, Kiyohara R, Kageyama H, Kitayama Y, Temamoto M, Yamaguchi A, Hijikata A, Go M, Iwasaki H, Takao T, Kondo T (2004) Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC7942. Proc Natl Acad Sci USA 101: 13927–13932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M (2004) Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell 15: 375–388 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS (1993) Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol 55: 16–54 [DOI] [PubMed] [Google Scholar]
- Satomi Y, Nishiwaki T, Nakajima M, Kiyohara R, Kageyama H, Kitayama Y, Temamoto M, Yamaguchi A, Hijikata A, Go M, Iwasaki H, Kondo T, Takao T (2005) Quantitative analysis of a singly and doubly phosphorylated peptides in the circadian clock-related protein (KaiC) by nano-flow LC/ESI-MS. Proceedings of the 53rd ASMS Conference on Mass Spectrometry & Allied Topics, San Antonio, Texas, June 5–9, 2005 [Google Scholar]
- Story RM, Weber IT, Steitz TA (1992) The structure of the E. coli recA protein monomer and polymer. Nature 355: 318–325 [DOI] [PubMed] [Google Scholar]
- Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A (2006) Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev 20: 2660–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SB, Vakonakis I, Golden SS, LiWang AC (2002) Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc Natl Acad Sci USA 99: 15357–15362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Johnson CH (2003) Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J 22: 2117–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Pattanayek R, Pattanayek S, Egli M, Johnson CH (2004) Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA 101: 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1