Abstract
Amyloid aggregates of the amyloid-β (Aβ) peptide are implicated in the pathology of Alzheimer's disease. Anti-Aβ monoclonal antibodies (mAbs) have been shown to reduce amyloid plaques in vitro and in animal studies. Consequently, passive immunization is being considered for treating Alzheimer's, and anti-Aβ mAbs are now in phase II trials. We report the isolation of two mAbs (PFA1 and PFA2) that recognize Aβ monomers, protofibrils, and fibrils and the structures of their antigen binding fragments (Fabs) in complex with the Aβ(1–8) peptide DAEFRHDS. The immunodominant EFRHD sequence forms salt bridges, hydrogen bonds, and hydrophobic contacts, including interactions with a striking WWDDD motif of the antigen binding fragments. We also show that a similar sequence (AKFRHD) derived from the human protein GRIP1 is able to cross-react with both PFA1 and PFA2 and, when cocrystallized with PFA1, binds in an identical conformation to Aβ(1–8). Because such cross-reactivity has implications for potential side effects of immunotherapy, our structures provide a template for designing derivative mAbs that target Aβ with improved specificity and higher affinity.
Keywords: amyloid, crystal structure, EFRH, monoclonal antibody, EFRHD
Normally soluble proteins and peptides can sometimes aggregate into insoluble, self-assembled filamentous aggregates, including amyloid and amyloid-like structures. Current interest in amyloid fibrils and related aggregates arises from their involvement in diseases such as Alzheimer's disease (AD), type 2 diabetes, prion diseases, and other protein misfolding disorders (1). Because aggregates of the amyloid-β (Aβ) peptide have been implicated in the molecular mechanism of AD, reversing or preventing Aβ aggregation is an important prospective approach to AD therapy. Antibodies capable of binding monomeric/low molecular weight forms of Aβ (in the periphery) or aggregated states of Aβ (in the brain) reduce the amyloid burden in animal studies (2). Passive immunization with humanized anti-Aβ monoclonal antibodies (mAbs) (3) is in phase II trials as an Alzheimer's therapy (www.elan.com/research_development/Alzheimers/). Because the EFRHD motif (Aβ residues 3–7) appears to be an immunodominant segment of Aβ (4, 5) and immunization to the EFRH sequence rescues cognitive function in mouse models of AD (6, 7), it is important to understand the structural basis of antibody recognition of this sequence.
At least three mechanisms for immunotherapy of AD have been proposed. Two mechanisms, microglial activation (8, 9) and catalytic dissolution (10), require that the antibody enter the central nervous system; the other mechanism, sometimes called the peripheral sink hypothesis, does not. In the microglial activation mechanism (8), antibodies bind to amyloid plaques, triggering microglia via their Fc receptors. As evidence for this mechanism, Aβ(1–42) immunization elicits synthesis of anti-Aβ antibodies and causes Aβ-reactive microglial cells to appear around plaques (8). In catalytic dissolution, antibodies are hypothesized to act as chaperones, catalyzing the structural change of the Aβ peptide from the β-strand to an alternative conformation less prone to aggregation (10). Consistent with this mechanism, the efficacy of a given mAb depends on the Aβ sequence element it binds; thus, the mAb 6C6, which recognizes the Aβ N terminus, is three times more effective in disaggregating Aβ fibrils than the mAb 1C2, which binds to the central residues of Aβ (10). In the peripheral sink hypothesis (3), antibodies bind to Aβ in the bloodstream, shifting the distribution of Aβ between the brain and the peripheral circulatory system and thereby leading to a net efflux of Aβ from the central nervous system to plasma, where it is degraded. Thus, DeMattos et al. (3) observed a 1,000-fold increase in the plasma Aβ levels in Aβ-overproducing transgenic mice after peripheral administration of m266, an mAb directed against the central domain of Aβ.
All three mechanisms require antibody binding to Aβ in either monomer or aggregated forms. Because it remains unclear which of these mechanisms might ultimately make contributions to AD therapy, the most appropriate antibodies are those equally capable of recognizing all assembly forms of Aβ peptides. Although anti-Aβ mAbs have been described [10D5 (11); 3D6 and 16C11 (9); m266 (12); NAB61 (13); and Ab42.2, Ab40.1, and Ab9 (14)], neither their structural details nor their specificities for different forms of Aβ have been reported. Thus, the structural basis of Aβ binding, which lies at the heart of all three mechanisms, remains unknown.
When immunizing mice to generate antibodies against Aβ, one may choose from several different states of Aβ: monomers (11, 12, 14, 15), oligomers (13), protofibrils, or fibrils (14, 16). Transient, metastable, spherical oligomers, and protofibrils are observed during the lag phase (an incubation period of up to several days preceding amyloid fibril growth phase) of spontaneous Aβ fibril growth in vitro from high concentrations of Aβ and, indeed, during amyloid growth by many peptides and proteins (17). Because the current wisdom is that spherical oligomers and/or protofibrils are the most important toxic species in AD and other amyloid-associated neurodegenerative diseases (18), the prevailing controversy about whether these intermediates are on- or off-pathway in amyloid formation (17) does not impact the validity of targeting these species therapeutically. Because isolated Aβ(1–40) protofibrils can, depending on conditions, either dissociate (19) or progress to mature fibrils, their use as immunogens is problematic. Protofibrils can be stabilized, however, by the small molecule calmidazolium (CLC), which allows the preparation of stable protofibrils for structural studies and for use as an antigen. These CLC-stabilized protofibrils (i) exhibit EM images and hydrogen–deuterium exchange kinetics essentially identical to normal Aβ protofibrils, (ii) share with protofibrillar assembly intermediates a poor ability to seed elongation of the monomer, and (iii) develop into amyloid fibrils, in analogy to normal protofibrils, but at a much slower rate than the latter (20).
We report here the isolation of two murine IgG2a mAbs, anti-protofibril antibodies (PFAs) PFA1 and PFA2, from mice challenged with a CLC-stabilized protofibril form of Aβ(1–40) and the structures of their antigen binding fragments (Fabs) in complex with the Aβ(1–8) peptide DAEFRHDS and with the AKFRHD peptide from the human glutamate receptor interacting protein, GRIP1 (21). The Fab fragments exhibit binding to Aβ monomers in the 20–40 nM range, and this binding is significantly impaired or eliminated in Aβ(1–40) mutants in which a single residue in the 3–7 segment is replaced with alanine. The full IgG molecules bind significantly better than Fab fragments to Aβ fibrils and protofibrils (0.1–0.2 nM), apparently because of an avidity effect. Our structures reveal that binding of both mAbs to the EFRHD sequence is mediated by a combination of salt bridges, hydrogen bonds, and hydrophobic contacts, including, in both cases, interactions with a striking WWDDD motif in the second heavy-chain (H) complementarity determining region (CDR) (CDR-H2). These structures provide a molecular basis for Aβ recognition that can be exploited in the design of derivative mAbs of enhanced affinity and specificity (22, 23).
Results
Antibody Isolation and Screening.
To isolate aggregate-specific mAbs (16), we injected mice with CLC-stabilized Aβ(1–40) protofibrils (20) and screened candidate hybridoma clones for persistent binding to protofibrils in the presence of large excesses of monomeric Aβ. A number of unique IgG and IgM mAbs, all capable of binding to Aβ protofibrils, were isolated. Two of the IgGs, PFA1 and PFA2, are described here. The mAbs were screened for protofibril affinity in competition assays with Aβ(1–40) monomers. PFA1 and PFA2 in IgG form both bind CLC-stabilized Aβ(1–40) protofibrils with EC50 (measured by ELISA) 0.15 nM. Binding to mature Aβ(1–40) amyloid fibrils is similar at 0.07 nM and 0.2 nM, respectively.
Surface Plasmon Resonance (SPR) Demonstrates Avidity Effects and Elucidates the Epitope.
SPR analysis for IgG binding to fibrils and protofibrils yields results comparable with those from ELISA (Table 1). ELISAs show that both mAbs bind Aβ(1–40) protofibrils equally well (Table 1), with EC50 values of 0.15–0.16 nM. ELISA indicates that PFA1 binds more tightly to fibrils than does PFA2 (0.07 vs. 0.2 nM), and SPR confirms this: the Kd values are 0.018 nM and 0.15 nM, respectively.
Table 1.
ELISA EC50 and SPR Kd values
| Antibody binding to Aβ(1–40) | Protofibrils, IgG ELISA EC50, nM | Fibrils |
Monomer, Fab SPR Kd, nM | ||
|---|---|---|---|---|---|
| IgG ELISA EC50, nM | IgG SPR Kd, nM | Fab SPR Kd, nM | |||
| PFA1 | 0.15 ± 0.12 | 0.07 ± 0.01 | 0.018 | 60 ± 10 | 39.0 ± 0.1; 21 ± 7 (IgG) |
| PFA2 | 0.16 ± 0.09 | 0.2 ± 0.2 | 0.150 ± 0.050 | 35 ± 1 | 25 ± 4 |
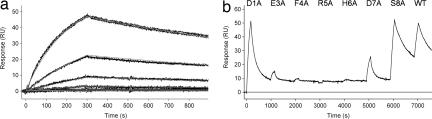
SPR experiments also revealed that the Fab fragments of PFA1 and PFA2 are capable of binding Aβ(1–40) monomers, with affinities of 39 ± 0.1 nM and 25 ± 4 nM, respectively (Fig. 1a and Table 1). This result indicates that avidity (5, 24) plays an important role in the binding of the bivalent IgGs to multivalent fibrils. Although the SPR Kd values are 0.018 nM (PFA1) and 0.15 nM (PFA2) for the polydentate binding of IgGs to fibrils, the corresponding Kd values for Fabs are 60 and 35 nM, representing decreased binding by 100- to 1,000-fold. These values closely match the Kd values for Fab binding to monomer: 39.0 nM (PFA1) and 25 nM (PFA2). The significantly enhanced binding of the IgG forms of PFA1 and PFA2 to Aβ aggregates, compared with the monovalent interaction with Aβ, may explain why competition with Aβ monomers was ineffective at eliminating these linear epitope mAbs during the hybridoma screening process.
Fig. 1.
SPR reveals Aβ peptide binding to PFA1 via the EFRHD epitope. (a) Kinetic analysis of Aβ(1–40) (WT) monomer at 0, 1.23, 3.70, 11.1, 33.3, 100, and 300 nM, binding to PFA1 Fab immobilized at densities of 2,720 response units (RU) (shown) and 1,280 RU (data not shown). Duplicate binding responses for each monomer concentration are overlaid with the global fit of a simple 1:1 interaction model (smooth lines), which yielded ka = (1.431 ± 0.003) × 104 M−1 s−1, kd = (5.58 ± 0.01) × 10−4 s−1, and Kd = 39.0 ± 0.1 nM. (b) WT and Ala-substituted mutants of Aβ(1–40) monomers were sequentially flowed over PFA1 IgG captured on anti-IgG flow cell surfaces (SPR). Significant RU peaks show good peptide binding, whereas the absence of a peak shows no binding. Results from PFA2 were essentially identical. The D7A mutation limits but does not completely eliminate binding. The numbering scheme is Aβ(1–40)-specific.
The binding epitope was determined by SPR analysis of previously described (25, 26) single Ala mutants of Aβ(1–40) (Fig. 1b). Ala mutation at peptide residue (PR) 8 (Fig. 1b) or higher (data not shown) consistently yields WT levels of binding in SPR experiments. The Asp-1→ Ala mutant peptide shows little reduction in binding, consistent with the crystal structures discussed below, in which Asp-1 is disordered. Binding is nearly eliminated for Ala mutations at positions 3, 4, 5, and 6. The Asp-7→ Ala mutant peptide exhibits significantly reduced binding to both mAbs. Thus, the Aβ binding epitope for PFA1 and PFA2 was shown by SPR to be the linear sequence EFRHD.
Overall Structure of PFA1 and PFA2 Fab apo and Complex Forms.
To explore the structural basis of binding, we solved the x-ray structures of the two Fab fragments both alone (apo) and bound to the peptide fragment Aβ(1–8) (“PFA1-pep” and “PFA2-pep”); the amino acid sequence of Aβ(1–8) is DAEFRHDS. The structures were determined by molecular replacement; the highest resolution structure is at 1.65 Å. Additionally, we crystallized PFA1 with a “mutant” Aβ(1–7) peptide AKFRHD, whose sequence is found in GRIP1 (residues 110–115), to illustrate the potential for cross reaction (PFA1–E3K). Abbreviated crystallographic data are presented in Table 2. Full crystallographic data collection and refinement parameters are presented in supporting information (SI) Table 5.
Table 2.
Abbreviated x-ray data measurement and refinement statistics
| Measurement | PFA1-apo | PFA1-pep | PFA1-E3K | PFA2-apo1 | PFA2-pep |
|---|---|---|---|---|---|
| Resolution, Å | 2.0 | 1.65 | 2.1 | 2.3 | 2.5 |
| Rsym | 0.05 (0.19) | 0.055 (0.467) | 0.2 (0.223)* | 0.072 (0.359) | 0.071 (0.473) |
| I/σI | 16.3 (4.5) | 21.9 (2.4) | 9.8 (4.1) | 10.8 (2.8) | 12.4 (1.5) |
| Completeness, % | 93.1 (85.1) | 90.7 (66.3) | 87.6 (65.6) | 93.0 (92.8) | 96.0 (89.8) |
| Rfac/Rfree | 0.177/0.223 | 0.182/0.225 | 0.201/0.261 | 0.211/0.278 | 0.208/0.277 |
| Bond RMSD, Å | 0.011 | 0.012 | 0.009 | 0.010 | 0.016 |
| Angle RMSD, ° | 1.429 | 1.436 | 1.169 | 1.332 | 1.573 |
*Rmerge. Data from two crystals of PFA1-E3K were scaled for higher completeness. Thus, we report Rmerge values in place of Rsym for this structure.
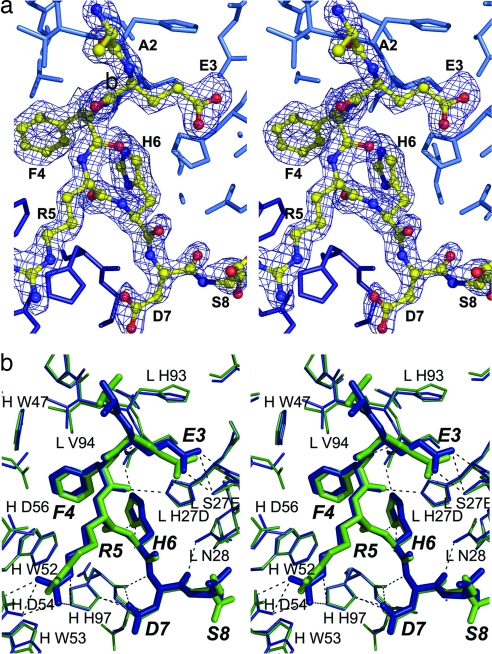
In our three complex structures, at least six residues of the peptide can be traced unambiguously from the electron density maps (Fig. 2a). In all structures, the electron density for the CDRs is readily traced. The antigen binding site, which lies in a cleft between the light-chain (L) and H variable domains, is formed by four of the six CDRs: CDR-L1, CDR-L3, CDR-H2, and CDR-H3 (sequences are shown in Table 2). The Aβ(1–8) peptide adopts nearly identical conformations when bound to the two Fabs (Cα RMS deviation for residues 2–8 is 0.48 Å) (Fig. 2b), which is not surprising given that PFA1-Fab and PFA2-Fab have identical L and similar H sequences.
Fig. 2.
PFA1 and PFA2 bind to the Aβ(1–8) peptide. (a) Stereoview of a simulated-annealing omit map contoured at 3σ shows the electron density for the free DAEFRHDS peptide bound to the CDR of PFA1. (b) Stereoview of the overlay of the peptides and CDRs highlights the similarity in binding. PFA1-pep is shown in blue, PFA2-pep is in green. Residues are numbered by the Kabat scheme.
Aβ(1–8) Binding to PFA1 and PFA2 Features the WWDDD Motif.
The structures of the antibody complexes reveal the key interactions for substrate binding. Small conformational changes in the two Fabs at the CDRs accompany peptide binding indicative of induced fit (SI Table 6). In both structures, the main chain of the CDR-H3 and CDR-L1 loops moves (maximum Cα displacement is 1.7 Å) to accommodate the peptide, alleviating steric clashes. Peptide binding also induces changes in side-chain conformations, with H His-97, Trp-53, and Asp-100C (Kabat numbering is used throughout) changing rotamers with peptide binding (SI Fig. 4).
The Aβ(1–8) peptide binds in a partially extended coil with no evident α or β structure. Complex formation between the Aβ(1–8) peptide and mAb buries 494 Å2 and 528 Å2 of PFA1 and PFA2, respectively. A striking feature of the CDR regions of both mAbs is the WWDDD motif in CDR-H2. The WWDDD motif is found in other mAbs such as AAO18227 (anti-human Fc), AAB02362 (anti-flu), AAQ24154 (anti-human CD16), AAS00782 (anti-DNA), and Fab28 (anti-HIV1-RT); it appears to be restricted to the IGHV8 subtype (Ig allele nomenclature from the IMGT database of Ig genes) (27) gene sequences, making it an important antigen recognition motif. Our structures provide the first example of the WWDDD motif binding to a positively charged residue and to an adjoining aromatic residue. In our structures, both Trp (W) residues and two of the three Asp (D) residues interact with the bound peptide, particularly residues Phe-4 and Arg-5, making this motif important in epitope recognition (Fig. 2b, SI Fig. 5, and SI Table 7).
The key residues of Aβs for binding PFA1 and PFA2 consist of PRs 3EFRHD7 at the Aβ N terminus (shown in Fig. 2b). They make bulk of the hydrogen bonds, ion-pair, and van der Waals interactions with the Fabs (SI Table 7). This EFRHD binding region is entirely consistent with the epitope derived by SPR, as discussed above. The carboxyl group of PR Glu-3 is within hydrogen bonding distance of the side-chain of L His-27D in both PFA1-pep and PFA2-pep. In both complexes, the side-chain of PR Glu-3 hydrogen bonds to L Ser27E (N and OG atoms) and makes some long-range (<5 Å) ion-pair interactions with the L His-93 side chain.
The electrostatic environment of the peptide-binding site is shown in Fig. 3. PR Phe-4 is buried deep within a binding pocket consisting of H Trp-47, and Trp-52 belonging to the aforementioned WWDDD motif (SI Fig. 5) and the L Leu-96 and H His-50 residues and makes edge-on hydrophobic interactions with these residues. PR Phe-4 also makes hydrogen bonds via its main-chain amide N and carbonyl O to the main-chain of L Ser-92 and the side chain of L His-27D, respectively. PR Arg-5 makes salt bridges with H Asp-54 and -56 belonging to the WWDDD motif, whereas the side chain of PR His-6 hydrogen bonds to the side chain of H Asp-100C. Furthermore, PR His-6 makes a parallel π-overlap with L Tyr-32. PR Asp-7 N hydrogen bonds to H His-97 O. PR Asp-7 makes hydrogen bonds to the side chain of H His-97 and to L Asn-28 ND2. No hydrogen bonds are made from the Fabs to the terminal residues PR Asp-1 and PR Ser-8. In fact, PR Asp-1 is not visible in the electron density maps in either complex. In both complexes, the amide of PR His-6 makes an intramolecular hydrogen bond to the carbonyl oxygen of Phe-4, stabilizing the peptide conformation.
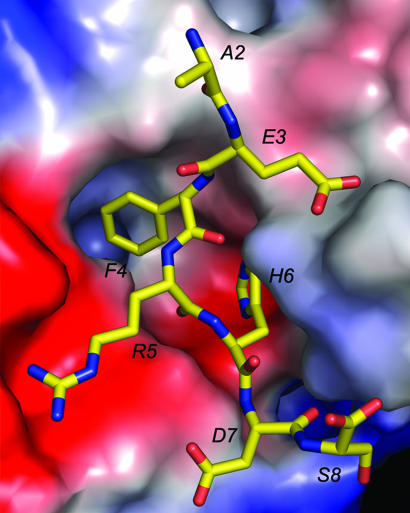
Fig. 3.
Electrostatics of binding. The electrostatic potential surface of PFA1 with bound peptide. Blue represents positive charge, red indicates negative charge, and the apolar surface is shown in white. The Aβ(1–8) peptide is drawn with carbon (yellow), nitrogen (blue), and oxygen (red). Although the Arg 5 residue sits in a pocket of strong negative charge, the Glu 3 residue has no correspondingly positive region around it. This position is susceptible to substitution and cross-reaction.
PFA1 and PFA2 Are Highly Homologous.
The similarity of mAbs in binding to Aβ reflects their sequence similarity: Their light chains are identical, expressing the kappa IGKV1–117 allele (27). The PFA1 and PFA2 heavy chains strongly resemble the product of the IGHV8S7 allele (27); they exhibit 92% sequence identity with one another, with four residue differences at the CDRs and one adjacent to CDR-H2 (Table 3). None of the divergent residues are directly involved in short-range (<3.5 Å) interactions to the peptide. However, the CDR-H3 regions of PFA1 and PFA2 differ from one another at two adjacent residues (TT vs. NV) that make 3.8–4.8 Å polar interactions with the bound peptide; such longer-range effects may contribute to the nearly 2-fold difference in binding affinity between these two mAbs and the Aβ monomer.
Table 3.
Sequence alignment of PFA1 and PFA2 CDRs
| Chain | CDR1 | CDR2 | CDR3 |
|---|---|---|---|
| PFA1_L | QSIVHSNGNTY | KVS | FQGSHVPLTF |
| PFA2_L | QSIVHSNGNTY | KVS | FQGSHVPLTF |
| Binding | 27+**++ | 50 | 89**** |
| PFA1_H | GFSLSTSGMG | IWWDDDR | VRRAHTTVLGDWFAY |
| PFA2_H | GFSLRTSGMG | IWWDDDK | VRRAHNVVLGDWFAY |
| Binding | 26 | 51++** | 93++*+* |
Italicized text indicates mismatches between PFA1 and PFA2. Bold text indicates binding to peptide. Asterisks indicate hydrogen bonding at <3.5 Å. + indicates a 3.5- to 5-Å distance between polar atoms.
Binding of PFA1 and PFA2 to Related Sequences.
Although the binding constants of these mAbs for Aβ are significant and within a therapeutically useful range, the shortness of the Aβ sequence epitope raises possible specificity issues. A BLAST search revealed that the only protein in the human genome containing the AEFRHD sequence is its membrane-associated amyloid precursor protein. Nonetheless, it is possible that human sequences related to AEFRHD might cross-react with antibodies, such as PFA1 and PFA2, causing side effects during passive immunotherapy. Indeed, the SPR studies showed that Ala replacements at residues 3 and 7 of this epitope do not fully eliminate binding. BLAST and FASTAS3 searches yielded six extracellular, secreted, or membrane-associated human proteins featuring sequences matching 5 of the 6 residues of AEFRHD (SI Table 8). If we limit our search to the EFRH motif, the dominant binding motif as revealed by our structures and SPR studies, we obtain additional hits (SI Table 8).
As a preliminary investigation of the specificity of PFA1 and PFA2 with respect to the human genome, the WT Aβ(1–7) peptide AEFRHD and mutants derived from proteins listed in SI Table 8, AKFRHD, AEIRHD, AEFRSD, and REEFRHEA, were synthesized, and their affinity for the two Fab fragments was determined by SPR. AEIRHD and AEFRSD showed no measurable binding to PFA1 and PFA2. However, we found that PFA1-Fab binds to AKFRHD (a sequence found GRIP1) with an affinity 28 times lower than that at which it binds to AEFRHD (Table 4). Similarly, SPR studies show that PFA2-Fab also binds to the GRIP1-derived peptide AKFRHD with 35-fold lower affinity than that at which it binds to AEFRHD (Table 4). Perhaps more significantly, the peptide sequence REEFRHEA, derived from the human receptor-related neurotrophic tyrosine kinase (ROR2), actually binds to PFA1 and PFA2 with approximately twice the affinity of the WT Aβ(1–7) peptide AEFRHD (Table 4). Thus, although the AEFRHD sequence is unique in the known mouse and human proteomes, it has near-matches with several other proteins in both species. These in vitro results do not necessarily imply that the full-length protein will interact with the mAb in vivo; however, they do illustrate the potential for complications from cross-reactivity.
Table 4.
SPR results for Aβ(1–7), Grip1(110–115), Ror2(518–525), and pyroglutamic Aβ(1–8)
| Peptide mAb | Aβ(1–7) AEFRHD ″WT,″ nM | Grip1(110–115) AKFRHD ″E3K,″ nM | Ror2(518–525) REEFRHEA ″Ror2,″ nM | Aβ(1–8) pGluFRHD ″pyro,″ nM |
|---|---|---|---|---|
| PFA1-FAB | 60–120 | 3,400 | 24 | 3,000 |
| PFA2-FAB | 160–280 | 9,880 | 93 | 12,200 |
Kd values were determined from kd/ka. pyro, pyroglutamate.
A significant portion of the Aβ found in the brain is N-terminally truncated, and the most prevalent of these modified forms is one lacking residues 1 and 2 and beginning with a pyroglutamate residue (pyroglutamate-Glu Aβ). This form is expected to be resistant to exopeptidase digestion and is also more amyloidogenic than full length peptides, suggesting that this Aβ breakdown product might play a significant role in pathology (28). Because E3 is part of the PFA1/PFA2 epitope, we expected these mAbs to exhibit diminished affinity for pyroglutamate-Glu forms of Aβ. In fact, we found that the peptide pGlu-FRHD is bound by the antibodies ≈50- to 70-fold less well than the corresponding WT sequence (Table 4). Thus, although the pathological roles of the pyroglutamate-Glu Aβ peptide and other truncated forms of Aβ is yet to be worked out, in the future it may also be necessary to develop antibodies that effectively bind to these forms.
PFA1 Complexed with the GRIP1 Peptide.
The AKFRHD peptide from GRIP1 (whose binding to PFA1 and PFA2 was discussed above) was cocrystallized with PFA1. We observed clear peptide electron density in a PFA1-AKFRHD cocrystal structure. Binding of the AKFRHD (“E3K”) peptide to PFA1 is nearly identical (Cα RMSD is 0.23 Å) to that of Aβ(1–8), except that it is the tail of the substituted lysine residue rather than the glutamate making a hydrogen bond with L Ser-32 (SI Fig. 6). The Glu-to- Lys substitution also results in replacing the favorable ion-pair interaction made by the Aβ peptide to His by a less favorable van der Waals contact formed between two basic residues. Quantitatively, AEFRHD of the Aβ peptide makes ≈30% more ion- pair interactions (12 vs. 9) at short range (<4 Å) and ≈60% more (18 vs. 11) at medium range (between 4 and 6 Å) than AKFRHD. Because they both make 18 hydrogen bonds, these ion-pairing differences may possibly account for the ≈30-fold reduction in binding of the GRIP1 peptide relative to the Aβ peptide.
Discussion
The structures of mAb Fab fragments in complex with an N-terminal Aβ peptide illustrate the molecular basis for Aβ recognition by these mAbs. The interface features a striking WWDDD motif. Residues in the CDR-L1, CDR-L3, CDR-H2, and CDR-H3 loops take part in binding.
The accumulated experience of many efforts to obtain antibodies to Aβ suggests that the N terminus may be the immunodominant epitope of this peptide. Furthermore, if aggregated forms of Aβ are to be targeted in therapy, antibodies to the N terminus will probably be required, given the poor accessibility of other portions of the sequence in aggregates (29, 30).
In summary, the structures and binding data presented here provide insights into at least one way in which the immune system generates antibodies to bind this epitope, and they also provide the necessary foundation for structure-based design of antibodies with enhanced properties. Structural modifications in the CDR residues, for example, may lead to derivative antibodies with improved binding to the WT sequence and its natural breakdown products (such as the pyroglutamate-Glu Aβ form) and reduced binding to related sequences, such as Ror2, found in the human genome that, if left unaddressed, might lead to significant side reactions in immunotherapy.
Methods
Preparation of disaggregated monomer (31), mature amyloid fibrils (31), and CLC-stabilized protofibrils (25) was as described. Preparation of hybridoma cell lines and antibody sequencing is described in SI Text. The mAb purification and papain fragmentation protocol was adapted from Goding (ref. 32, chap. 9).
IgG affinities for Aβ(1–40) fibrils and protofibrils were measured by using a microtiter plate assay as described in ref. 16. A full description of the SPR binding experiments is in SI Text. Briefly, binding studies were performed at 20°C by using BIAcore (Uppsala, Sweden) 2000 optical biosensors equipped with CM5 sensor chips.
Reagents 37–47 from the Hampton Index (33) were used for initial screening, and some initial apo hits were obtained by using the PHOENIX crystallization robot (Art Robbins Instruments, Sunnyvale, CA). Data were indexed, integrated, and scaled with the HKL2000 package (34). The original PFA2 triclinic apo structure was solved by molecular replacement, using the constant (CL+CH1) and variable portions of FAB4C6 (PDB ID code 1NCW) separately (35). Protein rebuilding, including water-picking, was performed in Coot (36); refinement was conducted with Refmac5 (37). A partially refined PFA2 apo model was idealized and used for solution of the other structures. Structures were validated with SFCHECK (38), PROCHECK (39), and MOLPROBITY (40).
Abbreviated data collection and refinement statistics appear in Table 2. Full crystallization conditions, validation information, and statistics appear in SI Table 5 and SI Text. Buried surface area was calculated with Areaimol with a 1.4-Å probe radius.
Supplementary Material
Acknowledgments
We thank Angela Williams (University of Tennessee) for providing disaggregated peptides and CLC-Aβ protofibrils, Dr. Elias Fernandez for useful suggestions, Dr. Hai Xu (University of Tennessee) for assistance with crystallography of PFA2-pep, and Lauren Cagliuso for assistance. We also thank BioCARS at the Advanced Photon Source. This work was supported by National Institutes of Health Grants NS46356 (to R.W., P.H.P., and C.D.) and R01 AG018416 (to R.W. and D.G.M.).
Abbreviations
- Aβ
amyloid-β
- AD
Alzheimer's disease
- CDR
complementarity determining region
- CLC
calmidazolium
- Fab
antigen binding fragment
- H
heavy-chain
- L
light-chain
- mAb
monoclonal antibody
- PR
peptide residue
- PFA
protofibril antibody
- SPR
surface plasmon resonance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2IPT, 2IPU, 2IQ9, 2IQA, 2ROZ, and 2ROW).
This article contains supporting information online at www.pnas.org/cgi/content/full/0705888104/DC1.
References
- 1.Dobson CM. Nature. 2002;418:729–730. doi: 10.1038/418729a. [DOI] [PubMed] [Google Scholar]
- 2.Weiner HL, Frenkel D. Nat Rev Immunol. 2006;6:404–416. doi: 10.1038/nri1843. [DOI] [PubMed] [Google Scholar]
- 3.DeMattos RB, Bales KR, Cummins DJ, Dodart JC, Paul SM, Holtzman DM. Proc Natl Acad Sci USA. 2001;98:8850–8855. doi: 10.1073/pnas.151261398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gevorkian G, Petrushina I, Manoutcharian K, Ghochikyan A, Acero G, Vasilevko V, Cribbs DH, Agadjanyan MG. J Neuroimmunol. 2004;156:10–20. doi: 10.1016/j.jneuroim.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Solomon B. Curr Alzheimer Res. 2004;1:149–163. doi: 10.2174/1567205043332126. [DOI] [PubMed] [Google Scholar]
- 6.Frenkel D, Dewachter I, Van Leuven F, Solomon B. Vaccine. 2003;21:1060–1065. doi: 10.1016/s0264-410x(02)00609-6. [DOI] [PubMed] [Google Scholar]
- 7.Lavie V, Becker M, Cohen-Kupiec R, Yacoby I, Koppel R, Wedenig M, Hutter-Paier B, Solomon B. J Mol Neurosci. 2004;24:105–113. doi: 10.1385/JMN:24:1:105. [DOI] [PubMed] [Google Scholar]
- 8.Schenk D, Barbour R, Dunn W, Gordon G, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, et al. Nature. 1999;400:173–177. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- 9.Bard F, Cannon C, Barbour R, Burke RL, Games D, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, et al. Nat Med. 2000;6:916–919. doi: 10.1038/78682. [DOI] [PubMed] [Google Scholar]
- 10.Solomon B, Koppel R, Frankel D, Hanan-Aharon E. Proc Natl Acad Sci USA. 1997;94:4109–4112. doi: 10.1073/pnas.94.8.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyman BT, Tanzi RE, Marzloff K, Barbour R, Schenk D. J Neuropathol Exp Neurol. 1992;51:76–83. doi: 10.1097/00005072-199201000-00009. [DOI] [PubMed] [Google Scholar]
- 12.Seubert P, Vigo-Pelfrey C, Esch F, Lee M, Dovey H, Davis D, Sinha S, Schlossmacher M, Whaley J, Swindlehurst C, et al. Nature. 1992;359:325–327. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- 13.Lee EB, Leng LZ, Zhang B, Kwong L, Trojanowski JQ, Abel T, Lee VM. J Biol Chem. 2006;281:4292–4299. doi: 10.1074/jbc.M511018200. [DOI] [PubMed] [Google Scholar]
- 14.Levites Y, Das P, Price RW, Rochette MJ, Kostura LA, McGowan EM, Murphy MP, Golde TE. J Clin Invest. 2006;116:193–201. doi: 10.1172/JCI25410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frenkel D, Solomon B, Benhar I. J Neuroimmunol. 2000;106:23–31. doi: 10.1016/s0165-5728(99)00232-5. [DOI] [PubMed] [Google Scholar]
- 16.O'Nuallain B, Wetzel R. Proc Natl Acad Sci USA. 2002;99:1485–1490. doi: 10.1073/pnas.022662599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kodali R, Wetzel R. Curr Opin Struct Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Walsh DM, Selkoe DJ. Protein Pept Lett. 2004;11:213–228. doi: 10.2174/0929866043407174. [DOI] [PubMed] [Google Scholar]
- 19.Harper JD, Wong SS, Lieber CM, Lansbury PT. Chem Biol. 1997;4:119–125. doi: 10.1016/s1074-5521(97)90255-6. [DOI] [PubMed] [Google Scholar]
- 20.Williams AD, Sega M, Chen M, Kheterpal I, Geva M, Berthelier V, Kaleta DT, Cook KD, Wetzel R. Proc Natl Acad Sci USA. 2005;102:7115–7120. doi: 10.1073/pnas.0408582102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruckner K, Pablo Labrador J, Scheiffele P, Herb A, Seeburg PH, Klein R. Neuron. 1999;22:511–524. doi: 10.1016/s0896-6273(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 22.Clark LA, Boriack-Sjodin PA, Eldredge J, Fitch C, Friedman B, Hanf KJ, Jarpe M, Liparoto SF, Li Y, Lugovskoy A, et al. Protein Sci. 2006;15:949–960. doi: 10.1110/ps.052030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Beuerlein G, Nie Y, Smith H, Lee BA, Hensler M, Huse WD, Watkins JD. Proc Natl Acad Sci USA. 1998;95:6037–6042. doi: 10.1073/pnas.95.11.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bard F, Barbour R, Cannon C, Carretto R, Fox M, Games D, Guido T, Hoenow K, Hu K, Johnson-Wood K, et al. Proc Natl Acad Sci USA. 2003;100:2023–2028. doi: 10.1073/pnas.0436286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams AD, Shivaprasad S, Wetzel R. J Mol Biol. 2006;357:1283–1294. doi: 10.1016/j.jmb.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 26.Wetzel R, Shivaprasad S, Williams AD. Biochemistry. 2007;46:1–10. doi: 10.1021/bi0620959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giudicelli V, Chaume D, Lefranc MP. Nucleic Acids Res. 2004;32:W435–W440. doi: 10.1093/nar/gkh412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schilling S, Lauber T, Schaupp M, Manhart S, Scheel E, Bohm G, Demuth HU. Biochemistry. 2006;45:12393–12399. doi: 10.1021/bi0612667. [DOI] [PubMed] [Google Scholar]
- 29.Kheterpal I, Williams A, Murphy C, Bledsoe B, Wetzel R. Biochemistry. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 30.Kheterpal I, Chen M, Cook KD, Wetzel R. J Mol Biol. 2006;361:785–795. doi: 10.1016/j.jmb.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 31.O'Nuallain B, Thakur AK, Williams AD, Bhattacharyya AM, Chen S, Thiagarajan G, Wetzel R. Methods Enzymol. 2006;413:34–74. doi: 10.1016/S0076-6879(06)13003-7. [DOI] [PubMed] [Google Scholar]
- 32.Goding JW. Monoclonal Antibodies: Principles and Practice. San Diego: Academic; 1996. [Google Scholar]
- 33.Harris LJ, Skaletsky E, McPherson A. Proteins. 1995;23:285–289. doi: 10.1002/prot.340230218. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. In: Macromolecular Crystallography, Part A. Carter CW, Sweet RM, editors. Vol 276. New York: Academic; 1997. pp. 307–326. [Google Scholar]
- 35.Zhu X, Heine A, Monnat F, Houk KN, Janda KD, Wilson IA. J Mol Biol. 2003;329:69–83. doi: 10.1016/s0022-2836(03)00406-6. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Vaguine AA, Richelle J, Wodak SJ. Acta Crystallogr D. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 40.Davis IW, Murray LW, Richardson JS, Richardson DC. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.