Abstract
Cystic fibrosis results from mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Premature termination codons represent a common minority of CFTR mutations, and are caused by base pair substitutions that produce abnormal stop codons in the coding sequence. Select aminoglycosides induce “translational readthrough” of premature stop codons and have been shown to restore full-length functional protein in a number of preclinical and clinical settings. We studied two well-described premature termination codons found in the distal open reading frame of CFTR, W1282X and R1162X, expressed in polarizing and nonpolarizing cells. Our findings indicate that W1282X CFTR–expressing cells demonstrate significantly greater CFTR activity when overexpressed compared with R1162X CFTR cells, even when truncated protein is the predominant form. In addition, our results show that the combination of stimulated expression and stop codon suppression produces additive effects on CFTR-mediated ion transport. These findings provide evidence that W1282X CFTR exhibits membrane localization and retained chloride channel function after enhanced expression, and suggest that patients harboring this mutation may be more susceptible to CFTR rescue.
Keywords: cystic fibrosis, cystic fibrosis transmembrane conductance regulator, premature termination codons, nonsense mutations, aminoglycosides
CLINICAL RELEVANCE
Evidence that W1282X cystic fibrosis transmembrane conductance regulator (CFTR) retains chloride channel function improves our understanding of CFTR biology, and may explain why subjects harboring W1282X CFTR appear more susceptible to the strategy of premature termination codon suppression.
Cystic fibrosis (CF) is caused by mutations in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR), and results in significant morbidity and early mortality in affected individuals. Over 1,400 disease-causing mutations in CFTR have been identified, and can be divided into six major classes (1). Nonsense mutations (a class I mutation type) are due to the presence of a premature termination codon (PTC or stop mutation), resulting in absent functional protein and typically a severe CF phenotype (2, 3). PTCs are relatively common in humans, accounting for approximately 5 to 10% of disease-causing CF mutations in white populations, and are the proximate cause of approximately 30% of human genetic diseases (4). Due presumably to a genetic founder effect, PTCs within CFTR are especially common in individuals of Ashkenazi Jewish decent, where they account for nearly 85% of CFTR mutations (5).
A potential treatment for conditions caused by nonsense mutations includes the use of aminoglycoside antibiotics or other agents to suppress the normal proofreading function of the ribosome, leading to insertion of a near-cognate amino acid at a PTC, and translation of full-length protein. This “translational readthrough” of premature stop mutations has been shown to restore protein function in a number of preclinical settings (6–12). Additional in vitro studies have demonstrated that aminoglycoside antibiotics can be used to suppress PTCs in genes involved in other diseases, including muscular dystrophy, Hurler's Syndrome, ceroid lipofuscinosis, nephropathic cystinosis, and expression of mutated P53 (6, 11, 13–15). Recently, Wilschanski and coworkers showed in two separate human studies performed on Israeli subjects that intranasal gentamicin induced bioelectric correction of nasal ion transport and membrane localization of CFTR specifically in patients with CF with premature stop mutations (16, 17). Importantly, all subjects with nonsense mutations in both of these studies carried at least one copy of W1282X CFTR, a PTC that is particularly common in this population (found in up to 40% of Israeli patients with CF) (5, 16, 17). These studies have led to a similar trial testing the nasal administration of tobramycin and gentamicin in the United States (18), as well as evaluation of a novel suppressive agent (PTC124) in clinical trials performed in the United States and Israel (19–21).
Linde and colleagues, and Shushi and coworkers, recently reported that the response to suppressive agents among patients with CF with PTCs depends in part upon basal CFTR mRNA levels (which are typically reduced due to the rapid degradation of nonsense transcripts by nonsense-mediated decay [NMD]) (22, 23). Other factors shown to influence CFTR rescue include the specific type of termination codon (UGA,UAA, or UAG) and the neighboring mRNA context (10, 24, 25). Each of these factors may induce variable responsiveness to stop codon suppression, and impact interpretation of ongoing clinical trials testing this treatment strategy.
Based on the promising clinical findings in patients possessing W1282X CFTR, coupled with variable in vivo responses to premature stop suppression, we designed experiments to examine the effects of stop codon site on two common PTCs found in CFTR (R1162X and W1282X). Our results indicate that enhanced expression is sufficient to restore measurable activity to W1282X CFTR but not R1162X CFTR. These findings have implications to strategies designed to activate mutant CFTR in vivo, and to the interpretation of studies examining this potential treatment modality.
MATERIALS AND METHODS
Development of Stable Cell Lines Expressing W1282X and R1162X CFTR
W1282X and R1162X CFTR cDNA were stably introduced into HeLa and CFBE41o- cells using TranzVector (Tranzyme, Inc., Birmingham, AL). TranzVector system represents an HIV-based lentiviral vector with unique safety features as described (26). To generate vector stock, CFTR cDNA was first cloned into the gene transfer component under the control of the human cytomegalovirus (hCMV) promoter. Use of the CMV promoter allowed the study of induced CFTR transcription using promoter-active agents. Expression of CFTR was also coupled to the puromycin-N-acetyltransferase gene (puro) gene via the internal ribosomal entry site (IRES) of encephalomyocarditis virus, allowing for rapid selection of cells expressing CFTR in media containing puromycin. HeLa and CFBE41o- cells were transduced at multiplicity of infection of one, followed by puromycin (4 μg/ml) selection. Puromycin-resistant cells were expanded to form a pool of stable CFTR expressors.
CFTR Immunoprecipitation
For each sample, total protein concentration was measured by absorbance at 760 nm and quantified by the relative standard curve method against bovine serum albumin as a protein standard. CFTR was immunoprecipitated from equal volume aliquots of cell lysate after normalizing for total protein concentration (typical final concentration is ∼ 1 mg/ml). CFTR was immunoprecipitated using 1 μg 24-1, anti–C-terminal antibody (#HB-11947; American Type Culture Collection [ATCC], Manassas, VA), anti–NBD-1 antibody (27), or H-182, anti–N-terminal antibody (SC#10747; Santa Cruz Biotechnology, Santa Cruz, CA) coupled to 20 μl Protein A agarose beads (Roche Molecular Biochemicals, Nutley, NJ). CFTR was labeled by in vitro phosphorylation with [γ32P] ATP (NEN) and PKA catalytic subunit (Promega, Madison, WI). Labeled CFTR was analyzed by SDS-PAGE (6% gel) and autoradiography as described previously (28).
Real-Time RT-PCR to Quantify CFTR Expression
A TaqMan One Step RT-PCR protocol (Applied Biosystems [ABI], Foster City, CA) was used to quantify CFTR mRNA transcripts using Assays on Demand Gene Expression Products, coupled with the ABI Prism 7500 sequence detection system (Applied Biosystems). Briefly, total RNA was isolated using the Qiagen RNeasy mini kit according to manufacturer's instructions. To prevent possible DNA contamination, the samples were pretreated with RNase-free DNase (Qiagen, Valencia, CA). Sequence-specific primers and probes for human CFTR and 18S rRNA were purchased from Assays on Demand (ABI); Assay ID for CFTR: Hs00357011_m1; the probe extends across the exon 21/22 boundary of the CFTR sequence. TaqMan One Step PCR Master Mix Reagents Kit (ABI) was used for reverse transcriptase and PCR. The reaction volume was 25 μl including 12.5 μl of 2× Master Mix without UNG, 0.625 μl of 40× MultiScribe and RNase Inhibitor Mix, 1.25 μl of 20× target primer and probe, 5.625 μl of Nuclease-free water (Ambion, Austin, TX), and 5 μl of RNA sample. The reaction plates were covered with an optical cap and centrifuged briefly to remove bubbles. The thermocycler conditions were as follows: Stage 1: 48°C for 30 minutes; Stage 2: 95°C for 10 minutes; Stage 3: 95°C for 15 seconds, repeat 40 cycles, 60°C for 1 minute. All experiments were run in triplicate on two separate days. The absolute value of the slope of log input amount versus ΔCt was > 0.1, implying that the efficiencies of CFTR and 18S rRNA amplification were not equal. Therefore, the relative quantification of transcript levels (CFTR compared with endogenous 18S rRNA) was performed using the standard curve method.
SPQ Studies of Halide Efflux in HeLa Cells
HeLa cells stably expressing W1282X, R1162X, or wild type CFTR were studied with the halide quenched dye 6-methoxy-N-(3-sulfopropyl)-quinolinium (SPQ; Molecular Probes, Inc., Eugene, OR) as previously described (27, 29). Briefly, cells were seeded at ∼ 5 × 105 cells/coverslip and grown in Dulbecco's modified Eagle's medium + 10% fetal bovine serum at 37°C for 24 to 48 hours. The media was changed, and cells were subsequently grown for 48 hours at 37°C. On the day of study, cells were loaded with hypotonic SPQ (3 mM) for 10 minutes, then placed in a NaI buffer to quench cellular fluorescence. The cells were then placed in a specially designed perfusion chamber and studied at room temperature. Fluorescence of individual cells was measured using a Zeiss inverted microscope (Thornwood, NY); excitation at 340 nm, emission at > 410 nm, a PTI imaging system (Monmouth Junction, NJ), and a Hamamatsu camera (Bridgewater, NJ). Baseline fluorescence was measured in isotonic NaI buffer, followed by perfusion with isotonic dequench buffer (NaNO3 replaced NaI) to measure unregulated efflux, and then NaNO3 buffer with CFTR agonists (20 μM forskolin and 50 μM genistein). At the end of each experiment, cells were returned to the NaI buffer for requench (1,100 s). Increase in fluorescence above the basal (NaI quenched) level is shown (% increase F > basal). The data are cumulative from multiple coverslips in each condition studied in a paired fashion (n = 30–90 cells/curve). All cells studied are reported in the data obtained from each condition (i.e., no “selection” of cells was performed) (27, 30).
Ussing Chamber Studies in CFBE41o- Cell Monolayers
Inserts were mounted in Ussing chambers, and Isc was measured under voltage clamp conditions as previously described (29, 31). Briefly, CFBE41o− cells expressing W1282X or R1162X CFTR were seeded on Costar 0.4-μm permeable supports (5 × 105 cells/filter, 6.5 mm diameter; Bethesda, MD) after coating with fibronectin as previously described (32). Cells were grown to confluence and then grown at an air–liquid interface (48 h) and mounted in modified Ussing chambers (Jim's Instruments, Iowa City, IA), and initially bathed on both sides with identical Ringers solutions containing (in mM): 115 NaCl, 25 NaHCO3, 2.4 KH2PO4, 1.24 K2HPO4, 1.2 CaCl2, 1.2 MgCl2, 10 D-glucose (pH 7.4). Bath solutions were vigorously stirred and gassed with 95%O2:5% CO2. Solutions and chambers were maintained at 37°C. Short-circuit current (Isc) measurements were obtained by using an epithelial voltage clamp (University of Iowa Bioengineering, Iowa City, IA). A 3-mV pulse of 1 second duration was imposed every 100 seconds to monitor resistance, which was calculated using Ohm's law. To measure stimulated Isc, the mucosal bathing solution was changed to a low Cl− solution containing (in mM): 1.2 NaCl, 115 Na gluconate, and all other components as above plus 100 μM amiloride. Agonists (20 μM forskolin and 50 μM genistein) were added to the bathing solutions as indicated (minimum 5 min of observation at each concentration). Glybenclamide (200 μM) was added to the mucosal bathing solution at the end of experiments to block CFTR-dependent Isc. The CFBE41o− W1282X and CFBE41o− R1162X cells were handled in exactly the same fashion during our studies. Specifically, chambers were maintained at 37°C, and agonist stimulation was initiated within 30 minutes of placement in the chambers.
Immunocytochemistry
HeLa cells were grown on glass cover slips for 24 to 48 hours as described above. After fixation and permeabilization in iced methanol for 10 minutes, nonspecific protein binding sites were blocked using goat serum diluted 1:20 in PBS for 1 hour. Anti–CFTR C-terminal monoclonal antibody (24-1, ATCC #HB-11947; 5 μg/ml for 2 h at room temperature) was used to detect full-length CFTR and anti–NBD-1 polyclonal rabbit antibody (5 μg/ml for 2 h at room temperature) was used to stain truncated CFTR. Cells were then incubated with AlexaFloor 596– or 488–labeled secondary antibody (anti-mouse or anti-rabbit, respectively; 2 μg/ml) for 1 hour at room temperature. Nuclei were stained with DAPI, and cells were mounted with Vectashield (Vector Labs, Burlingame, CA).
Microscopy
Images were captured on an Olympus IX170 inverted epifluorescence microscope equipped with step motor, filter wheel assembly (Ludl Electronics Products, Hawthorne, NY), and 83,000 filter set (Chroma Technology, Brattleboro, VT) and SenSys-cooled charge-coupled high-resolution camera (Photometrics, Tucson, AZ). Partial deconvolution of images was performed using IPLab software (Scanalytics, Fairfax, VA). The presence of absence of surface staining was classified for individual cells (at least 160 cells/condition) by a blinded investigator using a simplified protocol previously described (16).
Statistical Analysis
For Isc, florescence-based halide efflux, and RT- PCR measurements, descriptive statistics (mean, SD, and SEM) and paired and unpaired t tests were performed using SigmaStat (Jandel, CA) and Microsoft Excel (Seattle, WA). Immunofluorescence staining patterns were compared using two sample proportion tests and logistic regression (including mutation type, antibody staining, and treatment condition in the predictive model), and analyzed using SAS (Cary, NC). All statistical tests were two-sided and were performed at a 5% significance level (i.e., α = 0.05).
RESULTS
Preferential Enhancement of CFTR Activity in W1282X CFTR Expressing Cells by Sodium Butyrate
Previous studies have examined the function of several CFTR molecules containing clinically relevant premature stop codons in transient, high-level expression systems using nonpolarizing cell types (including G542X, R553X, R1162X, and W1282X CFTR), with variable levels of constitutive and regulated CFTR activity described (7, 8, 33). These are the four most common premature stop codon mutations found in patients with CF, and all are produced by a UGA (opal) premature stop codon. In the studies described here, we examined premature stop codons containing CFTR-constructs stably transduced into nonpolarizing and polarizing cell types (HeLa and CFBE41o− cell backgrounds, respectively). We limited our comparisons to R1162X and W1282X CFTR in readily comparable model systems widely used in CF research. These two mutants occur in the distal portion of CFTR (immediately after TMD-2 for R1162X CFTR, and approximately one third through NBD-2 for W1282X CFTR). Both mutations include translation of the regulatory (R) domain, which imparts PKA-dependent regulation to full-length CFTR (1).
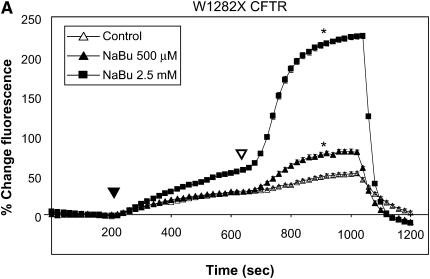
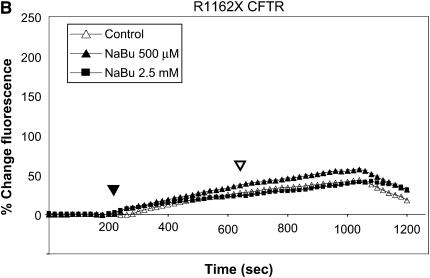
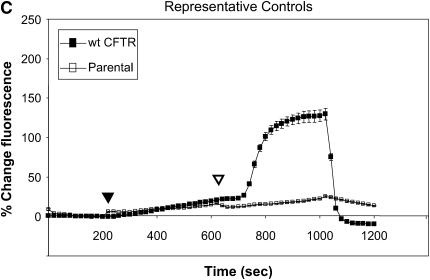
Figure 1 summarizes functional studies in HeLa cells expressing R1162X CFTR and W1282X CFTR. Cells were treated with two different concentrations of sodium butyrate (NaBu, 2.5 and 5 mM) overnight (approximately 16–24 h) at 37°C, and subsequently studied by SPQ. To enhance the sensitivity to detect mutant CFTR activity, cells were stimulated with a cocktail including both forskolin (20 μM, a concentration known to increase cAMP) and genistein (50 μM, a CFTR agonist needed to maximally activate mutant CFTR in both HeLa and CFBE41o− cells [32]). In Figure 1A, increasing concentrations of NaBu exposure led to dramatic increases in halide transport in W1282X CFTR–transduced cells. By contrast, NaBu had little effect on halide efflux in R1162X CFTR (Figure 1B). Representative fluorescence results of HeLa cells transduced with wild-type CFTR or no transgene expression (parental cells) are also shown for comparison (Figure 1C). We extended these studies to CFBE41o− monolayers mounted in Ussing chambers and studied under voltage clamp conditions (Figure 2). Representative tracings are shown in Figure 1A and are summarized in Figure 1B. W1282X CFTR–expressing cells had significantly increased agonist-stimulated Isc relative to R1162X CFTR–expressing cells at baseline (no NaBu treatment). We have previously shown that parental CFBE41o− cells (which express very low levels of ΔF508 CFTR) have no stimulated Isc under similar conditions (32). After treatment with NaBu, further enhancement of agonist-stimulated Isc was demonstrated in W1282X CFTR–transduced cells relative to R1162X CFTR. The negative results seen in the R1162X CFTR cells demonstrate that NaBu has no detectable functional effects on either the R1162X CFTR transgene or endogenous ΔF508 CFTR expressed in these cells (albeit at extremely low levels [32]). Together, the studies in both HeLa and CFBE41o− cell lines demonstrate similar results: NaBu treatment has unique effects on halide transport only in W1282X CFTR–expressing cells.
Figure 1.
Halide transport in W1282X and R1162X CFTR transduced HeLa cells. (A) Fluorescence-based halide efflux (SPQ) activity of W1282X CFTR HeLa cells after treatment with increasing concentrations of NaBu or control (vehicle media) for 24 h. Closed arrowhead represents basal CFTR activity after switch to dequenched buffer, and the open arrowhead signifies fluorescence after the addition of CFTR agonists forskolin (20 μM) and genistein (50 μM). * Halide efflux of W1282X compared with R1162X CFTR transduced cells was significantly increased after treatment with NaBu (P < 0.001, ± SEM).(B) Parallel experiments performed in R1162X CFTR HeLa cells showing no activation of halide efflux after CFTR stimulation. (C) Experiments performed on HeLa parental cells (no transduced CFTR expression) and HeLa transduced with wt CFTR show the expected fluorescence signature of negative (parental cells) and positive (wt CFTR) controls.
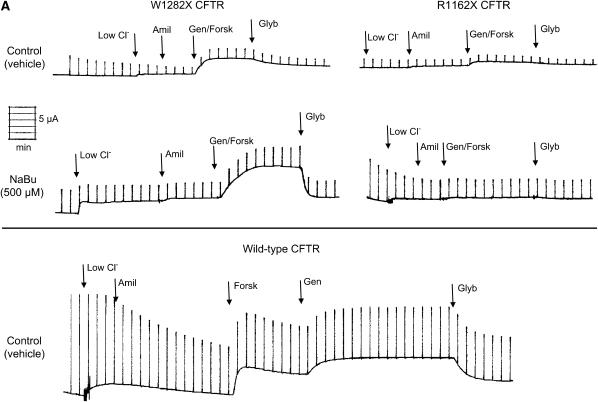
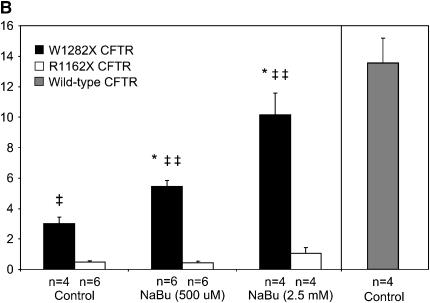
Figure 2.
NaBu treatment results in increased Isc in W1282X expressing cells. Short-circuit currents were measured in the presence of a serosal to mucosal Cl− gradient and 100 μM amiloride as described in Materials and Methods. Glybenclamide-inhibitable currents after forskolin (20 μM) and genistein (50 μM) stimulation are shown. (A) Representative tracings in CFBE41o− cells expressing W1282X CFTR (left) and R1162X CFTR (right) after treatment with NaBu (500 μM) or control conditions for 24 h. Experiments show increased Isc in W1282X CFTR–transduced CFBE41o− cells pretreated with NaBu after forskolin (forsk, 20 μM) and genistein (gen, 50 μM) stimulation. Lower panel shows a representative tracing from CFBE41o− cells expressing wild-type CFTR for comparison. Low Cl, change to chloride serosal to mucosal gradient; Amil, amiloride (100 μM); Glyb, glybenclamide (200 μM). (B) Summary data of CFBE41o- cells expressing W1282X (solid bars) or R1162X CFTR (open bars) exposed to various concentrations of NaBu versus control (vehicle media) for 24 h, then studied in modified Ussing chambers under voltage clamp conditions. Short-circuit currents were greater in W1282X compared with R1162X CFTR–transduced cells under control and NaBu-treated conditions. Isc in CFBE41o− cells transduced with wild-type CFTR are also shown for comparison (shaded bar). *P < 0.05 compared with W1282X control, ‡P < 0.05 compared with R1162X at same condition, ‡‡P < 0.01 compared with R1162X at same condition, (± SEM).
Enhancement of R1162X and W1282X CFTR Expression by Sodium Butyrate
NaBu has been shown to deacetylate the CMV promoter and enhance expression of transgenes in other model systems (34–36). To better understand the mechanism of the activating effects of NaBu in HeLa and CFBE41o− cells expressing W1282X CFTR, we measured mutant CFTR expression at the mRNA and protein level. Figure 3 demonstrates that under basal conditions, W1282X and R1162X CFTR–transduced cells exhibit slightly greater CFTR mRNA levels relative to endogenous CFTR in Calu-3 cells (Figure 3A); however, protein expression across the three cell lines are qualitatively similar, probably due to differences in processing efficiency between the native and transduced genes (32). Figure 4 summarizes real-time PCR studies comparing R1162X and W1282X CFTR expression in HeLa and CFBE41o− cells. Figure 4A shows dose–response studies with NaBu in transduced HeLa cells, and Figure 4B shows similar studies in CFBE41o− cells expressing R1162X or W1282X CFTR. NaBu led to dose-dependent increases in both R1162X and W1282X CFTR mRNA levels in HeLa cells, with similar enhancing effects seen in CFBE41o− cells. Figure 5 compares truncated CFTR protein levels in the transduced HeLa (Figure 5A) and CFBE41o− (Figure 5B) cells as detected by immunoprecipitation and in vitro phosphorylation (IP). Lysates were probed for truncated CFTR with antibodies directed against the N-terminus or NBD-1, respectively. Truncated CFTR expression was enhanced by NaBu in both cell types. When cell lysates were probed using antibody directed against the C-terminus of CFTR (24-1 antibody), no fully glycosylated full-length CFTR protein was detected under control conditions in either cell line (note absence of band C between 150–200 kD; see Figure 6, control lanes). Together, the results from Figures 1–6 provide evidence that NaBu enhanced the expression of truncated CFTR (without significantly increasing the level of full-length protein) in both W1282X and R1162X CFTR–expressing cells; moreover, only W1282X CFTR–expressing cells demonstrated increased agonist stimulated ion transport after NaBu treatment.
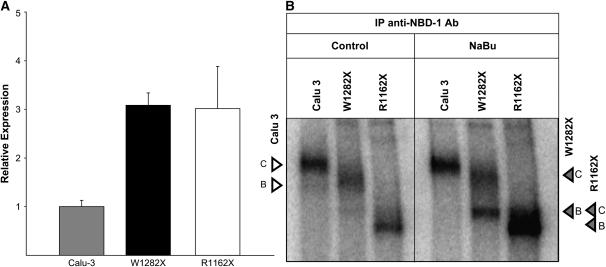
Figure 3.
Relative expression of R1162X and W1282X CFTR in transduced HeLa cells is similar to the Calu-3 type. (A) R1162X and W1282X CFTR transduced HeLa cells grown to confluence and then assayed by RT-PCR after RNA isolation showed mRNA expression within 3-fold compared with endogenously expressing Calu-3 cells. All samples are normalized to endogenous ribosomal (18S) RNA levels, and presented relative to Calu-3. (B) Confluent HeLa and Calu-3 cells probed for total CFTR with N-terminal antibody (H-182) demonstrated qualitatively similar CFTR protein expression levels among all cell types. Arrowheads represent full-length (open arrowheads) and truncated (shaded arrowheads) CFTR bands B and C in each respective cell line.
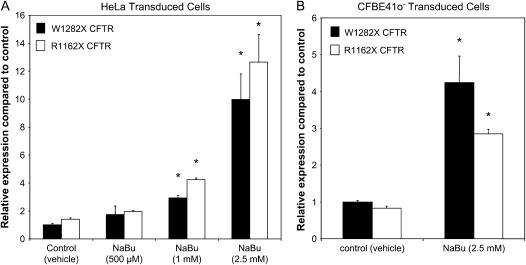
Figure 4.
Relative transcript levels of R1162X and W1282X CFTR in transduced HeLa (A) and CFBE41o− (B) cells. (A) HeLa cells were grown to confluence, treated with various concentrations of NaBu or control (vehicle media) overnight, then assayed by RT-PCR after RNA isolation. All samples are normalized to endogenous ribosomal (18S) RNA levels, and presented relative to controls. W1282X CFTR mRNA levels under control conditions were 72% of R1162X CFTR expression. *P < 0.05 compared with control; n = 6 per condition (± SEM). (B) CFBE41o- cells were grown on permeable supports at an air–liquid interface before RNA isolation and RT-PCR. Cells were then treated with NaBu (2.5 mM) or control (vehicle) conditions. 18S RNA was used as endogenous control. R1162X CFTR mRNA levels under control conditions were 83% of W1282X expression levels. *P < 0.05, n = 3 per condition (± SEM).
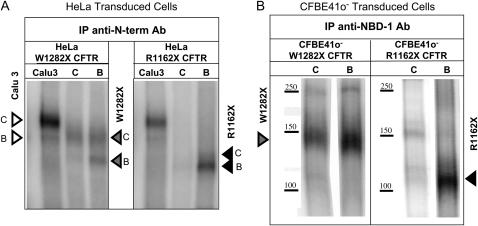
Figure 5.
Detection of truncated R1162X and W1282X CFTR in HeLa (A) and CFBE41o− (B) cells. (A) HeLa Cells were selectively probed for truncated CFTR using an antibody directed against NBD-1 in W1282X and R1162X CFTR–transduced cells. Truncated protein expression (core and fully glycosylated CFTR represented by open arrowheads) was increased after 24 h exposure to NaBu (lane B, 500 μM) compared with control conditions (lane C). Calu-3 cells are also shown as a control for wt protein expression. (B) Similar analysis in CFBE41o− cells. NaBu treatment significantly enhanced W1282X (∼ 140 kD, left panel) or R1162X (∼ 110 kD, right panel) CFTR protein expression. The absence of identifiable band C likely reflect more stringent control of glycosylation in the polarizing cell type (32).
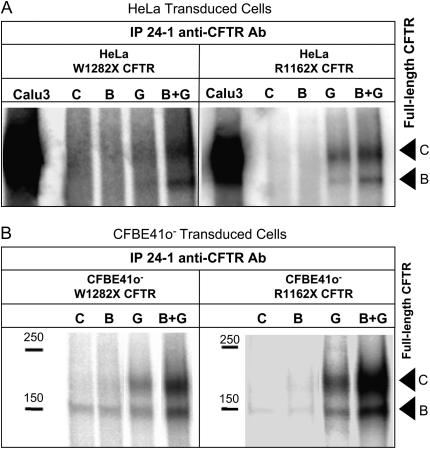
Figure 6.
Full-length CFTR expression after NaBu, G418, and combination treatment in W1282X or R1162X CFTR–transduced HeLa (A) and CFBE41o− (B) cells. (A) Immunoprecipitation with C-terminus–specific antibody (24–1) followed by in vitro phosphorylation of W1282X and R1162X CFTR–transduced HeLa cells is shown. Full-length CFTR (arrows represent bands B and C, respectively) is seen after treatment with NaBu (2.5 mM) and G418 (500 μg/ml) (lane B+G) compared with control (lane C) conditions. Lower levels were also seen with G418 treatment alonge (lane G), but not NaBu (lane B). (B) Similar analysis on CFBE41o- transduced cells demonstrate synergistic effects of stop codon suppression with G418 (500 μg/ml) and increased expression with NaBu (500 μM) in full-length CFTR expression (CFTR band B and band C are demonstrated by arrows). In control cell lysates the faint ∼ 150-kD band likely represents endogenous ΔF508 CFTR expressed in this cell type, as previously described (32).
Synergistic Effects of Stop Codon Suppression and Enhanced Expression in R1162X and W1282X CFTR–Expressing Cells
We next examined the effects of NaBu combined with G418 (an aminoglycoside and previously described potent suppressor of premature stop mutations) on full-length CFTR production in HeLa and CFBE41o− cells transduced with either of the two CFTR premature stop codons of interest. Figure 6 summarizes the effect of the two agents on full-length CFTR protein expression (IP completed with anti-CFTR C-terminal [24-1] antibody). Treatment with the combination of NaBu and G418 for 16 hours led to increased full-length core glycosylated (band B) and fully glycosylated (band C) CFTR production in HeLa (Figure 6A) and CFBE41o− (Figure 6B) cells transduced with R1162X or W1282X CFTR. As noted above, no full-length CFTR was detected under control conditions. Treatment with G418 alone confirmed the stop codon suppressive effect of this agent, producing mature (band C) CFTR, but at lower levels than combination treatment. No readthrough was detected in transduced cells treated with NaBu only.
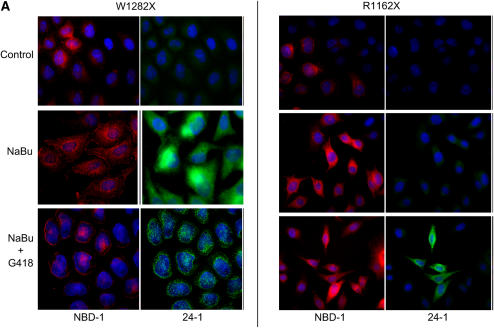
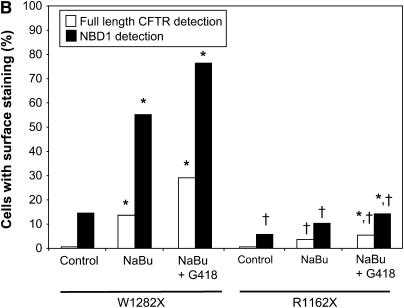
To confirm the presence of truncated W1282X CFTR at the cell surface compared with R1162X-expressing cells, immunofluorescence studies were performed. Results in HeLa cells using anti–NBD-1 and anti–C-terminal CFTR antibodies are summarized in Figure 7. A high proportion of cells positive for surface-localized truncated W1282X CFTR was detectable after treatment with NaBu alone (55% of cells, P < 0.001 versus control, probed with anti–NBD-1 antibody). As anticipated, the greatest number of cells positive for surface localized full-length CFTR was seen after co-treatment with NaBu and G418 (29% of cells, versus < 1% with control and 13% with NaBu treatment alone, P < 0.001 for both comparisons; probed with anti–C-terminal antibody). In contrast, R1162X CFTR–transduced cells demonstrated overall lower surface expression of both full-length and truncated protein (controlled for treatment condition), reflecting less efficient surface localization of truncated protein relative to W1282X CFTR (P < 0.001 by logistic regression analysis). Moreover, the combination of NaBu and G418 produced 5.4% of cells with detectable full-length surface localized CFTR (versus < 0.6% of cells at control conditions [P < 0.01] and 3.7% with NaBu alone [P = NS]).
Figure 7.
NaBu increases surface-localized truncated CFTR expression in W1282X CFTR–expressing cells as detected by immunofluorescence. (A) Immunofluorescence staining of W1282X (left) and R1162X CFTR (right)–expressing HeLa cells after 24 h exposure to NaBu (2.5mM), NaBu + G418 (500 μg/ml), or untreated (vehicle control) conditions. Staining with anti–NBD-1 antibody for truncated CFTR is noted in red, while selective full-length CFTR staining is noted in green (anti–C-terminus antibody, 24–1). W1282X CFTR–transduced cells demonstrate significantly greater number of cells with surface localized truncated CFTR after 24 h treatment with NaBu compared with R1162X cells (55.2% versus 10.4% of cells with surface staining, respectively; P < 0.001). A significant number of cells with surface localized full-length CFTR is detected in both cell lines after the addition of G418, although a greater percentage was seen in W1282X-transduced cells (29.1% of W1282X cells exhibit surface staining after G418 + NaBu versus 0.5% under control conditions, P < 0.001; 5.4% and 0.06% surface staining was seen in R1162X cells, respectively, P < 0.01). (B) Summary figure representing percentage of cells with surface staining evident. Open bars represent full-length CFTR detection, and solid bars represent detection of CFTR with anti–NBD-1 antibody (truncated protein). Between 160 and 190 cells were scored for CFTR surface staining per treatment condition, and totaled over three separate experiments. *P < 0.05 versus control condition for each cell type; †P < 0.001 comparing W1282X versus R1162X cells at exact same treatment condition.
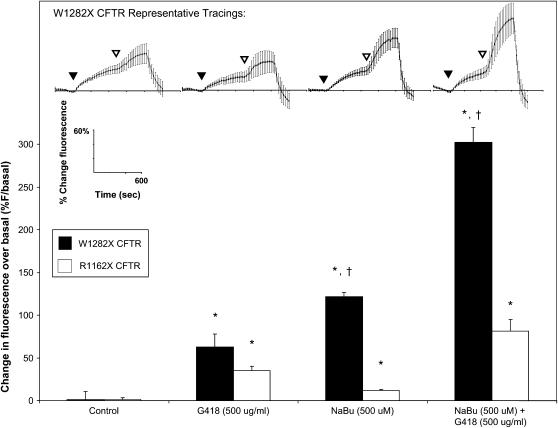
The synergistic effects of G418 and NaBu on W1282X and R1162X CFTR activity were also measured by SPQ (Figure 8). The top portion shows examples of experiments in W1282X CFTR–expressing cells, and the bottom portion summarizes results from multiple paired studies comparing R1162X and W1282X CFTR–expressing cells. Control conditions (no G418 or NaBu treatment) demonstrate minimal agonist-stimulated halide transport. G418 treatment had positive effects on halide transport in both R1162X and W1282X CFTR–expressing cells (reflecting readthrough of PTCs), and NaBu treatment had significantly stronger effects on agonist stimulated halide transport in the W1282X CFTR–expressing cells relative to cells transduced with R1162X CFTR. Combination treatment with G418 and NaBu dramatically increased halide efflux in cells expressing W1282X CFTR, with smaller synergistic effects seen in the R1162X CFTR–transduced cells. Together, the results from Figures 6–8 provide evidence for synergistic effects on CFTR activity and surface localization produced by enhanced expression (NaBu treatment) coupled with stop codon suppression (G418 treatment) in W1282X and R1162X CFTR–transduced cells, and further emphasize apparent differences in the relative susceptibility of these CFTR mutations to functional rescue.
Figure 8.
Additive effects of increased expression (NaBu) and stop codon suppression (G418) on CFTR activity measured by halide transport. CFTR-mediated halide transport in W1282X and R1162X CFTR–transduced HeLa cells studied by SPQ. Top: representative experiments in W1282X CFTR–transduced cells at various conditions. Bottom: change in halide transport after stimulation with forskolin (20 μM) and genistein (50 μM) in W1282X or R1162X CFTR–transduced HeLa cells. Conditions include overnight exposure to G418 (500 μg/ml), NaBu (500 μM), or G418 plus NaBu. Results demonstrate CFTR activity in W1282X CFTR–transduced cells after treatment with NaBu that is not present in R1162X CFTR–transduced cells. *P < 0.001 for treated conditions compared with untreated controls, †P < 0.001 for W1282X versus R1162X under identical conditions, n = 30 cells/condition, ± SEM.
DISCUSSION
In these studies, we examined two proximate, disease-causing mutations caused by PTCs in the distal coding region of CFTR that were expressed in polarizing and nonpolarizing cell systems. We identified distinct differences in the responses of R1162X and W1282X CFTR to treatment designed to enhance transgene expression (NaBu). NaBu increased truncated mRNA levels of both CFTR mutations in a similar fashion within each of the cell lines (Figure 3–4). However, immunohistochemical detection using anti–NBD-1 antibody revealed significantly greater cell surface localization of W1282X CFTR under control and NaBu-treated conditions, even with very low levels of readthrough of full-length CFTR (Figure 7). Furthermore, W1282X (but not R1162X) CFTR activity was partially restored in both cell lines after treatment with NaBu alone (Figures 1, 2, and 8). Treatment with an agent to suppress premature stop codons (G418) led to proportionate increases in full-length CFTR protein from both CFTR mutations in either cell type that was further enhanced by the addition of NaBu (Figures 6–8). Together, our results suggest that increasing the expression of W1282X CFTR was sufficient to restore partial CFTR activity, and this effect was likely due to retained function of truncated W1282X CFTR that localizes to the plasma membrane. In contrast, increasing expression of the truncated R1162X CFTR did not restore functional activity.
The findings reported here raise important questions regarding the post-transcriptional processes responsible for the differences in protein translation and function demonstrated between the two premature stop mutations. Enhanced expression alone (as measured by mRNA levels) was not likely the source of these differences, as the levels of mRNA derived from both cDNA constructs were similar within each of the two cell lines. Because mRNA levels were similar after NaBu treatment for both mutations, enhanced efficiency of cellular processing and localization of W1282X CFTR relative to R1162X likely account for the observed differences.
Recent studies provide evidence that NBD-1 and NBD-2 interact during ATP binding and hydrolysis as part of activation of the CFTR Cl− channel (37, 38), and that NBD-1 and -2 intramolecular interactions are important to the Cl− channel function of full-length CFTR (39, 40). Furthermore, reports by Du and colleagues suggest that the NBD-1 ΔF508 mutation impairs NBD-2 folding, emphasizing the interdependence of the NBD domains to overall protein stability (41). Our studies extend these observations to clinically relevant CFTR mutations. As R1162X CFTR is truncated immediately after the TMD-2 domain and W1282X CFTR is truncated within the NBD-2 domain, our results suggest that critical sequences between R1162X and W1282X may be responsible for functional differences between the mutations. Supporting this hypothesis, Mickle and coworkers reported that a disease-causing CFTR mutation produced by a PTC located 26 amino acids from the C-terminus (S1455X CFTR) retains partial function and demonstrates a mild pulmonary phenotype (but abnormal ion transport in the sweat gland [42]). Our findings are also in agreement with single channel studies recently reported by Wang and colleagues, in which low-level activity was retained in W1282X CFTR expressing HeLa cells without treatments to enhance protein expression (43). Moreover, surface localization of W1282X CFTR in HeLa cells was confirmed using a biotinylation assay. Cui and coworkers have also reported that a similar nonsense mutant (1248X) exhibits evidence of CFTR activity and surface localization, and provide evidence that mutations occurring after 1164–1168 satisfy ER quality control mechanisms (44). Whether a functional equivalent exists to the sharp processing cutoff seen between 1164X and 1168X requires further study, and may identify specific motifs that are critical to the stabilization and function of CFTR.
In previously published studies, our laboratory has reported that high-level expression of the two most common CFTR premature stop mutations G542X and R553X CFTR (using transient vaccinia-based expression in non-polarizing cells) led to constitutive halide transport function lacking cAMP-dependent regulation (8, 27). The current studies extend those findings to two mutations that are inclusive of the R-domain, indicating that negative regulation (by the R-domain) is retained, and CFTR halide transport function requires the addition of CFTR-activating agonists. The studies reported here also used more sophisticated systems, including CFTR cDNAs expressed in paired polarizing and nonpolarizing cell types that could be externally regulated (by NaBu). The similar findings between the two cell types studied here (airway and nonairway) suggest that fundamental differences in cellular processing of R1162X and W1282X CFTR are retained across both model systems. Studies examining PKA-dependent and independent regulation of the two mutations will be needed to determine if cellular differences are also seen in regards to cAMP activation of truncated W1282X CFTR (as is seen with ΔF508 CFTR expressed in these two cell types) (32).
It has also been demonstrated that a PDZ binding motif found at the C-terminus of CFTR (DTRL, positions 1476–1480 of the full-length protein) plays an important role governing apical membrane targeting and retention of CFTR in polarized cells (45). In addition, efficient coupling between CFTR and membrane receptors (β2-adrenergic receptors) are believed to be mediated through PDZ-binding motif interactions between the two membrane proteins and EBP50 (45, 46). Benharouga and colleagues (47) and Gentzsch and Riordan (48) have reported that late CFTR truncations (lacking the PDZ binding motif) may be capable of localizing to the cell membrane, although this finding is controversial (45). Our results are in agreement with these reports, suggesting that additional motifs not located at the C-terminus of CFTR play important roles in localizing CFTR to the cell membrane. In the context of adequate expression, W1282X CFTR function could be demonstrated at the cell membrane of both polarizing and nonpolarizing cell types.
While our studies demonstrate distinct differences in truncated protein activity between R1162X and W1282X CFTR, they do not rule out the possibility that NaBu treatment leads to enhanced levels of background translational readthrough of W1282X CFTR relative to R1162X CFTR. In fact, this is suggested by a higher proportion of W1282X cells with detectable full-length CFTR at the cell surface of NaBu-treated HeLa cells (13.6 versus 3.7% of cells, respectively; Figure 7). Background readthrough is not likely to account for all the findings presented here, as combination treatment with NaBu and G418 led to similar levels of total (surface and nonsurface) full-length CFTR in cell lines expressing either R1162X or W1282X CFTR (Figures 6A and 6B), but NaBu alone led to much larger increases in truncated W1282X CFTR activity (Figures 1 and 2). Further studies directly comparing protein expression and function from CFTR cDNAs containing premature stop codons with and without distal CFTR open reading frames will be necessary to definitively determine the contribution of low-level background readthrough to the activity of W1282X CFTR. In addition, evaluation of primary airway cells isolated from subjects with CF harboring each CFTR mutation would provide additional confirmation of our results obtained in heterologous expression systems.
Taken together, our results are important to the interpretation of clinical studies examining the capacity of small molecule agents to suppress PTCs in human subjects. To date, the strongest suppressive and corrective effects have been seen in studies completed within the Israeli population (16, 17, 20), which is enriched with patients possessing a limited repertoire of premature stop mutations (particularly W1282X CFTR). At least three interpretations may be responsible for the treatment effects seen in these studies relative to others in patients with CF, including (1) enhancement of truncated but functionally active W1282X CFTR expression through inhibition of mRNA degradation by aminoglycoside-induced suppression of nonsense-mediated decay; (2) inherent, CFTR-independent population differences influencing CFTR mRNA stability among different study populations; or (3) differences in the relative suppressibility of Israeli subjects (particularly those subjects harboring W1282X CFTR). For example, although both W1282X and R1162X CFTR are caused by an abnormal UGA stop codon, W1282X is flanked by an adenine (as opposed to guanine in R1162X), a factor that enhances susceptibility to translational readthrough (8). Regardless of the contributor(s), the differences between R1162X and W1282X CFTR activity after enhancement of expression should be considered when interpreting the results of related clinical trials designed to restore CFTR function through suppression of premature stop mutations.
In summary, the results reported here demonstrate that W1282X CFTR function can be partially restored after enhancement of transcription relative to R1162X CFTR. Our findings point toward critical sequences between positions 1,162 and 1,282 in CFTR that are responsible for these differences, and that the activity of distal CFTR stop mutations may be restored by maneuvers that increase expression. The results suggest W1282X may be more susceptible to corrective therapies than other mutations, and should be considered in the interpretation of clinical trials examining agents to suppress premature stop codons as a treatment strategy for genetic diseases.
Acknowledgments
The authors thank Ms. Lijuan Fan, Ms. Meena Statham, and Mr. James Fortenberry for their assistance performing a number of the experiments presented here. They also acknowledge Dr. John Wakefield (Tranzyme, Inc.) and Dr. David Bedwell for facilitating these experiments and numerous useful discussions.
This study was supported by Cystic Fibrosis Foundation grants ROWE05D0, CLANCY06A0, CLANCY96LO, and R464-CR02; Cystic Fibrosis Foundation Therapeutics, Inc. grant CLANCY02Y0; and NIH grants K23 DK075788–01 (to S.M.R.) and P50DK053090 (to E.J.S.)
Originally Published in Press as DOI: 10.1165/rcmb.2006-0176OC on May 31, 2007
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med 2005;352:1992–2001. [DOI] [PubMed] [Google Scholar]
- 2.The Cystic Fibrosis Genotype-Phenotype Consortium. Correlation between genotype and phenotype in patients with cystic fibrosis. N Engl J Med 1993;329:1308–1313. [DOI] [PubMed] [Google Scholar]
- 3.Shoshani T, Kerem E, Szeinberg A, Augarten A, Yahav Y, Cohen D, Rivlin J, Tal A, Kerem B. Similar levels of mRNA from the W1282X and the delta F508 cystic fibrosis alleles, in nasal epithelial cells. J Clin Invest 1994;93:1502–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 1999;8:1893–1900. [DOI] [PubMed] [Google Scholar]
- 5.Kerem E, Kerem B. Genotype-phenotype correlations in cystic fibrosis. Pediatr Pulmonol 1996;22:387–395. [DOI] [PubMed] [Google Scholar]
- 6.Keeling KM, Brooks DA, Hopwood JJ, Li P, Thompson JN, Bedwell DM. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum Mol Genet 2001;10:291–299. [DOI] [PubMed] [Google Scholar]
- 7.Howard M, Frizzell RA, Bedwell DM. Aminoglycoside antibiotics restore CFTR function by overcoming premature stop mutations. Nat Med 1996;2:467–469. [DOI] [PubMed] [Google Scholar]
- 8.Bedwell DM, Kaenjak A, Benos DJ, Bebok Z, Bubien JK, Hong J, Tousson A, Clancy JP, Sorscher EJ. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat Med 1997;3:1280–1284. [DOI] [PubMed] [Google Scholar]
- 9.Tok JB, Bi L. Aminoglycoside and its derivatives as ligands to target the ribosome. Curr Top Med Chem 2003;3:1001–1019. [DOI] [PubMed] [Google Scholar]
- 10.Bonetti B, Fu L, Moon J, Bedwell DM. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol 1995;251:334–345. [DOI] [PubMed] [Google Scholar]
- 11.Barton-Davis ER, Cordier L, Shoturma DI, Leland SE, Sweeney HL. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest 1999;104:375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA 2000;6:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helip-Wooley A, Park MA, Lemons RM, Thoene JG. Expression of CTNS alleles: subcellular localization and aminoglycoside correction in vitro. Mol Genet Metab 2002;75:128–133. [DOI] [PubMed] [Google Scholar]
- 14.Sleat DE, Sohar I, Gin RM, Lobel P. Aminoglycoside-mediated suppression of nonsense mutations in late infantile neuronal ceroid lipofuscinosis. Eur J Paediatr Neurol 2001;5:57–62. [DOI] [PubMed] [Google Scholar]
- 15.Keeling KM, Bedwell DM. Clinically relevant aminoglycosides can suppress disease-associated premature stop mutations in the IDUA and P53 cDNAs in a mammalian translation system. J Mol Med 2002;80:367–376. [DOI] [PubMed] [Google Scholar]
- 16.Wilschanski M, Yahav Y, Yaacov Y, Blau H, Bentur L, Rivlin J, Aviram M, Bdolah-Abram T, Bebok Z, Shushi L, et al. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med 2003;349:1433–1441. [DOI] [PubMed] [Google Scholar]
- 17.Wilschanski M, Famini C, Blau H, Rivlin J, Augarten A, Avital A, Kerem B, Kerem E. A pilot study of the effect of gentamicin on nasal potential difference measurements in cystic fibrosis patients carrying stop mutations. Am J Respir Crit Care Med 2000;161:860–865. [DOI] [PubMed] [Google Scholar]
- 18.Clancy JP, Rowe SM, Bebok Z, Aitken ML, Gibson R, Zeitlin P, Berclaz P, Moss R, Knowles MR, Oster RA, et al. No detectable improvements in cystic fibrosis transmembrane conductance regulator by nasal aminoglycosides in patients with cystic fibrosis with stop mutations. Am J Respir Cell Mol Biol 2007;37:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clancy JP, Konstan MW, Rowe SM, Accurso F, Zeitlin P, Moss R, Bebok Z, Northcutt VJ, Elfring GL, Miller L, et al. A phase II study of PTC124 in CF patients harboring premature stop mutations [abstract]. Ped Pulmonol Suppl 2006;41:A269. [Google Scholar]
- 20.Kerem E, Hirawat S, Armoni S, Yaacov Y, Blau H, Rivlin J, Aviram M, Shoseyov D, Cohen M, Northcutt VJ, et al. PTC124 activity in CF patients carrying stop mutations: interim results of a phase II study [abstract]. Ped Pulmonol Suppl 2006;41:A242. [Google Scholar]
- 21.Ainsworth C. Nonsense mutations: running the red light. Nature 2005;438:726–728. [DOI] [PubMed] [Google Scholar]
- 22.Linde L, Boelz S, Nissim-Rafinia M, Oren YS, Wilschanski M, Yaacov Y, Virgilis D, Neu-Yilik G, Kulozik AE, Kerem E, et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest 2007;117:683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shushi L, Nissim-Rafinia M, Natan C, Kerem E, Wilschanski M, Yaacov Y, Rivlin J, Yahav Y, Blau H, Bentur L, et al. NMD pathway regulates the response to aminoglycoside treatment. Pediatr Pulmonol Suppl 2004;38:175–176. [Google Scholar]
- 24.Kopelowitz J, Hampe C, Goldman R, Reches M, Engelberg-Kulka H. Influence of codon context on UGA suppression and readthrough. J Mol Biol 1992;225:261–269. [DOI] [PubMed] [Google Scholar]
- 25.Poole ES, Brown CM, Tate WP. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J 1995;14:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Wakefield JK, Liu H, Xiao H, Kralovics R, Prchal JT, Kappes JC. Development of a novel trans-lentiviral vector that affords predictable safety. Mol Ther 2000;2:47–55. [DOI] [PubMed] [Google Scholar]
- 27.Clancy JP, Hong JS, Bebok Z, King SA, Demolombe S, Bedwell DM, Sorscher EJ. Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain 1 (NBD-1) and CFTR truncated within NBD-1 target to the epithelial plasma membrane and increase anion permeability. Biochemistry 1998;37:15222–15230. [DOI] [PubMed] [Google Scholar]
- 28.Bebok Z, Venglarik CJ, Panczel Z, Jilling T, Kirk KL, Sorscher EJ. Activation of DeltaF508 CFTR in an epithelial monolayer. Am J Physiol 1998;275:C599–C607. [DOI] [PubMed] [Google Scholar]
- 29.Cobb BR, Ruiz F, King CM, Fortenberry J, Greer H, Kovacs T, Sorscher EJ, Clancy JP. A(2) adenosine receptors regulate CFTR through PKA and PLA(2). Am J Physiol Lung Cell Mol Physiol 2002;282:L12–L25. [DOI] [PubMed] [Google Scholar]
- 30.Clancy JP, Ruiz FE, Sorscher EJ. Adenosine and its nucleotides activate wild-type and R117H CFTR through an A2B receptor-coupled pathway. Am J Physiol 1999;276:C361–C369. [DOI] [PubMed] [Google Scholar]
- 31.Hentchel-Franks K, Lozano D, Eubanks-Tarn VL, Cobb BR, Fan L, Oster R, Sorscher EJ, Clancy JP. Activation of airway Cl- secretion in human subjects by adenosine. Am J Respir Cell Mol Biol 2004;31:140–146. [DOI] [PubMed] [Google Scholar]
- 32.Bebok Z, Collawn JF, Wakefield J, Parker W, Li Y, Varga K, Sorscher EJ, Clancy JP. Failure of cAMP agonists to activate rescued deltaF508 CFTR in CFBE41o- airway epithelial monolayers. J Physiol 2005;569:601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clancy JP, Bebok Z, Ruiz F, King C, Jones J, Walker L, Greer H, Hong J, Wing L, Macaluso M, et al. Evidence that systemic gentamicin suppresses premature stop mutations in patients with cystic fibrosis. Am J Respir Crit Care Med 2001;163:1683–1692. [DOI] [PubMed] [Google Scholar]
- 34.Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol 1997;137:1211–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wacker I, Kaether C, Kromer A, Migala A, Almers W, Gerdes HH. Microtubule-dependent transport of secretory vesicles visualized in real time with a GFP-tagged secretory protein. J Cell Sci 1997;110:1453–1463. [DOI] [PubMed] [Google Scholar]
- 36.Dorner AJ, Wasley LC, Kaufman RJ. Increased synthesis of secreted proteins induces expression of glucose-regulated proteins in butyrate-treated Chinese hamster ovary cells. J Biol Chem 1989;264:20602–20607. [PubMed] [Google Scholar]
- 37.Ikuma M, Welsh MJ. Regulation of CFTR Cl- channel gating by ATP binding and hydrolysis. Proc Natl Acad Sci USA 2000;97:8675–8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger AL, Ikuma M, Welsh MJ. Normal gating of CFTR requires ATP binding to both nucleotide-binding domains and hydrolysis at the second nucleotide-binding domain. Proc Natl Acad Sci USA 2005;102:455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Csanady L, Chan KW, Nairn AC, Gadsby DC. Functional roles of nonconserved structural segments in CFTR's NH2-terminal nucleotide binding domain. J Gen Physiol 2005;125:43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vergani P, Nairn AC, Gadsby DC. On the mechanism of MgATP-dependent gating of CFTR Cl- channels. J Gen Physiol 2003;121:17–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du K, Sharma M, Lukacs GL. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol 2005;12:17–25. [DOI] [PubMed] [Google Scholar]
- 42.Mickle JE, Macek M Jr, Fulmer-Smentek SB, Egan MM, Schwiebert E, Guggino W, Moss R, Cutting GR. A mutation in the cystic fibrosis transmembrane conductance regulator gene associated with elevated sweat chloride concentrations in the absence of cystic fibrosis. Hum Mol Genet 1998;7:729–735. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Bernard K, Li G, Kirk KL. Curcumin opens cystic fibrosis transmembrane conductance regulator channels by a novel mechanism that requires neither ATP binding nor dimerization of the nucleotide-binding domains. J Biol Chem 2007;282:4533–4544. [DOI] [PubMed] [Google Scholar]
- 44.Cui L, Aleksandrov L, Chang XB, Hou YX, He L, Hegedus T, Gentzsch M, Aleksandrov A, Balch WE, Riordan JR. Domain interdependence in the biosynthetic assembly of CFTR. J Mol Biol 2007;365:981–994. [DOI] [PubMed] [Google Scholar]
- 45.Moyer BD, Duhaime M, Shaw C, Denton J, Reynolds D, Karlson KH, Pfeiffer J, Wang S, Mickle JE, Milewski M, et al. The PDZ-interacting domain of cystic fibrosis transmembrane conductance regulator is required for functional expression in the apical plasma membrane. J Biol Chem 2000;275:27069–27074. [DOI] [PubMed] [Google Scholar]
- 46.Naren AP, Cobb B, Li C, Roy K, Nelson D, Heda GD, Liao J, Kirk KL, Sorscher EJ, Hanrahan J, et al. A macromolecular complex of beta 2 adrenergic receptor, CFTR, and ezrin/radixin/moesin-binding phosphoprotein 50 is regulated by PKA. Proc Natl Acad Sci USA 2003;100:342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benharouga M, Sharma M, So J, Haardt M, Drzymala L, Popov M, Schwapach B, Grinstein S, Du K, Lukacs GL. The role of the C terminus and Na+/H+ exchanger regulatory factor in the functional expression of cystic fibrosis transmembrane conductance regulator in nonpolarized cells and epithelia. J Biol Chem 2003;278:22079–22089. [DOI] [PubMed] [Google Scholar]
- 48.Gentzsch M, Riordan JR. Localization of sequences within the C-terminal domain of the cystic fibrosis transmembrane conductance regulator which impact maturation and stability. J Biol Chem 2001;276:1291–1298. [DOI] [PubMed] [Google Scholar]