Abstract
Expression of transforming growth factor α (TGF-α) in the respiratory epithelium of transgenic mice caused pulmonary fibrosis, cachexia, pulmonary hypertension, and altered lung function. To identify genes and molecular pathways mediating lung remodeling, mRNA microarray analysis was performed at multiple times after TGF-α expression and revealed changes consistent with a role for TGF-α in the regulation of extracellular matrix and vasculogenesis. Transcripts for extracellular matrix proteins were augmented along with transcripts for genes previously identified to have roles in pulmonary fibrosis, including tenascin C, osteopontin, and serine (or cysteine) peptidase inhibitor, clade F, member 1. Transcripts regulating vascular processes including endothelin receptor type B, endothelial-specific receptor tyrosine kinase, and caveolin, caveolae protein 1 were decreased. When TGF-α expression was no longer induced, lung remodeling partially reversed and lung function and pulmonary hypertension normalized. Transcripts increased during resolution included midkine, matrix metalloproteinase 2, and hemolytic complement. Hierarchical clustering revealed that genes regulated by TGF-α were similar to those altered in the lungs of patients with idiopathic pulmonary fibrosis. These studies support a role for epithelial cell–derived TGF-α in the regulation of processes that alter the airway and vascular architecture and function.
Keywords: epidermal growth factor receptor, idiopathic pulmonary fibrosis, vasculogenesis, angiogenesis, interstitial lung disease
CLINICAL RELEVANCE
This study demonstrates that pulmonary fibrosis induced through epidermal growth factor receptor (EGFR) activation is reversible. Targeting the EGFR may prove to be a target to reverse fibrotic disease.
Interstitial lung disease (ILD), characterized by fibroblast proliferation and excessive collagen deposition, is a heterogeneous group of severe pulmonary disorders (1). Idiopathic pulmonary fibrosis (IPF) is the most common form of ILD that affects ∼ 90,000 people in the United States, with an estimated 34,000 new diagnoses occurring each year (2, 3). The mortality for IPF is 60–70% within 5 years of diagnosis (2). Currently, no effective treatments exist for IPF or other fibrotic lung diseases, emphasizing the need to identify additional therapeutic targets.
Previous studies suggested that pulmonary fibrosis in diseases such as IPF results from a chronic inflammatory process that initiates focal accumulation of extracellular matrix in the interstitium (4). The role of inflammation in initiating and propagating pulmonary fibrosis has been debated as clinical measurements of inflammation fail to correlate with disease severity in IPF and anti-inflammatory therapy does not improve disease outcomes. An alternative hypothesis proposes that pulmonary epithelial injury is an initiating event and leads to abnormal wound healing with excessive extracellular matrix formation that is mediated by epithelial–mesenchymal interactions. Several murine models have been used to study the pathogenesis of pulmonary fibrosis, including intratracheal bleomycin-induced lung fibrosis and the expression of various injurious or inflammatory proteins in the lung.
The epidermal growth factor receptor (EGFR) belongs to a receptor tyrosine kinases protein family with the related proteins: HER2/neu, HER3, and HER4. Depending on the activating ligand, EGFR family members form various homo- or heterodimers with different biological capacities (5, 6). Transforming growth factor-α (TGF-α) is a ligand for the EGFR, and constitutive or conditional expression of TGF-α in the respiratory epithelium of transgenic mice caused peribronchial, perivascular, and pleural fibrosis in the absence of inflammation (7–9). Fibrosis in constitutive TGF-α transgenic mice is prevented in mice in which the EGFR is genetically disrupted, supporting TGF-α as inducing fibrotic lesions primarily through the EGFR signaling pathway (10). To identify genes and pathways influenced by TGF-α during the progression of pulmonary disease in the mouse, we correlated changes in mRNA levels with histologic and functional changes in fibrosis and pulmonary hypertension. Finally, we compared mRNAs altered by TGF-α to those altered by intratracheal bleomycin in mouse lung or to those altered in idiopathic pulmonary fibrosis in human patients.
MATERIALS AND METHODS
Transgenic Mice
We used CCSP-rtTA activator mice, which express the reverse tetracycline-responsive transactivator (rtTA) fusion protein under control of the 2.3-kb rat Clara Cell Secretory Protein (CCSP, a.k.a. secretoglobin, family 1A, member 1 [uteroglobin]) gene promoter (11). CCSP-rtTA activator mice were mated to conditional doxycycline (Dox)-regulated transgenic mice containing the human TGF-α cDNA under the control of seven copies of the tetracycline Operon ((TetO)7-cmv TGF-α) plus a minimal CMV promoter (8). Single transgene (CCSP-rtTA+/−) and bitransgenic (CCSP-rtTA+/−/(TetO)7-cmv TGFα+/−) mice were produced within the same litter by mating homozygous CCSP-rtTA+/+ mice to hemizygous (TetO)7-cmv TGFα+/− mice. All mice were derived from the FVB/NJ inbred strain. Mice were maintained in virus-free containment and handled in accordance with the Institutional Animal Use and Care Committee of the Children's Hospital Research Foundation. To induce TGF-α expression, Dox (Sigma, St. Louis, MO) was administered in the drinking water at a final concentration of 0.5 mg/ml with 1% ethanol and in food (62.5 mg/kg). Water was replaced three times per week. Mice were treated with Dox supplemental water and food for up to 56 d and then placed on regular water and food for up to an additional 210 d. Body weights were measured and weekly values (means ± SEM) were considered different from time-matched controls not receiving Dox using a significance (P < 0.05) determined by testing for normality and using a Kruskal-Wallis one way ANOVA followed by an all pairwise multiple comparison procedure (Dunn's Method).
Mice were killed with pentobarbital sodium (65 mg/ml) euthanasia solution (Fort Dodge Animal Health, Fort Dodge, IA) for analysis at selected intervals on and off Dox supplemental water and food.
Lung Histology and Immunohistochemistry
To assess lung morphology and collagen deposition, lungs were inflation-fixed using a phosphate-buffered saline (PBS) solution containing 4% paraformaldehyde at 25 cm H2O pressure (overnight, 4°C), washed with PBS, dehydrated through a graded series of ethanol washes, and embedded in paraffin. Sections (5 μm) were placed onto polysine slides for immunohistochemistry. Sections were stained with hematoxalin and eosin, Gomori's trichrome stain, or pentachrome for detection of collagen and extracellular matrix deposition (12).
Protein Analyses
Mice were killed and lungs were removed and homogenized (Tissue Tearor; Biospec Products, Bartlesville, OK) in 2 ml PBS (pH 7.4) containing protease inhibitors (“Complete” protease inhibitor cocktail; Roche, Indianapolis, IN) then centrifuged (10 min, 1000 × g; 4°C) to remove insoluble debris. Supernatants were diluted 1:2 in PBS containing protease inhibitors and concentrations of TGF-α in the lungs of transgenic mice on Dox, and the times for TGF-α concentration to return to control levels after Dox was removed were determined by a human TGF-α enzyme-linked immunosorbent assay (ELISA) (Oncogene Research Products, San Diego, CA). Lung homogenate protein levels were determined with a bicinchoninic acid assay (Sigma). TGF-α concentrations in lung homogenates (pg/ml) were normalized to total lung protein levels (mg/ml) and values (mean ± SEM n = 4–5 mice/group) were assessed by a Kruskal-Wallis One Way Analysis of Variance on Ranks followed by an All Pairwise Multiple Comparison Procedure using the Student-Newman-Keuls Method.
Total lung collagen was determined by quantifying total soluble collagen using the Sircol Collagen Assay kit (Biocolor, Newtownabbey, Ireland). The right lung was homogenized in PBS with protease inhibitors. Sircol dye reagent (1 ml) that binds to collagen was added to 100 μl of each sample and mixed for 30 min. After centrifugation the pellet was suspended in 1 ml of alkali reagent (0.5 M NaOH) and optical density measured at 540 nm with a spectrophotometer. The optical density in the test samples were compared with the values obtained with collagen standard solutions provided by the manufacturer. The values (means ± SEM) in the test samples on and off Dox were compared with initial control using ANOVA followed by a Student-Newman-Keuls all pairwise comparison to identify significant differences (P < 0.05).
Pulmonary Mechanics
Lung mechanics were assessed on mice with a computerized Flexi Vent system (SCIREQ, Montreal, PQ, Canada), as previously described (13). Mice were anesthetized intraperitoneally with 0.1 ml /10 g body weight PBS solution containing 178 mM (40 mg/ml) ketamine and 7.8 mM (2 mg/ml) xylazine. To determine dynamic lung compliance, mice were tracheostomized and ventilated with a tidal volume of 8 ml/kg at a rate of 450 breaths/min and positive end-expiratory pressure (PEEP) of 2 cm H2O (13). To determine respiratory impedance, the ventilation mode was changed to forced oscillatory signal (0.5–19.6 Hz). Tissue resistance or damping and tissue elastance was obtained for mice at 2 cm H2O PEEP by fitting a model to each impedance spectrum (13). Values were corrected for the impedance of the equipment and tracheal tube. Hysteresivity describes the mechanical coupling between tissue damping and elastance and was calculated as tissue damping/tissue elastance (13). All pulmonary mechanism measurements were compared between groups using ANOVA with Tukey-Kramer multiple comparison test to identify significant differences (P < 0.05).
Microarray Hybridization and Data Analysis
Differential gene expression was assessed at Days 1, 4, 21, and 42 after Dox was added and Days 1, 4, 12, 21, and 42 during recovery when Dox was discontinued. For each time point of the experiment, a total of five Dox animals were analyzed (except 4 d and 6 wk off, when four mice were assessed). Data were normalized and compared with mice not receiving Dox. There were five independent control mice samples for the first half of the experiment, and five separate control mice samples for the second half (recovery period). Each microarray compared the lung RNA from one Dox-treated and one control mouse; thus, biological replicates were used throughout the study. Microarrays (n = 4–5/time) were performed using a mouse library consisting of 13,443 70-mer oligonucleotides (Version 1.1; Qiagen; Alameda, CA). The library was suspended in 3× saline-sodium citrate (SSC: 20× = 0.3 M sodium citrate, 3 M sodium chloride, pH 7.0, Cat. No. BP1325–4; FisherBiotech, Hampton, NH) at 30 μM and printed (Omnigrid; GeneMachines, San Carlos, CA equipped with Stealth SMP3 pins; Telechem, Sunnyvale, CA) onto aminosilane-coated slides (Cel Associates, Inc., Pearland, TX) (22°C, 65% relative humidity). Target volumes were 0.5 nl and spot diameters are 75–85 μm. The oligonucleotides were cross-linked to the slide substrate by exposure to 600 mJ of ultraviolet light.
Fluorescence-labeled cDNAs were synthesized from lung RNA using an indirect amino allyl labeling method via an oligo(dT)-primed, reverse transcriptase reaction. The cDNA was decorated with monofunctional reactive Cyanine-3 (Cy3) or Cyanine-5 (Cy5) dyes (Amersham, Piscataway, NJ). The microarrayed DNA probes were incubated in prehybridization buffer (5× SSC, 0.1% sodium dodecyl sulfate [SDS: Cat. No. 0837; Amresco, Solon, OH], and 1% bovine serum albumin) (42°C; 90 min). For hybridization, the microarray slides were spotted with 42 μl hybridization buffer (25% formamide, 5× SSC, 0.1% SDS, 5 μg/μl highly repetitive calf thymus DNA [COT-1: Cat. No. D-8661; Sigma], 5 μg/μl poly(A)-DNA [Cat. No. P 9403; Sigma] and 2 μg/μl yeast tRNA [Cat. No. R 8759; Sigma]) and the fluorescence-labeled target cDNA, covered with glass coverslips (Fisher Scientific, Pittsburgh, PA), and placed in humidified hybridization chambers (Corning, Acton, MA) set in a water bath (48°C; 66 h). The slides were serially washed with agitation in 1× SSC with 0.2% SDS (48°C; 4 min), 0.1× SSC and 0.2% SDS (23°C; 4 min), and twice 0.1× SSC (23°C; 4 min each). The slides were spun-dried immediately after washing. The microarray slides were scanned with dual lasers with wavelength frequencies to excite Cy3 and Cy5 fluorescence emittance; images were captured in JPEG and TIFF files, and DNA spots were captured by the adaptive circle segmentation method (GenePix 4000A and 4000B; Axon Instruments; Union City, CA with software from Axon Instruments, Inc.; Foster City, CA). Information extraction for a given spot is based on the median value for the signal pixels minus the median value for the background pixels to produce a gene set data file for all the DNA spots (GenePix Pro version 5.0 software).
Data normalization was performed in two steps for each microarray separately (14). First, background-adjusted intensities were log-transformed and the differences (R) and averages (A) of log-transformed values were calculated as R = log2(X1) − log2(X2) and A = [log2(X1) + log2(X2)]/2, where X1 and X2 denote the Cy5 and Cy3 intensities after subtracting local backgrounds, respectively. Second, data centering was performed by fitting the array-specific local regression model of R as a function of A. The difference between the observed log-ratio and the corresponding fitted value represented the normalized log-transformed gene expression ratio. Normalized log-intensities for the two channels were calculated by adding half of the normalized ratio to A for the Cy5 channel and subtracting half of the normalized ratio from A for the Cy3 channel. The statistical analysis was performed for each gene separately by fitting the following mixed effects linear model (15, 16): Yijk = μ + Ai + Sj + Ck+ εijk, where Yijk is the normalized log-intensity on the ith array, with the jth treatment time, and labeled with the kth dye (k = 1 for Cy5, and 2 for Cy3), μ is the overall mean log-intensity, Ai is the effect of the ith array, Sj is the effect of the jth treatment, and Ck is the effect of the kth dye. Assumptions about model parameters are the same as described (15), with array effects assumed to be random and treatment and dye effects assumed to be fixed. Differential transcript levels were calculated from the mixed model by estimating the appropriate contrasts (treated versus control at each time-point). T-statistics were also obtained from these contrasts to calculate P values. Statistical significance of differential expression was assessed by calculating P values, and adjusting for multiple hypotheses testing by calculating False Discovery Rates (FDR) and Expected False Positives (EFP) (17). Estimates of fold-change were considered significant when ≥ 2-fold different from control with EFP ≤ 5, and average spot intensity level ≥ 150. Data normalization and statistical analyses were performed using SAS statistical software package (SAS Institute Inc., Cary, NC).
Bayesian infinite mixture (BIM) model–based clustering (18) was performed using expression ratios of experimental replicates (19) for each time comparison after adjusting for the gene-specific dye effects (estimated from the linear model described above). BIM model–based clustering allows fitting of the statistical mixture model without knowing the number of clusters in the data. The statistical model was fitted and hierarchical clustering was produced by treating pairwise posterior probabilities as the similarity measure and applying the traditional complete-linkage principle. In this situation, the BIM clustering produced higher quality clusters than the traditional heuristic methods. The clustering results were displayed using TreeView (20).
Genes in individual clusters (four with increased expression levels and two clusters with decreased expression levels) were tested independently for overrepresentation of annotation classes using Expression Analysis Systematic Explorer (EASE) (21). EASE analyses tests each subset list against the population of all genes detected, and calculates the likelihood of overrepresentation in the Gene Ontology Consortium annotation categories GO biological process, GO cell component, and GO molecular function, as well as KEGG pathway and SwissProt keyword. Fisher's Exact probability, using the Benjamini FDR adjustment, was calculated for each category (21), and categories that contained > 2 members and had a Benjamini FDR < 0.05 were considered significant. Subsets with fewer members of larger categories were combined into the larger category. The overall temporal behavior of significant categories was displayed as number of significant members (independent variable) versus time (dependent variable).
Quantitative Real-Time Polymerase Chain Reaction
Fifteen transcripts identified by microarray were also measured by quantitative real-time polymerase chain reaction (qRT-PCR). Total RNA was isolated from cells with TRIzol reagent (Invitrogen, Carlsbad, CA), and quantity was assessed by A260/A280 spectrophotometric absorbance (SmartSpec 3000; Bio-Rad, Hercules, CA). RNA quality was assessed by separation with a denaturing formaldehyde/agarose/ethidium bromide gel, and quantified by analysis with an Agilent Bioanalyzer (Quantum Analytics, Inc., Foster City, CA). For qRT-PCR, 100 ng lung RNA from mouse lung mice (n = 3–4 mice/group) was reverse transcribed into first-strand cDNA using a High Capacity cDNA Archive Kit (Applied Biosytems, Foster City, CA) in a 100-μl reaction volume. cDNA (10 μl) was used in a subsequent PCR reaction using 25 μl TaqMan Universal PCR Master Mix (Applied Biosystems), 2.5 μl of each primer mixture, and 12.5 μl RNAse-free water. Primer mixtures for COL1A1 (Cat. No. Mm00801666_g1), COL3A1 (Cat. No. Mm00802331_m1), TIMP1 (Cat. No. Mm00441818_m1), and TNC (Cat. No. Mm00495662_m1) SPP1 (Cat. No. Mm00436767_m1), ROR2 (Cat. No. Mm00443470_m1), FZD1 (Cat. No. Mm00445405_s1), MDK (Cat. No. Mm00440279_m1) CAV1 (Cat. No. Mm00483057_m1), EDNRB (Cat. No. Mm00432989_m1), PECAM1 (Cat. No. Mm00476702_m1), FOXF1A (Cat. No. Mm00487497_m1), SOX18 (Cat. No. Mm00656049_gH), SOD3 (Cat. No. Mm00448831_m1), and TEK (Cat. Mm00607939_s1) were purchased from Applied Biosystems, and analysis performed with an Applied Biosystems 7900HT System using the following conditions: 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. For relative quantization of expression of each transcript between the 42 Days On were compared with nontreated controls (No Dox), the comparative Ct method (ΔΔCt) was used. ΔCt = Ct (gene of interest) − Ct (ACTB), and this value was calculated for each sample where Ct = cycle number threshold. The comparative ΔΔCT calculation involved finding the difference between each sample's ΔCt and the mean ΔCt for the control sample. These values were transformed to absolute values using the formula: comparative expression level (fold change) = 2−ΔΔCt. An ANOVA was performed to evaluate the fold change comparing 42 Days On to No Dox followed by a Student-Newman-Keuls all pairwise multiple comparison to identify those differences that were significant (P < 0.05).
Pulmonary Vascular Pressure and Ventricular Hypertrophy
Pulmonary hypertension was assessed by measuring both right ventricular hypertrophy (RVH) and pulmonary systolic pressures. RVH was assessed as previously described (22). Briefly, hearts were removed and dissected to isolate the free wall of the RV from the left ventricle and septum (LV + S). The ratios of RV weight to LV + S weight (RV/LV + S) were used as an index of RVH, which develops as a result of pulmonary hypertension. Values (mean ± SEM) for RV/LV + S were compared between groups using ANOVA followed by a Student-Newman-Keuls all pair-wise multiple comparison to identify significant differences (P < 0.05).
To measure pulmonary pressures, mice were anesthetized with intraperitoneal ketamine (6 mg/100 g) and xylazine (1 mg/100 g). The trachea was cannulated, and the lungs were ventilated with room air at a tidal volume of 0.2 ml and a rate of 90 breaths/min. A 26-gauge needle was then introduced percutaneously into the right ventricle via the subxiphoid approach. Right ventricular systolic pressure was measured using a Gould P10 EZ pressure transducer connected to pressure modules and a Gould TA 550 recorder. The heart rate under these conditions was between 300 and 500 beats per minute (bpm). If the heart rate fell below 300 bpm, measurements were excluded from analysis.
Identification of Transcripts Altered in Mice Treated with TGF-α, Bleomycin, and in Patients with IPF
Average linkage, hierarchical clustering using the uncentered correlation similarity metric, among TGF-α mice, bleomycin-treated mice, and human patients with IPF was performed on homologous transcripts. Performed with Affymetrix GeneChip Murine Genome U74 Version 2 Set MG-U74A, the Gene Expression Omnibus (GEO) dataset (GDS251), compared the lung RNA levels following bleomycin treatment of susceptible C57BL/6J (C57) and resistant BALB/cJ (Balb) mouse strains. Sampling was performed at 1, 3, and 14 d after treatment. Performed with Amersham Biosciences CodeLink Uniset Human I Bioarray, the GDS1252 dataset consisted of archived lung tissues of patients with IPF that were obtained from the tissue bank of the Department of Pathology at the University of Pittsburgh (23). The diagnosis of IPF was confirmed by open lung biopsy. These patients fulfilled the criteria of the American Thoracic Society and European Respiratory Society for the diagnosis of IPF (n = 6). Normal histology lung tissues resected from patients with lung cancer were used as controls (n = 11). To calculate differential gene expression in the human IPF and mouse bleomycin experiments, the two expression datasets were downloaded directly from GEO and analyzed using Bioconductor in R statistical software. Estimated fold changes were calculated for both datasets using the average log-base-2 difference. To ensure an unbiased comparison of the two mouse models to human IPF, only transcripts that were differentially expressed in the human IPF study and that had known mouse orthologs in the bleomycin and TGF-α datasets were included in the clustering. To calculate the differentially expressed genes in the human IPF dataset and thus identify transcripts for clustering, we used a simple ANOVA model using the limma package of Bioconductor, with an inclustion criteria of an FDR < 0.05 (17). This resulted in a total of 142 transcripts for clustering. Ortholog transcripts were identified by comparing the National Center for Biological Information (NCBI) Entrez Gene IDs for elements identified as significantly different for GEO dataset GDS1252.
RESULTS
Conditional Regulation of Pulmonary Fibrosis
TGF-α protein levels were controlled by the administration of Dox (Figure 1). Pulmonary TGF-α protein concentrations were increased by 6 wk of Dox treatment and decreased within 4 d after removal from Dox. Expression of TGF-α caused fibrotic lesions in the peribronchial and perivascular adventitia (Figure 2A) that began to resolve 4 d after removal from Dox. Twelve days after discontinuing Dox, fibrosis continues to diminish and is almost completely resolved in the perivascular regions 42 d after discontinuation of Dox. Peribronchial and perivascular fibrosis resolved completely 126 d after removal from Dox (data not shown). While pleural fibrosis was decreased 21 d after Dox withdrawal, pleural fibrosis did not completely resolve (Figure 2B).
Figure 1.
Conditional regulation of TGF-α expression. Bitransgenic mice expressing human TGF-α under control of the reverse tetracycline-responsive transactivator (rtTA) fusion protein were treated with Dox supplemental water and food for 42 d (On Dox) and then provided Dox-free water and food for up to 42 d (Off Dox: days −1 to −42). At indicated times, lungs were removed and homogenized, and TGF-α protein levels measured by ELISA. TGFα levels were normalized to total protein and are expressed as means (± SEM). Values were compared with untreated control (No Dox) by a Kruskal-Wallis one-way ANOVA on Ranks followed by an All Pairwise Multiple Comparison Procedure using the Student-Newman-Keuls method. *Greater than no Dox; **greater than controls and less than Day 42, on Dox; ***less than no Dox.
Figure 2.
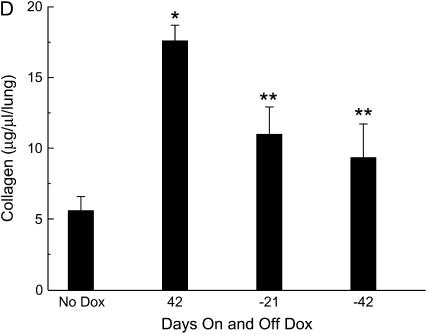
Reversibility of TGF-α–induced pulmonary fibrosis. (A) Reversible peribronchial and perivascular fibrosis. In regulatable TGF-α mice after 6 wk of Dox, extensive peribronchial, peribronchiolar, and perivascular fibrosis developed in the lung. Within 4 d of discontinuing Dox, less fibrosis is detected in the perivascular and peribronchial regions. Twelve days after discontinuing Dox, fibrosis continues to diminish and is almost completely resolved in the perivascular regions 42 d off Dox with minimal residual thickening seen adjacent to the airway (arrow). Photomicrographs are representative of 3–6 mice per time point. (B) Partial reversal of pleural fibrosis. After 6 wk of Dox extensive pleural fibrosis developed in the lungs of regulatable TGF-α mice. Twenty-one days after removing Dox, the pleural fibrosis is reduced. Pleural fibrosis is further diminished 42 d off Dox, but persists 126 d off Dox. Photomicrograph is representative of 3–6 mice per time point. All photomicrographs were taken at the same magnification. (C) Pentachrome staining of pleural fibrosis. After 6 wk of Dox, pleural fibrotic regions demonstrate intense blue staining consistent with young, immature matrix-rich collagen. After removal of Dox, staining of the pleural fibrosis becomes predominantly yellow at 126 d off Dox, indicative of established collagen. Photomicrograph is representative of three mice per time point. (D) Reversal of lung collagen. Lung collagen per whole lung was measured by Sircol Red collagen assay. After 42 d of Dox, collagen was increased compared with no Dox controls. Mice receiving 42 d of Dox followed by 21 and 42 d off Dox had less total lung collagen compared with mice receiving 42 d of Dox only. Values (means ± SEM, n = 4 mice/group) were compared with initial collagen (no Dox) using ANOVA followed by a Student-Newman-Keuls all pair-wise multiple comparison test to identify significant differences: *P < 0.01 compared with no Dox; **P < 0.02 compared with 6 wk of Dox.
Pentachrome staining demonstrated intense blue staining, indicative of immature matrix-rich collagen deposition induced by TGF-α. After removal of Dox, the pleural lesions stained predominantly yellow, indicating the irreversibility of the fibrotic pleural lesions (Figure 2C). Lung collagen content was increased by expression of TGF-α (Figure 2D). Total collagen concentration significantly decreased after removal from Dox.
Reversibility of Lung Mechanics
Consistent with the structural change of pulmonary fibrosis, lung function was changed by expression of TGF-α. Pulmonary compliance decreased 54%, and airway elastance and tissue dampening increased 300% from control measurements (Figure 3A). Airway resistance, tissue elastance, and hysteresivity were not statistically different from pretreatment controls (P > 0.05). Forty-two days after removal of Dox, lung function returned to control values.
Figure 3.
Reversibility of abnormalities in lung mechanics and body weight. (A) Reversible lung mechanics changes. Compliance, tissue elastance, tissue damping, hysteresivity, airway elastance, and airway resistance were measured in adult regulatable mice either receiving no Dox, 42 d of Dox, or 42 d on and 42 d off Dox (n = 4, n = 4, and n = 5, respectively). In mice receiving 42 d of Dox, compliance was decreased over 50%, while airway elastance and tissue damping were increased over 300%. Removal of Dox restored pulmonary mechanics changes. *P < 0.05 versus no Dox group using ANOVA followed by Tukey-Kramer multiple comparison test. (B) Restoration of body weight loss. Regulatable transgenic mice were divided into two groups and either provided water and food alone (control; squares) or water and food containing Dox (Dox; diamonds). Mice that were treated with Dox-supplemented water and food for 42 d (On Dox: Weeks 0–6) and then provided Dox-free water and food for up to 42 d (Off Dox: Weeks 6–12). Groups were sex-matched at the initiation of the test. Body weights were measured and weekly values (means ± SEM) were considered different from time-matched controls using a significance (*P < 0.05) determined by testing for normality and using a Kruskal-Wallis one-way ANOVA followed by an all pairwise multiple comparison procedure (Dunn's Method).
Weight Loss
Bitransgenic mice were provided food and water with or without Dox for 42 d. The groups were matched by sex and body weight at time zero (23.1 ± 3.0 g n = 15 doxycycline-treated versus 24.7 ± 3.2 g n = 14 water control). Doxycycline-treated mice lost on average 13% of their baseline body weight, while control mice gained 10% (Figure 3B). Two weeks after discontinuation of Dox, body weights returned to pretreatment levels, but remained decreased from controls at the end of the study period (23.4 ± 4.0 g versus 28.9 ± 4.1 g, respectively).
mRNA Microarray Analysis during TGF-α–Induced Pulmonary Fibrosis
Transcript levels were assessed 1, 4, 21, and 42 d after administration of Dox and 1, 4, 12, 21, and 42 d after its removal. A total of 484 mRNAs were identified as differentially expressed at any time using the statistical threshold of EFP < 5 (equivalent to an error probability of ∼ 0.01). The stringency of the threshold of significance was further increased to ≥ 2-fold difference and an average intensity of ≥ 150 fluorescence units. This filtering resulted in the identification of 389 unique transcripts (i.e., 12 mRNAs were duplicates or triplicates) and included 259 annotated genes, 36 nonannotated RIKEN cDNAs, 2 pseudogenes, and 1 withdrawn RIKEN cDNA. These transcripts were analyzed using BIM model–based clustering, and genes in individual clusters were tested for overrepresentation of annotation classes using EASE (21). The Benjamini FDR multiplicity correction was used to test each subset list against the population of all genes detected for Gene Ontology categories enriched with differentially expressed genes. This analysis yielded 6 distinct clusters with mRNAs that increased or decreased with treatment (Figure 4). Complete microarray data is available at the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/; accession number: E-MEXP-966).
Figure 4.
Bayesian infinite mixture (BIM) model based clustering of significant changes in transcripts after TGF-α induction in the mouse lung. BIM model based clustering was performed using log-expression ratios of experimental replicates for each time comparison after adjusting for the gene-specific dye effects. The statistical model was fitted and hierarchical clustering was produced by treating pair-wise posterior probabilities as the similarity measure and applying the traditional complete-linkage principle. The clustering results were displayed using TreeView. Genes in individual clusters (four increased and two decreased expression levels) were tested for overrepresentation of annotation classes using Expression Analysis Systematic Explorer (EASE), which tests each subset list against the population of all genes detected, and calculates the likelihood of overrepresentation in the Gene Ontology Consortium annotation categories of biological process, cell component, and molecular function, as well as KEGG pathway and SwissProt keyword. Fisher's Exact probability, using the Benjamini FDR adjustment, was calculated for each category, and a category was considered significant when it contained > 2 members and had a Benjamini value < 0.05. Subsets with fewer members of larger categories were combined into the larger category. The overall temporal behavior of significant categories was displayed as number of significant members (independent variable) versus time (dependent variable).
Cluster I
Differentially displayed transcripts that were increased by expression of TGF-α were divided into four clusters (Figure 4). Cluster I transcripts increased within 1–4 d, returning to control levels within 12 d after Dox removal. Cluster I contained 19 different annotated transcripts and was enriched in mRNAs encoding proteins related to extracellular matrix and cell adhesion. TGF-α mRNA was included in this cluster as expected (see Figure E1 in the online supplement). Included in this cluster were several extracellular matrix structural proteins, including procollagens type I, α 1 (COL1A1), COL3A1, COL5A2, and COL15A1, and elastin (ELN). Also increased in this cluster is the collagenase matrix metalloproteinase 14 (membrane-inserted) (MMP14).
Cluster II
Transcripts in this temporal cluster had increased levels that were delayed when compared with Cluster I, increasing primarily after 21–42 d of Dox treatment and returning to control levels within 12 d of Dox removal. Cluster II consisted of 47 transcripts (Figure E2) with overrepresentation associated with metal ion binding, including metallothionein 2 (MT2), fibrillin-1 (FBN1), matrix Gla protein (MGP), and SPARC-like 1 (mast9, hevin) (SPARCL1). Other metal-binding transcripts in Cluster II included two cysteine proteases with elastinolytic activity, cathepsin C and S (CTSC and CTSS). Previous studies demonstrated that CTSH and CTSS have roles in pulmonary fibrosis (24) and emphysema (25). Also represented in this cluster were transcripts for receptor tyrosine kinase-like orphan receptor 2 (ROR2), a receptor involved in Wnt/β-catenin signaling pathway, and members of protein-lysine 6-oxidase activity family including: lysyl oxidase (LOX) and related enzyme protein family members, lysyl oxidase-like 1 (LOXL1), LOXL2, and LOXL3. Additional extracellular matrix structural proteins were also represented including: fibrillin 1 (FBN1), procollagen, type XVI, α 1 (COL16A1), and elastin microfibril interfacer 1 (EMILIN1).
Cluster III
Cluster III contained 24 transcripts primarily characterized as increasing while on Dox and persisting as long as 42 d after its discontinuation. Several of the transcripts in this cluster have emerging roles in pulmonary fibrosis, including tissue inhibitor of metalloproteinase 1 (TIMP1), tenascin C (TNC), secreted phosphoprotein 1 (SPP1), and frizzled homolog 1 (Drosophila) (FZD1) (26, 27) (Figure E3). Other increased transcripts included: serine (or cysteine) peptidase inhibitor, clade F, member 1 (SERPINF1), and lumican (LUM). These and other transcripts, including latent TGF-β–binding protein 2 (LTBP2) and decorin (DCN), produce peptides that are involved in TGF-β latency/activation in the lung.
Cluster IV
Cluster IV contained elements that increased primarily during the recovery period after TGF-α was no longer expressed (Figure E4). Increases were noted for transcripts associated with vasculogenesis, including midkine (MDK), somatostatin receptor 4 (SSTR4), and aldehyde dehydrogenase family 1, subfamily A2 (ALDH1A2) (28, 29). Other transcripts in this cluster included members of the defense/immunity category, including hemolytic complement (HC), coagulation factor VII (F7), TGF-β 3 (TGFB3), apolipoprotein E (APOE), and serine (or cysteine) proteinase inhibitor, clade E, member 2 (SERPINE2). Eight of 43 mRNAs in this cluster were immunoglobulins. Also increased in this cluster is matrix metalloproteinase 2 (MMP2).
Cluster V
Clusters V and VI contained genes that decreased while on Dox. Cluster V contains transcripts that are more intensely decreased with many remaining decreased up to 6 wk off Dox. Of the 50 different transcripts in cluster V, 16 encoded genes are associated with development and 6 with cell motility (Figure E5). Transcripts previously associated with branching morphogenesis included bone morphogenetic proteins 4 (BMP4), BMP6, and Wnt inhibitory factor 1 (WIF1) (30). Transcripts critical to vascular formation and function included kinase insert domain protein receptor (KDR). Other vascular transcripts included: endothelial-specific receptor tyrosine kinase (TEK), kruppel-like factor 2 (KLF2), angiotensin receptor-like 1 (AGTRL1), and endothelin receptor type B (EDNRB). Also included in this cluster are transcripts associated with vascular structural proteins including: actin, α, cardiac (ACTC1), troponin I, cardiac (TNNI3) and troponin T2 (TNNT2), plasmalemma vesicle–associated protein (PLVAP), and ankyrin repeat domain 1 (cardiac muscle) (ANKRDI). These decreases are consistent with a decrease in pulmonary vascular formation and function.
Cluster VI
This cluster contained 158 different transcripts that included members of blood vessel development GO category, which involves the progression of blood vessel from formation to mature structure. Similar to the decreased transcripts in Cluster V, many of the decreased transcripts in Cluster VI were associated with endothelial cell function and VEGF signaling. Among these was vascular endothelial growth factor A (VEGFA), which is the ligand for the KDR, a member of Cluster V, that mediates endothelial proliferation and motility in vasculogenesis (vasculature formation) and angiogenesis (new vessels emerging of pre-existing blood vessels) (Figure E6). Another decreased transcript was tyrosine kinase receptor 1 (TIE1), an orphan receptor expressed predominantly in endothelial cells, where it physically associates with the related receptor TEK, a member of Cluster V. Related to these decreases was decreased platelet/endothelial cell adhesion molecule 1 (PECAM1), which directly transmits mechanical force in the vasculature. PECAM1 interacts with cadherin 5 (CDH5), also decreased in this cluster, and the formed complex functions as an adaptor to KDR. Also decreased in this cluster was caveolin, caveolae protein 1 (CAV1), which has roles in endothelial-dependent relaxation, contractility, and maintenance of myogenic tone (31). Also found in Cluster VI, superoxide dismutase 3, extracellular (SOD3) is believed to function to protect NO and NO-mediated signaling over its entire diffusion route (from site of production within endothelium to its major molecular target in vascular muscle) (32).
Transcription factors associated with vascular formation that were decreased in this cluster included forkhead box F1a (FOXF1A) (33). Notch gene homolog 4 (Drosophila) (NOTCH4), a related receptor, and its downstream transcription factor, hairy/enhancer-of-split related with YRPW motif 1 (HEY1), were decreased, which is consistent with an overall decrease in the Delta-Notch signaling pathway and altered vascular development (34). Other decreased transcripts key to blood vessel development include SRY-box containing gene 18 (SOX18), endoglin (ENG), activin A receptor, type II-like 1 (ACVRL1), and endothelial cell–specific molecule 1 (ESM1).
qRT-PCR Validation of Candidate Transcripts Altered during the Development of Pulmonary Fibrosis
Supportive of the microarray analysis, changes in 15 transcripts from Clusters I–VI were similar when measured by qRT-PCR (Figure 5). In this analysis, mouse lung transcripts treated with Dox 42 d were compared with controls. Eight transcripts (COL1A1, COL3A1, ROR2, TIMP1, TNC, SPP1, FZD1, MDK) increased, and seven transcripts (EDNRB, TEK, PECAM1, CAV1, SOD3, FOXF1A, SOX18) decreased significantly (P < 0.05); although the exact values varied somewhat (e.g., COL1A1 qRT-PCR = 4.9- ± 0.9- and microarray = 3.5- ± 0.6-fold), the magnitude of change was similar (i.e., the slope of a plot of the qRT-PCR to microarray fold change equaled 1.3, indicating that the microarray underreported fold changed slightly).
Figure 5.
Candidate transcripts altered during the development of pulmonary fibrosis after TGF-α induction in mouse lung. Lung mRNA from mice with TGF-α transgene induction for 42 d of Dox treatment (42 Days On Dox qRT-PCR) were compared with mice not treated with Dox (control qRT-PCR) using qRT-PCR with the comparative Ct method. Also included are results obtained by dual hybridization microarray that compares control to 42 d on Dox (42 Days On Dox Microarray). (A) Eight transcripts were increased and (B) seven decreased similarly measured by qRT-PCR or by microarray. Values present are from qRT-PCR that were normalized to ACTB and transformed to absolute values using 2−ΔΔCt formula to yield fold change. *Significantly different from control as determined by ANOVA followed by a Student-Newman-Kuels all pair-wise multiple (P < 0.05) (n = 3–4). †Statistical significance of differential expression was assessed by calculating P values, and adjusting for multiple hypotheses testing by calculating FDR and Expected False Positives (EFP) (17). Estimates of fold-change were considered significant when ≥ 2-fold different from control with EFP ≤ 5, and average spot intensity level ≥ 150.
Pulmonary Vascular Pressure and Ventricular Hypertrophy
Because several transcripts associated with vascular development were altered with TGF-α induction, RVH was assessed by measuring the right ventricular: left ventricular + septal weight. Expression of TGF-α for 56 d caused increased RVH compared with controls (Figure 6). Right ventricular systolic pressure in mice treated with Dox for 56 d also increased significantly (transgenic: 32.4 ± 2.1 versus control: 25.3 ± 0.4 mm Hg, P < 0.05). When Dox was administered for 42 d and then removed for 42 d, RVH remained increased compared with control. When Dox was administered for 42 d and then removed for 210 d, RVH was not different from control.
Figure 6.
Right ventricular hypertrophy induced by TGF-α induction. Mice were treated with Dox-supplemented water and food for 42 d (On Dox) and then provided Dox-free water and food for up to 210 d. An additional group of mice were treated with Dox-supplemented water and food for 56 d. Right ventricular hypertrophy (RVH) was assessed as an index of pulmonary hypertension. Hearts were removed and dissected to isolate the free wall of the right ventricle from the left ventricle and septum. The ratio of right ventricular to left ventricular and septum weight (RV/LV+S) was used as an index of RVH. Values are means ± SEM (n = 4–15 mice/group). *P < 0.05 compared with control using ANOVA followed by a Student-Newman-Keuls all pair-wise multiple comparison test.
Comparisons of Transcripts Altered in Mice Treated with TGF-α, Bleomycin, and in Patients with IPF
To compare mRNA profile after TGF-α induction to those of mice treated with bleomycin and those of patients with IPF, hierarchical clustering was performed on 142 homologous transcripts. The results displayed as a dendrogram indicate that the transcriptional profiles most similar to IPF were those of mice after TGF-α induction at a number of times starting with 21 d of Dox treatment and including all periods of Dox withdrawal (Figure 7). Significant correlation between the responses of patients with IPF and TGF-α induction from 21 d after treatment to the remainder of the study was observed (seven of the nine sampling periods; significant P values ranged from 0.008 to < 0.0001). Distant to the patients with IPF were TGF-α induction at 1 and 4 d and the 1 and 14 d bleomycin-treated susceptible C57 mice and 3 and 14 d bleomycin-treated resistant Balb mice. Significant correlation between the bleomycin-treated mice and the patients with IPF were noted for C57 mice at 72 h (P ≤ 0.006) and unexpectedly in the Balb mice at 24 h (P ≤ 0.01).
Figure 7.
Comparative transcriptional profiling of patients with IPF with mouse models of pulmonary fibrosis. Hierarchical clustering was performed on homologous transcripts that were identified by comparing the National Center for Biological Information (NCBI) Entrez Gene IDs for elements identified as significantly different for Gene Expression Omnibus (GEO) dataset GDS1252 using the inclusion criteria of a FDR < 0.05. Performed with Affymetrix GeneChip Murine Genome U74 Version 2 Set MG-U74A, the mouse dataset compared the lung mRNA levels after bleomycin treatment of susceptible C57BL/6J (C57) and resistant BALB/cJ (Balb) mouse strains. Sampling was performed at 1, 3, and 14 d after treatment. The IPF data set was performed with Amersham Biosciences CodeLink Uniset Human I Bioarray, and consisted of archived lung tissues of patients with IPF. IPF samples were obtained from the tissue bank of the Department of Pathology at the University of Pittsburgh. The diagnosis of IPF was confirmed by open lung biopsy (n = 6). Normal histology lung tissues resected from patients with lung cancer were used as controls (n = 11). The results displayed as a dendrogram indicate that the transcriptional profiles most similar to IPF were those of mice after TGF-α induction at a number of times starting with 21 d of Dox treatment and including all periods of Dox withdrawal.
Many of the decreased transcripts associated with blood vessel development in TGF-α–induced fibrosis in mice were also altered in patients with IPF. Of the 25 mRNAs that markedly decreased from TGF-α exposure (Cluster V and VI), human homologs were detected for 22 transcripts, and of these 11 also were decreased in patients with IPF (Table E1). These included: CAV1, EDNRB, TEK, PECAM1, TIE1, and CDH5. Other notable decreased transcripts in both TGF-α mice and patients with IPF included transcripts associated with cell signaling including HEY1, ENG, quaking (QK), roundabout homolog 4 (Drosophila) (ROBO4), and laminin, α 3 (LAMA3). Comparison of the increased transcripts revealed that of 19 increases noted during TGF-α induction in mice (Clusters I-III), 12 had homologous transcripts in humans, and of these, 6 also increased in patients with IPF, including TNC, SPP1, COL5A2, COL15A1, FBN1, and COL18A1. The induction of multiple cytokines after intratracheal bleomycin in mice was not observed in lungs from patients with IPF or the mice expressing TGF-α.
DISCUSSION
The Development and Partial Reversal of TGF-α–Induced Lung Remodeling
Our data demonstrated that conditional expression of TGF-α induction in the respiratory epithelium caused progressive pleural, interstitial, airway, and vascular remodeling accompanied by decreased lung function, weight loss, increased total lung collagen, and later, pulmonary hypertension. Reversal of TGF-α expression resulted in improved lung and pulmonary vascular function, weight gain, and substantial resolution of parenchymal fibrosis, while pleural surface thickening persisted. A number of other experimental studies support a role for TGF-α in transient fibroproliferative processes. Madtes and coworkers demonstrated TGF-α mRNA and protein increased from control rats after bleomycin injury with the highest expression noted at times of cellular proliferation and collagen deposition with decreases seen at later time points (35). Van Winkle and colleagues demonstrated Clara cell injury to mice induced by naphthalene injury is associated with increased TGF-α expression at cellular sites of cell proliferation and injury (36). Increased TGF-α protein expression is also detected in rodents following exposure to asbestos and hyperoxia (37, 38). Collectively, these studies support augmented TGF-α expression after epithelial injury, which is consistent with a role in wound repair proliferation and extracellular matrix production. Our data demonstrate persistent TGF-α expression in the absence of an inflammatory insult can lead to progressive pulmonary fibrosis, consistent with the hypothesis that fibrogenesis may be secondary to epithelial injury and repair.
Transcriptional Changes Associated with Lung Remodeling
Recently it has become apparent that inflammation may not be the defining event in pulmonary fibrosis and that inflammatory mediator and reactive oxygen species release may be a sufficient but not necessary event in the initiation of fibrosis (3). Overall, remarkably few changes occurred in transcript levels of chemokines, cytokines (including interferon and interleukins), or their receptors throughout the induction or reversal of TGF-α expression, consistent with the overall lack of inflammation observed in this model. Cytokine expression (including CXCl4, CXCl7, and IL12A) was decreased after chronic exposure to TGF-α. Thus, in the transcriptional analysis of the TGF-α transgenic mouse model, the influences of extensive inflammation are not detected.
Consistent with the TGF-α–induced lung pathology, expression of several members of extracellular matrix structural proteins were increased. Cluster I identified several structural protein transcripts, including procollagens (COL1A1, COL3A1, COL5A2, COL15A1) and elastin, that were increased 24 h into TGF-α induction and returned to control levels within 12 d of discontinuing Dox, suggesting a direct TGF-α effect on transcription. Treatment with EGF, an EGFR-ligand like TGF-α, can stimulate lung epithelial cells to directly secrete type I and V collagen supporting a direct role of EGFR activation and extracellular matrix production in the lung (39).
Expression of TGF-α is associated with progressive deposition of extracellular matrix with weight loss detected after 21 d and significant changes in lung mechanics detected after 42 d. Cluster II contained transcripts that were delayed in onset increasing between 21 and 42 d after initiating Dox treatment, but like cluster I, returned to control levels within 12 d of discontinuing Dox. This cluster included a number of transcripts that increased encoded protein-lysine 6-oxidase activity proteins, which also are metal binding proteins. These transcripts included LOX1 and related enzyme protein family members, LOXL1, LOXL2, LOXL3, that can catalyze oxidative deamination of the epsilon-amino group in certain lysine and hydroxylysine residues of collagens and lysine residues of elastin, thereby leading to the cross-linking of fibrillar collagens and elastin (40). Also in this cluster was MGP, a mineral-, extracellular matrix–binding protein synthesized by vascular smooth muscle cells and chondrocytes involved in extracellular matrix mineralization, and SPARCL1, a collagen type I binding protein (41). Thus, genes in this cluster could be considered important in propagating and maturing the progressive fibrotic process.
Cluster III contained transcripts which increased on Dox treatment and persisted as long as 42 d after TGF-α extinction. The transcripts in this cluster may represent a cascade of fibrogenic factors that remain elevated for a significant period of time after the inciting “trigger” is removed. Progressive fibrosis in mice after adenoviral-mediated gene transfer of TGF-β1 has been measured for up to 56 d after levels of detectable active TGF-β1 are returned to baseline, further supporting genes or pathways that can remain activated after injury or insult (42). Several of the transcripts in this cluster already have emerging roles in human and animal models of pulmonary fibrosis and may also directly contribute to the increased extracellular matrix deposition. SPP1 (a.k.a. osteopontin) increases in the lungs of mice after bleomycin (24) and is elevated in patients with IPF (23), as well as in the serum of patients with pleural mesothelioma (43). SERPINF1 (a.k.a. PEDF) is also increased in the lungs of patients with IPF. Similarly, TNC has been associated with fibrotic lung diseases (44), and LUM, an extracellular matrix protein, has been demonstrated to mediate epithelial–mesenchymal transition and human fibrosis (45).
Degradation of collagen is a complex process involving a group of zinc-containing proteinases, the MMPs. MMP14 is well characterized as a collagenase which cleaves collagens types I, II, and III and degrades a range of extracellular macromolecules including fibronectin, vitronectin, and fibrin (46). MMP14 is significantly increased 4 d after initiating Dox when fibrosis is first detected by histology and remains increased up to 12 d off Dox. The induction of MMP14 during the development of fibrosis would appear to be contradictory. However MMP activity is closely regulated through endogenous inhibitors, mainly through TIMPs, and an increase in TIMPs over MMPs has been hypothesized to support a “nondegrading” environment. In the TGF-α mice, TIMP1 expression increased over 10-fold within 24 h of Dox induction and remained increased throughout the study period, potentially inhibiting MMP activity. Alternatively, the increased MMP14 during the development of fibrosis may be required to remodel the extracellular matrix and allow the progression of fibrosis. Adenoviral gene transfer of TGF-β1 induced high levels of TIMP1 expression in pulmonary fibroblasts as well as in lungs of fibrosis-prone mouse strains compared with fibrosis-resistant strains, supporting Timp1 as a candidate gene conferring increased fibrosis in susceptible strains (47). Our data, taken together with these other studies, are consistent with the hypothesis that TIMP1 expression supports a nondegradative fibrillar collagen microenvironment that prevails in the development and incomplete resolution of pulmonary fibrosis and pulmonary hypertension.
Inasmuch as lung function was restored when TGF-α expression was reversed, transcripts remaining differentially altered could be considered secondary to injury, rather than drivers of the response. In contrast, transcripts altered only during the recovery period in cluster IV may have critical roles in restoration of lung function. Included in this cluster is MMP2 which increased 2.9 fold following withdrawal of TGFα, and remained elevated 1.9-fold 6 wk off Dox. MMP2 has been suggested to play a role in the mechanism of reversibility of fibrotic changes in bronchiolitis obliterans organizing pneumonia, although the experimental role of MMP2 has yet to be demonstrated in vivo (48). Interestingly, the resolution of pulmonary hypertension has been associated with an increase in MMP2 in the vascular wall. (49) Because TIMP1 can also inhibit MMP2 (50), the interactions between these proteins in the development and resolution of fibrosis is complex and implies a need for further investigation of the possible role of MMP2 in the resolution of pulmonary fibrosis and hypertension.
Also increased only during recovery, HC appears to be protective in bleomycin-induced acute lung injury, and has been suggested to be linked to the regulation of matrix metalloproteinases and their involvement in cell migration (51). Although not yet associated with pulmonary fibrosis, a third candidate increased during this period was SERPINE2. SERPINE2 is thought to be critical in tissue remodeling in the brain during development, in control of thrombin clearance (52), and possibly the development of COPD (53). Also in cluster IV is MDK, which increases from control at 42 d on Dox and remains increased up to 21 d off Dox (Figures 5A and E4). MDK overexpression in the lung epithelium of transgenic mice resulted in increased muscularization of small pulmonary arteries, and the prolonged recovery of RVH after removal of TGF-α overexpression could thereby potentially be mediated from persistent MDK expression (29).
While extracellular matrix and lung remodeling transcripts increased early (Day 1) and throughout the progression of lung fibrosis, at later times (3–6 wk) we measured progressive decreases in transcripts associated with vascular development and vascular proteins in clusters V and VI. KDR decreased along with its complementary ligand VEGFA, and these decreases in the VEGF-signaling pathway were accompanied with decreased PECAM1 and CDH5, which interact together to control a mechanosensing pathway that is required for early atherogenesis (54).
Other decreased transcripts important to blood vessel development were TIE1, ENG, ACVRL1, and QK. TIE1 is critical to endothelial cell migration in vasculogenesis, and mice lacking TIE1 die of hemorrhage and pulmonary edema (55). ENG is an accessory co-receptor that specifically modulates ACVRL1 signaling (56). These genes are expressed during vasculogenesis and have been implicated in hereditary hemorrhagic telangiectasia (HHT), a vascular dysplasia characterized by the inappropriate fusion of arterioles with venules (57). QK is a nonsecreted protein expressed in embryonic endoderm, adjacent to mesodermal sites where differentiation of endothelial and smooth muscle cells first occurs (58).
Transcript levels of caveolin family members, CAV1 in Cluster VI and in Cluster V, CAV2, were both decreased with induction of TGF-α. Previously, decreased CAV1 levels were noted in irradiation-induced pulmonary fibrosis and in lung fibroblasts isolated from patients with scleroderma with pulmonary fibrosis (59). In mice, gene targeting of CAV1 impairs nitric oxide and calcium signaling in the cardiovascular system, causing aberrant endothelium-dependent maintenance of myogenic tone. Moreover, the lungs of these mice display thickening of alveolar septa caused by altered endothelial cell proliferation and fibrosis (31, 60). Analysis of receptor transcripts altered by TGF-α induction demonstrated changes in a number of receptors associated with vascular function and development (Table 1). Together, these findings support a role for epithelial-derived TGF-α in the regulation of epithelial–vascular signaling that alter the vascular architecture and function during the progression of pulmonary fibrosis.
TABLE 1.
FUNCTIONAL ANALYSIS OF RECEPTORS ALTERED BY TRANSFORMING GROWTH FACTOR-α INDUCTION
| Receptor | Ligand | Antagonist | Related Protein | Possible Consequence |
|---|---|---|---|---|
| EGFR | TGFA*‡ | DCN*‡ | COL3A1*‡, 5A2*‡, 18A1*‡ | Extracellular matrix remodeling |
| FZD1*‡ | WNT2 | WIF1†, SOX18† | CDH5†‡, PECAM1†‡ | Epithelial/mesenchymal disruption |
| ROR2* | WNT2 | WIF1†, SOX18† | CDH5†‡, PECAM1†‡ | Dysregulated airway branching |
| TGFBR2 | TGFB1 | LTBP2†, LTBP3† | SPP1*‡, TNC*‡, FBN1*‡ | Extracellular matrix remodeling |
| KDR†‡ | VEGFA†‡ | EPAS1†‡ | Inhibited endothelial proliferation | |
| TEK†‡ (TEK:TIE1)† | ANGPT1 | Disrupted vascular mechanosening | ||
| EDNRB†‡ | EDN1 | Altered pulmonary vascular tension | ||
| NOTHCH4†‡ | DLL1 | HEY1†‡, FOXF1A†‡, FOX2† | Altered vascular development | |
| TGFBR2 | TGFB2 | LTBP2†, LTBP3† | ENG†‡, ACVRL1†‡ | Arterial-venous malformation |
| ROBO4†‡ | SLIT2†‡ | CDC42EP2† | Abnormal vascular guidance | |
| CD36†‡, CD47 | THBS1*‡, THBS2* | Anti- and pro-angiogensis |
Definition of abbreviations: ACVRL1, activin A receptor, type II-like 1; ANGPT1, angiopoietin 1; CD36, CD36 antigen; CD47, CD47 antigen (Rh-related antigen, integrin-associated signal transducer); CDC42EP2, CDC42 effector protein (Rho GTPase binding) 2; CDH5, Cadherin 5; COL3A1, procollagen, type III, alpha 1; COL5A1, procollagen, type V, alpha 2; COL18A1, procollagen, type XVIII, alpha 2; DCN, deorin; DLL1, delta-like homolog (Drosophila); EDN1, endothelin 1; EDNRB, endothelin receptor type B; EGFR, epidermal growth factor receptor; ENG, endoglin; EPAS1, endothelial PAS domain protein 1; FBN1, fibrillin 1; FOXF1A, forkhead box F1a; FOXF2, forkhead box F2; FZD1, frizzeled homolog 1 (Droshilia); HEY1, hairy/enhancer-of-split related with YRPW motif 1; KDR, kinase insert domain protein receptor (Vegfr2); LTBP2, latent transforming growth factor beta binding protein 2; LTBP3, latent transforming growth factor beta binding protein 3; NOTCH4, notch gene homolog 4 (Drosophila); PECAM1, platelet/endothelial cell adhesion molecule 1; ROBO4, roundabout homolog 4 (Drosophila); ROR2, receptor tyrosine kinase-like orphan receptor 2; SLIT2, slit homolog 2 (Drosophila); SOX18, SRY (sex determining region Y)-box 18; SPP1, secreted phosphoprotein 1; TEK, endothelial-specific receptor tyrosine kinase (Tie2); TGFA, transforming growth factor, alpha; TGFB1, transforming growth factor, beta 1; TGFB2, transforming growth factor, beta 2; TGFBR2, transforming growth factor, beta receptor type II; THBS1, thrombospondin 1; THBS2, thrombospondin 2; TIE1, tyrosine kinase receptor 1; TNC, tenascin C; VEGFA, vascular endothelial growth factor A; WIF1, WNT inhibitory factor 1; WNT2, wingless-related MMTV integration site 2.
Increased transcripts during TGF-α induction in mouse lung.
Decreased transcripts during TGF-α induction in mouse lung.
Transcripts altered in these mice and in patients with idiopathic pulmonary fibrosis.
Transcript Similarities Induced by TGF-α and Human IPF
A comparison of transcripts altered in IPF identified that the transcriptional profiles of IPF were more similar to TGF-α induction than bleomycin injury, largely because of the extensive cytokine increase in bleomycin injury not detected in either IPF or TGF-α induction. In addition, many of the decreased transcripts associated with vascular development after TGF-α induction were also altered in patients with IPF, and the persistent decrease in many of the vascular genes up to 42 d off TGF-α induction accounts for the continued correlation in transcriptional changes with IPF at this late time point. The activation and resolution of angiogenesis is a fundamental aspect of wound healing, and abnormal angiogenesis has been linked to the development of lung fibrosis. However, the precise relationship between angiogenesis and fibrosis in the lung remains controversial. Increased angiogenesis has been reported in bleomycin-induced murine pulmonary fibrosis, and increased angiogenesis is also reported with airway remodeling and fibrosis in chronic asthma and IPF. Conversely, recent reports in IPF lungs found a direct relationship between vascular remodeling and regression and parenchymal remodeling. Cosgrove and coworkers (61) found homogenates from IPF lungs inhibited angiogenesis in vitro, and within fibroblastic foci of IPF lungs vascular density was regionally decreased and associated with increased angiostatic protein SERPINF1 (a.k.a. PEDF) while VEGF was decreased. Our findings support the observations of Cosgrove and colleagues, not only by including elevations of SERPINF1 with concomitant decreases in VEGFA, but also by the decreases in the VEGF receptors and signaling proteins supporting a decreased angiogenesis in the progression of pulmonary fibrosis.
Two receptors involved in Wnt/β-catenin signaling pathway, FZD1 and ROR2, were increased. FZD1 is expressed primarily in the developing lung mesenchyme (62) and ROR2 is critical to appropriate cardiovascular and respiratory (proximal-distal branching) development (63). The increase in these receptors was accompanied by decreases in pathway inhibitors of Wnt-signaling, WIF1 and SOX18. Thus, the increase in receptors and decrease in an intracellular inhibitor is consistent with augmentation of Wnt/β-catenin signaling. Increased Wnt/β-catenin activation has been identified in the fibroblastic foci and proliferative bronchiolar lesions of patients with IPF (64).
In addition to the transcriptional similarities, there are several histological features of fibrosis in the TGF-α transgenic models which are also seen in the pathologic lesions of IPF, including interstitial alveolar wall thickening, type II cell hyperplasia, pleural-based fibrosis migrating into the interstitium, differentiation of myofibroblasts, pulmonary hypertension, and areas of fibroblastic foci (7–9, 65). In human studies, TGF-α was detected in the lung lavage fluid of all 10 patients with IPF, but in none of 13 normal volunteers (66). Baughman and coworkers also demonstrated an increase in TGF-α in IPF by immunohistochemistry (IHC) with increased TGF-α localized to type II epithelial cells, fibroblasts, and the vascular endothelium compared with controls (67). Increased EGFR ligands have also been demonstrated in fibrotic areas from patients with asthma, CF, ARDS, and BPD (66, 68, 69). If TGF-α overexpression is a significant mediator in the pathogenesis of fibrosis in human disease, our findings support targeting TGF-α overexpression to prevent progression or reverse fibrotic consequences.
Summary
To develop the desired architectures during organogenesis, appropriate communication must be established between the vascular, extracellular matrix, and epithelial compartments. In this study, alterations were noted in transcripts for receptors and signaling pathways during the progression of pulmonary fibrosis that supported a role for disruption in epithelial–mesenchymal–vascular signaling, reminiscent of developmental program critical to vasculogenesis. With TGF-α induction, transcript levels for multiple extracellular matrix genes markedly increased, including several products of fibroblasts, especially multiple procollagens, elastin enzymes associated with elastin processing, and transcripts produced by vascular smooth muscle. This period was followed by decreased transcript levels for genes encoding endothelial cell growth factors, receptors, and markers of differentiated function. Remarkably, few cytokines or other signs of inflammation were changed after expression of TGF-α. Together, the transcriptional profile observed in this mouse model was comparable to that of human IPF and implicates a disruption in cell–cell communication that altered the airway and vascular architecture in the pathogenesis of this disease. Progression of fibrosis and secondary pulmonary hypertension in the absence of inflammation supports EGFR as a signaling pathway influencing the pathogenesis of lung remodeling in IPF.
This work was supported by National Institutes of Health grants HL086598 (W.D.H.), HL077763 (G.D.L.), ES015675 (G.D.L.), HL061646 (M.I.), HL058795 (T.R.K.), and HL72894 (T.D.L.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2006-0455OC on May 11, 2007
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Crystal RG, Gadek JE, Ferrans VJ, Fulmer JD, Line BR, Hunninghake GW. Interstitial lung disease: current concepts of pathogenesis, staging and therapy. Am J Med 1981;70:542–568. [DOI] [PubMed] [Google Scholar]
- 2.Coultas DB, Zumwalt RE, Black WC, Sobonya RE. The epidemiology of interstitial lung diseases. Am J Respir Crit Care Med 1994;150:967–972. [DOI] [PubMed] [Google Scholar]
- 3.King TE Jr. Clinical advances in the diagnosis and therapy of the interstitial lung diseases. Am J Respir Crit Care Med 2005;172:268–279. [DOI] [PubMed] [Google Scholar]
- 4.Katzenstein AL, Myers JL. Idiopathic pulmonary fibrosis: clinical relevance of pathologic classification. Am J Respir Crit Care Med 1998;157:1301–1315. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter G. ErbB-4: mechanism of action and biology. Exp Cell Res 2003;284:66–77. [DOI] [PubMed] [Google Scholar]
- 6.Citri A, Skaria KB, Yarden Y. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res 2003;284:54–65. [DOI] [PubMed] [Google Scholar]
- 7.Hardie WD, Piljan-Gentle A, Dunlavy MR, Ikegami M, Korfhagen TR. Dose-dependent lung remodeling in transgenic mice expressing transforming growth factor-alpha. Am J Physiol Lung Cell Mol Physiol 2001;281:L1088–L1094. [DOI] [PubMed] [Google Scholar]
- 8.Hardie WD, Le Cras TD, Jiang K, Tichelaar JW, Azhar M, Korfhagen TR. Conditional expression of transforming growth factor-alpha in adult mouse lung causes pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2004;286:L741–L749. [DOI] [PubMed] [Google Scholar]
- 9.Korfhagen TR, Swantz RJ, Wert SE, McCarty JM, Kerlakian CB, Glasser SW, Whitsett JA. Respiratory epithelial cell expression of human transforming growth factor-alpha induces lung fibrosis in transgenic mice. J Clin Invest 1994;93:1691–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardie WD, Kerlakian CB, Bruno MD, Huelsman KM, Wert SE, Glasser SW, Whitsett JA, Korfhagen TR. Reversal of lung lesions in transgenic transforming growth factor alpha mice by expression of mutant epidermal growth factor receptor. Am J Respir Cell Mol Biol 1996;15:499–508. [DOI] [PubMed] [Google Scholar]
- 11.Tichelaar JW, Lu W, Whitsett JA. Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J Biol Chem 2000;275:11858–11864. [DOI] [PubMed] [Google Scholar]
- 12.Garvey W, Fathi A, Bigelow F, Carpenter B, Jimenez C. Improved Movat pentachrome stain. Stain Technol 1986;61:60–62. [DOI] [PubMed] [Google Scholar]
- 13.Schuessler TF, Bates JH. A computer-controlled research ventilator for small animals: design and evaluation. IEEE Trans Biomed Eng 1995;42:860–866. [DOI] [PubMed] [Google Scholar]
- 14.Sartor M, Schwanekamp J, Halbleib D, Mohamed I, Karyala S, Medvedovic M, Tomlinson CR. Microarray results improve significantly as hybridization approaches equilibrium. Biotechniques 2004;36:790–796. [DOI] [PubMed] [Google Scholar]
- 15.Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS. Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 2001;8:625–637. [DOI] [PubMed] [Google Scholar]
- 16.Sartor MA, Zorn AM, Schwanekamp JA, Halbleib D, Karyala S, Howell ML, Dean GE, Medvedovic M, Tomlinson CR. A new method to remove hybridization bias for interspecies comparison of global gene expression profiles uncovers an association between mRNA sequence divergence and differential gene expression in Xenopus. Nucleic Acids Res 2006;34:185–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc [Ser A] 1995;B57:289–300. [Google Scholar]
- 18.Medvedovic M, Sivaganesan S. Bayesian infinite mixture model based clustering of gene expression profiles. Bioinformatics 2002;18:1194–1206. [DOI] [PubMed] [Google Scholar]
- 19.Medvedovic M, Yeung KY, Bumgarner RE. Bayesian mixture model based clustering of replicated microarray data. Bioinformatics 2004;20:1222–1232. [DOI] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998;95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosack DA, Dennis G Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol 2003;4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Cras TD, Hardie WD, Fagan K, Whitsett JA, Korfhagen TR. Disrupted pulmonary vascular development and pulmonary hypertension in transgenic mice overexpressing transforming growth factor-alpha. Am J Physiol Lung Cell Mol Physiol 2003;285:L1046–L1054. [DOI] [PubMed] [Google Scholar]
- 23.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2005;2:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaminski N, Allard JD, Pittet JF, Zuo F, Griffiths MJ, Morris D, Huang X, Sheppard D, Heller RA. Global analysis of gene expression in pulmonary fibrosis reveals distinct programs regulating lung inflammation and fibrosis. Proc Natl Acad Sci USA 2000;97:1778–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng T, Zhu Z, Wang Z, Homer RJ, Ma B, Riese RJ, Chapman HA Jr, Shapiro SD, Elias JA. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J Clin Invest 2000;106:1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly MM, Leigh R, Gilpin SE, Cheng E, Martin GE, Radford K, Cox G, Gauldie J. Cell-specific gene expression in patients with usual interstitial pneumonia. Am J Respir Crit Care Med 2006;174:557–565. [DOI] [PubMed] [Google Scholar]
- 27.Kaarteenaho-Wiik R, Mertaniemi P, Sajanti E, Soini Y, Paakko P. Tenascin is increased in epithelial lining fluid in fibrotic lung disorders. Lung 1998;176:371–380. [DOI] [PubMed] [Google Scholar]
- 28.Sumi Y, Muramatsu H, Takei Y, Hata K, Ueda M, Muramatsu T. Midkine, a heparin-binding growth factor, promotes growth and glycosaminoglycan synthesis of endothelial cells through its action on smooth muscle cells in an artificial blood vessel model. J Cell Sci 2002;115:2659–2667. [DOI] [PubMed] [Google Scholar]
- 29.Reynolds PR, Mucenski ML, Le Cras TD, Nichols WC, Whitsett JA. Midkine is regulated by hypoxia and causes pulmonary vascular remodeling. J Biol Chem 2004;279:37124–37132. [DOI] [PubMed] [Google Scholar]
- 30.Wissmann C, Wild PJ, Kaiser S, Roepcke S, Stoehr R, Woenckhaus M, Kristiansen G, Hsieh JC, Hofstaedter F, Hartmann A, et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J Pathol 2003;201:204–212. [DOI] [PubMed] [Google Scholar]
- 31.Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC, et al. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science 2001;293:2449–2452. [DOI] [PubMed] [Google Scholar]
- 32.Faraci FM, Didion SP. Vascular protection: superoxide dismutase isoforms in the vessel wall. Arterioscler Thromb Vasc Biol 2004;24:1367–1373. [DOI] [PubMed] [Google Scholar]
- 33.Kalinichenko VV, Gusarova GA, Kim IM, Shin B, Yoder HM, Clark J, Sapozhnikov AM, Whitsett JA, Costa RH. Foxf1 haploinsufficiency reduces Notch-2 signaling during mouse lung development. Am J Physiol Lung Cell Mol Physiol 2004;286:L521–L530. [DOI] [PubMed] [Google Scholar]
- 34.Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev 2004;18:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madtes DK, Busby HK, Strandjord TP, Clark JG. Expression of transforming growth factor-alpha and epidermal growth factor receptor is increased following bleomycin-induced lung injury in rats. Am J Respir Cell Mol Biol 1994;11:540–551. [DOI] [PubMed] [Google Scholar]
- 36.Van Winkle LS, Isaac JM, Plopper CG. Distribution of epidermal growth factor receptor and ligands during bronchiolar epithelial repair from naphthalene-induced Clara cell injury in the mouse. Am J Pathol 1997;151:443–459. [PMC free article] [PubMed] [Google Scholar]
- 37.Liu JY, Morris GF, Lei WH, Corti M, Brody AR. Up-regulated expression of transforming growth factor-alpha in the bronchiolar-alveolar duct regions of asbestos-exposed rats. Am J Pathol 1996;149:205–217. [PMC free article] [PubMed] [Google Scholar]
- 38.Waheed S, D'Angio CT, Wagner CL, Madtes DK, Finkelstein JN, Paxhia A, Ryan RM. Transforming growth factor alpha (TGF(alpha)) is increased during hyperoxia and fibrosis. Exp Lung Res 2002;28:361–372. [DOI] [PubMed] [Google Scholar]
- 39.Federspiel SJ, DiMari SJ, Howe AM, Guerry-Force ML, Haralson MA. Extracellular matrix biosynthesis by cultured fetal rat lung epithelial cells. III. Effects of chronic exposure to epidermal growth factor on growth, differentiation, and collagen biosynthesis. Lab Invest 1991;64:463–473. [PubMed] [Google Scholar]
- 40.Hornstra IK, Birge S, Starcher B, Bailey AJ, Mecham RP, Shapiro SD. Lysyl oxidase is required for vascular and diaphragmatic development in mice. J Biol Chem 2003;278:14387–14393. [DOI] [PubMed] [Google Scholar]
- 41.Brekken RA, Sullivan MM, Workman G, Bradshaw AD, Carbon J, Siadak A, Murri C, Framson PE, Sage EH. Expression and characterization of murine hevin (SC1), a member of the SPARC family of matricellular proteins. J Histochem Cytochem 2004;52:735–748. [DOI] [PubMed] [Google Scholar]
- 42.Sime PJ, Xing Z, Graham FL, Csaky KG, Gauldie J. Adenovector-mediated gene transfer of active transforming growth factor-beta1 induces prolonged severe fibrosis in rat lung. J Clin Invest 1997;100:768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pass HI, Lott D, Lonardo F, Harbut M, Liu Z, Tang N, Carbone M, Webb C, Wali A. Asbestos exposure, pleural mesothelioma, and serum osteopontin levels. N Engl J Med 2005;353:1564–1573. [DOI] [PubMed] [Google Scholar]
- 44.Kuhn C, Mason RJ. Immunolocalization of SPARC, tenascin, and thrombospondin in pulmonary fibrosis. Am J Pathol 1995;147:1759–1769. [PMC free article] [PubMed] [Google Scholar]
- 45.Saika S, Miyamoto T, Tanaka S, Tanaka T, Ishida I, Ohnishi Y, Ooshima A, Ishiwata T, Asano G, Chikama T, et al. Response of lens epithelial cells to injury: role of lumican in epithelial-mesenchymal transition. Invest Ophthalmol Vis Sci 2003;44:2094–2102. [DOI] [PubMed] [Google Scholar]
- 46.Itoh Y, Ito N, Nagase H, Evans RD, Bird SA, Seiki M. Cell surface collagenolysis requires homodimerization of the membrane-bound collagenase MT1-MMP. Mol Biol Cell 2006;17:5390–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Min LJ, Cui TX, Yahata Y, Yamasaki K, Shiuchi T, Liu HW, Chen R, Li JM, Okumura M, Jinno T, et al. Regulation of collagen synthesis in mouse skin fibroblasts by distinct angiotensin II receptor subtypes. Endocrinology 2004;145:253–260. [DOI] [PubMed] [Google Scholar]
- 48.Fukuda Y, Ishizaki M, Kudoh S, Kitaichi M, Yamanaka N. Localization of matrix metalloproteinases-1, -2, and -9 and tissue inhibitor of metalloproteinase-2 in interstitial lung diseases. Lab Invest 1998;78:687–698. [PubMed] [Google Scholar]
- 49.Crossno JT Jr, Morris KG Jr, Klemm DJ. Attenuation of hypoxia-induced pulmonary artery remodeling by a peroxisome proliferator-activated receptor-gamma agonist. Chest 2005;128:580S. [DOI] [PubMed] [Google Scholar]
- 50.Baker AH, Edwards DR, Murphy G. Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J Cell Sci 2002;115:3719–3727. [DOI] [PubMed] [Google Scholar]
- 51.Addis-Lieser E, Kohl J, Chiaramonte MG. Opposing regulatory roles of complement factor 5 in the development of bleomycin-induced pulmonary fibrosis. J Immunol 2005;175:1894–1902. [DOI] [PubMed] [Google Scholar]
- 52.Crisp RJ, Knauer MF, Knauer DJ. Protease nexin 1 is a potent urinary plasminogen activator inhibitor in the presence of collagen type IV. J Biol Chem 2002;277:47285–47291. [DOI] [PubMed] [Google Scholar]
- 53.Demeo DL, Mariani TJ, Lange C, Srisuma S, Litonjua AA, Celedon JC, Lake SL, Reilly JJ, Chapman HA, Mecham BH, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet 2006;78:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, Engelhardt B, Cao G, DeLisser H, Schwartz MA. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature 2005;437:426–431. [DOI] [PubMed] [Google Scholar]
- 55.Sato TN, Tozawa Y, Deutsch U, Wolburg-Buchholz K, Fujiwara Y, Gendron-Maguire M, Gridley T, Wolburg H, Risau W, Qin Y. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature 1995;376:70–74. [DOI] [PubMed] [Google Scholar]
- 56.Oh SP, Seki T, Goss KA, Imamura T, Yi Y, Donahoe PK, Li L, Miyazono K, ten Dijke P, Kim S, et al. Activin receptor-like kinase 1 modulates transforming growth factor-beta 1 signaling in the regulation of angiogenesis. Proc Natl Acad Sci USA 2000;97:2626–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorensen LK, Brooke BS, Li DY, Urness LD. Loss of distinct arterial and venous boundaries in mice lacking endoglin, a vascular-specific TGFbeta coreceptor. Dev Biol 2003;261:235–250. [DOI] [PubMed] [Google Scholar]
- 58.Li Z, Takakura N, Oike Y, Imanaka T, Araki K, Suda T, Kaname T, Kondo T, Abe K, Yamamura K. Defective smooth muscle development in qkI-deficient mice. Dev Growth Differ 2003;45:449–462. [DOI] [PubMed] [Google Scholar]
- 59.Tourkina E, Gooz P, Pannu J, Bonner M, Scholz D, Hacker S, Silver RM, Trojanowska M, Hoffman S. Opposing effects of protein kinase Calpha and protein kinase Cepsilon on collagen expression by human lung fibroblasts are mediated via MEK/ERK and caveolin-1 signaling. J Biol Chem 2005;280:13879–13887. [DOI] [PubMed] [Google Scholar]
- 60.Razani B, Engelman JA, Wang XB, Schubert W, Zhang XL, Marks CB, Macaluso F, Russell RG, Li M, Pestell RG, et al. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J Biol Chem 2001;276:38121–38138. [DOI] [PubMed] [Google Scholar]
- 61.Cosgrove GP, Brown KK, Schiemann WP, Serls AE, Parr JE, Geraci MW, Schwarz MI, Cool CD, Worthen GS. Pigment epithelium-derived factor in idiopathic pulmonary fibrosis: a role in aberrant angiogenesis. Am J Respir Crit Care Med 2004;170:242–251. [DOI] [PubMed] [Google Scholar]
- 62.Wang Z, Shu W, Lu MM, Morrisey EE. Wnt7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd1, Fzd10, and LRP5. Mol Cell Biol 2005;25:5022–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takeuchi S, Takeda K, Oishi I, Nomi M, Ikeya M, Itoh K, Tamura S, Ueda T, Hatta T, Otani H, et al. Mouse Ror2 receptor tyrosine kinase is required for the heart development and limb formation. Genes Cells 2000;5:71–78. [DOI] [PubMed] [Google Scholar]
- 64.Chilosi M, Zamo A, Doglioni C, Reghellin D, Lestani M, Montagna L, Pedron S, Ennas MG, Cancellieri A, Murer B, et al. Migratory marker expression in fibroblast foci of idiopathic pulmonary fibrosis. Respir Res 2006;7:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hardie WD, Bruno MD, Huelsman KM, Iwamoto HS, Carrigan PE, Leikauf GD, Whitsett JA, Korfhagen TR. Postnatal lung function and morphology in transgenic mice expressing transforming growth factor-alpha. Am J Pathol 1997;151:1075–1083. [PMC free article] [PubMed] [Google Scholar]
- 66.Madtes DK, Rubenfeld G, Klima LD, Milberg JA, Steinberg KP, Martin TR, Raghu G, Hudson LD, Clark JG. Elevated transforming growth factor-alpha levels in bronchoalveolar lavage fluid of patients with acute respiratory distress syndrome. Am J Respir Crit Care Med 1998;158:424–430. [DOI] [PubMed] [Google Scholar]
- 67.Baughman RP, Lower EE, Miller MA, Bejarano PA, Heffelfinger SC. Overexpression of transforming growth factor-alpha and epidermal growth factor-receptor in idiopathic pulmonary fibrosis. Sarcoidosis Vasc Diffuse Lung Dis 1999;16:57–61. [PubMed] [Google Scholar]
- 68.Stahlman MT, Orth DN, Gray ME. Immunocytochemical localization of epidermal growth factor in the developing human respiratory system and in acute and chronic lung disease in the neonate. Lab Invest 1989;60:539–547. [PubMed] [Google Scholar]
- 69.Strandjord TP, Clark JG, Guralnick DE, Madtes DK. Immunolocalization of transforming growth factor-alpha, epidermal growth factor (EGF), and EGF-receptor in normal and injured developing human lung. Pediatr Res 1995;38:851–856. [DOI] [PubMed] [Google Scholar]