Abstract
Human gene RFP2 is a candidate tumor suppressor located at 13q14.3 and deleted in multiple tumor types. To explore regulation of RFP2, we determined structure of the 5′-untranslated region of RFP2 gene and its promoter. RFP2 promoter area is TATA-less, highly enriched in G and C nucleotides, and contains multiple quadruplex forming GGGGA-repeats. Deletion analysis of 5′-flanking sequences demonstrated that repeat containing fragment possesses activity seven times exceeding that of the combined SV40 promoter/enhancer. Other unusual features of the RFP2 promoter include anomalously high electrostatic fields induced by sequence-dependent dipoles and very low nucleosome forming potential. A “minimized” version of the RFP2 promoter could be used for overexpression of the various transgenes in the mammalian cells.
Keywords: Chronic lymphocytic leukemia, Candidate tumor suppressor gene RFP2, Promoter, Quadruplex, Nucleosome forming potential, Luciferase assay
Human gene Ret finger protein 2 (RFP2), also known as TRIM13 and RNF77, encodes a protein that contains a tripartite RING finger-B-box-coiled-coil domain (RBCC) and, therefore, belongs to a subgroup of RING finger proteins often involved in developmental, lymphogenic, and oncogenic processes [1–3]. In human tissues RFP2 is represented by at least three mRNA isoforms of 1.6, 2.4, and 7.5 kb in size with recognizable tissue specific difference in the prominence of those mRNAs. The smallest mRNA isoform is expressed at the highest level in testis, while the 2.4 kb transcript is most abundant in skeletal muscle, and the largest transcript, 7.5 kb in size, is weakly present or absent in most human tissues except skeletal muscle, prostate, spleen, thymus, and small intestine [1]. RFP2 occupies 25 kb on human chromosome 13 in a region q14.3 region frequently deleted in a number of malignancies including chronic lymphocytic leukemia (CLL) [4] and multiple myeloma (MM) [5]. It has been shown that expression of RFP2 is downregulated in CLL cells at the advanced stage of disease in comparison with the CLL cells from the same patient at diagnosis [1].
Human RFP2 consists of three exons, with exon 1 included in mRNA as longer or shorter variant (exon 1a and exon 1b) [1]. Complete open-reading frame of RFP2 (407 aa) is located inside exon 3 the only coding exon. Although the overall organization of RFP2 is relatively simple, some features point to the unusual complexity of its regulation (Fig. 1). First, 5′-untranslated exons of RFP2 overlap with the untranslated opposite strand transcript RFP2OS [1]. Second, the 3′-untranslated part of RFP2 exon 3 overlaps with the promoter area and exon 1 of another gene, KCNRG, encoding a protein with high homology to tetramerization domain of voltage-gated K+ channels that suppresses K+ channel activity [6]. As these genes are encoded by the same DNA strand, it is possible that expression of RFP2 interferes with the initiation of KCNRG transcription. Third, comparative studies of the promoter area and the 5′-untranslated area of RFP2 in rodents revealed DNA rearrangements that lead to an absence of any sequence homology to human RFP2 in the non-coding parts of mouse and rat orthologues [1].
Fig. 1.
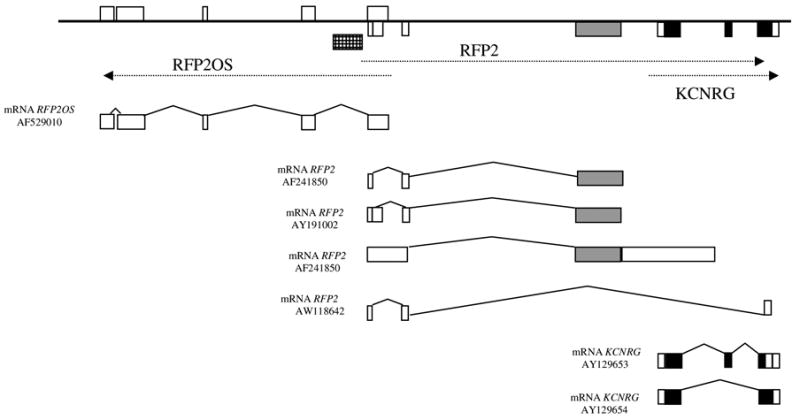
Schematic representation of the RFP2/RFP2OS/KCNRG locus. Non-coding exons are shown in white, RFP2 open-reading frame is in black, KCNRG open-reading frame is in grey. RFP2 promoter area studied here is shown in checkered box.
To investigate the regulation of the human RFP2, the genomic region adjacent to the transcription start site was isolated and a detailed map of the transcripts encoded by this complex genomic sequence was created. In describing the unusual structure of RFP2 promoter region, computational predictions of putative sites for transcription factor binding were performed and the relative activity of putative promoter elements using a luciferase reporter gene assay was studied. In the course of these studies, it was found that isolated fragments of the RFP2 promoter demonstrate extremely high activity in luciferase assays, exceeding that of widely used SV40 promoter. We believe that hyperactive fragments of RFP2 could be useful for improving current vector systems aimed at production of various proteins in mammalian cells.
Materials and methods
Cloning and sequence analysis of the 5′-genomic region of the RFP2 gene
Human cosmid clone LANL116c1 was identified previously by Southern hybridization as one that contains RFP2 [1]. Clone LANL116c1 was digested by HindIII, EcoRI, and BamHI digestion enzymes separately and in combinations. Obtained products of digestion were probed with radioactively labeled cDNA fragment corresponding to 5′-untranslated region of human RFP2. Labeling was performed with Prime-A-Gene Labeling system (Promega) and [α-32P]ATP. Hybridization was carried out in standard conditions [7]. The 4-kb promoter area of a human RFP2 gene has been identified by hybridization screening and subcloned in pUC18 and pGEM3zf- vectors (Promega). Entire DNA inserts within the described subclones have been sequenced manually with the fmol DNA Cycle Sequencing System (Promega) in the presence of betaine as described earlier [8]. Resulting nucleotide sequence is registered in GenBank (AF363782). The search for binding sites of putative transcriptional factors was performed using software PWMatcher [9] against the TRANS-FAC professional database [10].
Construction of luciferase constructs
To generate luciferase constructs with the various parts of the putative promoter region of RFP2, we first generated a DNA fragment containing 5′-promoter region with untranslated exon 1 by PCR amplification using Taq DNA polymerase in the presence of betaine (Promega, Madison, WI). The PCR products were purified by Wizard SV Gel and PCR Clean-up System (Promega). For directed cloning, the cutting sites for XhoI/HindIII restriction enzyme (Fermentas, Lithuania) were included in the sequences of the PCR primers. Series of DNA fragments of various lengths were cloned into a promoterless pGL3-Basic plasmid in front of the luciferase gene luc+ using a combination of cutting with digestion enzymes RsaI, NheI, and HindIII (Promega, USA), and generating a progressive 5′-nested deletions in an Erase-a-Base system (Promega). Escherichia coli NM522 cells were transformed with the resulting constructs by heat-shock according to the standard protocol [11]. Plasmids for the transfection of eukaryotic HEK293 cells were purified with Qiagen Plasmid Maxi Kit (Qiagen, Hilden, Germany). All luciferase constructs, including pGL3-Control, were sequenced with the Bio-Rad Sequi-Gen equipment and the fmol DNA Cycle Sequencing System (Promega).
Luciferase and β-galactosidase reporter gene assays
The HEK293 cells were cultured in six-well plates (Corning, NY) at 70–80% confluency and transfected with 5 mkg of each luciferase construct and pCMVb (Clon-tech, USA) using the LipofectAMINE 2000 Reagent (Invitrogen) as described in the manufacturer’s protocol. To assay lumeniscense the culture medium was removed and the cells were washed in PBS twice, then removed from plates in 500 mkl of lysis buffer (25 mM Tris-phosphate, pH 7.8, 10% glycerol, 2 mM trans-1,2-diaminocyclohexane-N,N,N′,N′-tetra-aceticacid, 2 mM DTT, and 1% Triton X-100) after 5 min incubation. Ten mkl of lysate was assayed for luciferase activity in Luciferase Assay System (Promega) at room temperature using a luminometer (Turner Designs, Sunnyvale, CA) by adding 100 mkl of the luciferase assay buffer (40 mM Tris–HCl, pH 7.8, 0.5 mM ATP, 10 mM MgSO4, 0.5 mM EDTA, 10 mM DTT, 0.5 mM coenzyme A, and 0.5 mM luciferin). Luciferase content was assessed by measuring the light emitted during the initial 30 s of the reaction and the values expressed in arbitrary light units. Ninety-six-well plate (PS LUMITRAC 200, Greiner Bio-one, Frickenhausen, Germany) pGL3-Control vector containing SV40 promoter and enhancer sequences that provide strong expression of luc+ in many types of mammalian cells has been used to assay baseline luciferase activity [12].
At the same time cellular lysates were assayed for the activities of the β-galactosidase contained in cotransfected plasmid pCMVb. For this purpose, 30 mkl cellular lysate was mixed with 500 mkl of Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 50 mM β-mercaptoethanol, pH 7.0) and 100 mkl o-nitrophenyl-β-D-galactopyranoside (ONPG) solution (4 mg/ml). After 2 h of incubation at 37 °C reactions were stopped by adding 250 mkl of 1 M Na2CO3, the optical densities were measured with an ELISA reader at 420 nm.
Three independent co-transfection experiments were performed for each tested construct and assayed for luciferase/β-galactosidase activities. A negative control of non-transfected cells was always included and assayed both for luciferase and for β-galactosidase, and the resulting background values were subtracted. In all experiments averaged Luc/Gal values for pGL-Basic were by at least two orders of magnitude less than Luc/Gal values for pGL3-Control and pGL vectors with cloned inserts. For each of the construct tested and for pGL3-Control vector, activity of luciferase was normalized to the β-galactosidase activity in the same lysate and expressed as luciferase/β-galactosidase ratio for every lysate studied. After that, luciferase activities of constructs containing various fragments of RFP2 promoter were compared to that of pGL3-Control vector and expressed as percentages.
Computational predictions of the promoter elements
To predict location of the promoter and the start of the transcription, we employed publicly available tools Human First Exon Finder (http://rulai.cshl.edu/software/index1.htm), Promoter 2.0 Prediction Server (http://www.cbs.dtu.dk/services/Promoter/), Promoter Scan (http://bimas.dcrt.nih.gov/molbio/proscan/), Markov Chain Promoter Finder McPromoter MM:II (http://genes.mit.edu/instructionsMMII.html), and Neural Network Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html).
Calculations of the dipole moments on DNA surface and nucleosome binding profiles
Sequence-dependent electrostatic fields for free DNA double helix were calculated by the vector summation of the electrostatic fields of the electrostatic dipole moments according to DB Property (http://wwwmgs.bionet.nsc.ru/mgs/systems/bdnavideo/). For every nucleotide position of the analyzed sequence electrostatic field on double stranded DNA surface was calculated as an additive sum of the partial electrostatic fields induced by electrostatic dipole moment in every stacked nucleotide pair between positions −10 and +10 relative to the reference nucleotide.
where i represents a position of the nucleotide pair according to the reference point (i = 0), ri is a vector of particular nucleotide pair and reference point, and di is a electrostatic dipole momentum of nucleotide pair.
Nucleosome binding profile for the RFP2 promoter was built with B-DNA-VIDEO tool (http://wwwmgs.bionet.nsc.ru/mgs/programs/bdna/na_bdna.html) using default set of parameters as recommended by [13].
Formation of the DNA quadruplexes was predicted under the Folding Rule by quadparser software available at http://www-shankar.ch.cam.ac.uk/quadparserframeset.html. Quadparser algorithm is built upon the Folding Rule: ‘A sequence of the form d(G3+N1–7G3+N1–7G3+N1–7G3+) will fold into a quadruplex under near-physiological conditions.’ In this rule, N refers to any base, including guanine, and the near-physiological conditions are 100 mM KCl and 10 mM Tris–HCl, pH 7.4 [14].
Results
In our previous paper [1], we described gap-free genomic sequence overlapping the entire RFP2-containing region (GenBank Accession No. AF241848). The first exon of RFP2 and its promoter region is embedded within a CpG island (CpG score 0.87%, 69.6% G + C) with no consensus TATA motif detected. In the several 50 nt blocs contained within RFP2 promoter, GC content reaches 82%. One of the promoter fragments represents an imperfect GGGGA repeat with coordinates −195 to −1 related to the most upstream starting point of RFP2 mRNA transcription according to primer extension study (data not shown) and to the results of an alignment of RFP2 EST sequences with the human genome sequence (Fig. 2A). Multiple GnA-repeats preceding the first exon of human RFP2 resemble those found in the genes KIT [15] and MYC [16]. These GnA-repeats have proven to be capable of the formation of stacked planar tetrads known as quadruplexes [15,16].
Fig. 2.
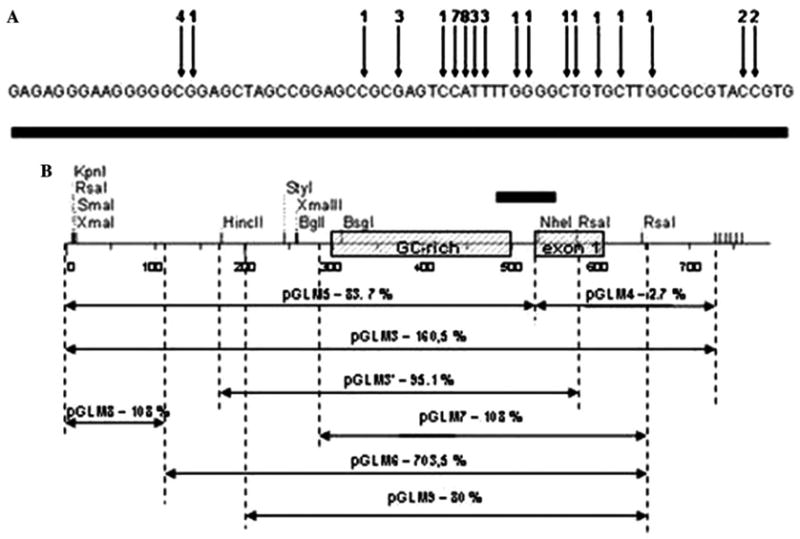
(A) Results of the alignment of RFP2 EST sequences with human genome sequence. Arrows indicate 5′-ends of individual cDNA clones primed on RFP2 mRNA, numbers above arrows correspond to amount of individual EST clones. (B) Transcriptional activity of the human RFP2 promoter. Fragments of indicated sizes in the 5′-region of the RFP2 gene were cloned in the pGL-Basic and transfected in HEK293 cells. The highest activity was observed with pGLM6 construct. The results are means of at least three independent experiments per construct.
Functional characterization of the promoter region
To investigate the functional activity of the RFP2 promoter, we constructed plasmid pGLM3 with 750 nt insert covering 530 nt of the promoter area of RFP2 including the imperfect GGGGA repeat described above, as well as the first exon of RFP2 and part of its first intron. To our surprise, this construct led to unusually high activity of the luciferase reporter corresponding to 160.5% of that measured for pGL3-Control vector containing SV40 promoter and enhancer sequences. We used the advantage of NheI site present at the beginning of RFP2 exon 1 to divide the pGLM3 insert onto fragments of 530 nt located upstream of the major start of RFP2 transcription, and 220 nt overlapping exon 1 and part of the intron 1 of RFP2. These fragments were assayed as inserts in sub-clones pGLM5 and pGLM4, respectively, and their promoter like activities were found to be 88.7% and 2.7% of pGL3-Control, respectively. To further delineate a minimal promoter, a variety of deletion mutants of the promoter were constructed by combination of DNA digestion with enzymes RsaI, NheI, and HindIII and generation of the progressive 5′-nested deletions (see Fig. 2B). Removal of the sequences located at the position −300 to −150 nt upstream of the imperfect GGGGA repeat and most of the RFP2 intron generated the most active plasmid construct pGLM6 (708.6% of that generated by pGL3-Control). The insert from pGLM6 corresponds to the DNA sequence with coordinates from 2489 to 3038, according to AF241848. This stretch of DNA was found to be seven times stronger as a promoter than the combined promoter/enhancer of SV40 in pGL3-Control vector. Further reduction of pGLM6 insert resulted in generation of minimized versions of the RFP2 promoter represented by constructs pGLM7, pGLM9, and pGLM3′ (see Fig. 2B). Latter constructs were characterized by somewhat reduced activity estimated as 108%, 80%, and 95% of that measured for pGL3-Control vector.
Computational analysis of RFP2 promoter region
Extensive analysis of the sequence AF241848 that contains promoter area of RFP2 was performed. No TATA or CAAT boxes were found upstream of the first exon of RFP2. All software applications tested indicated the presence of the putative promoter located within studied fragment of human genome, although with widely varying confidence of the prediction (Fig. 3). Human First Exon Finder software successfully pinpointed a large transcriptionally active fragment with coordinates 2339–2908 associated with the predicted first exon within the location 2839–3041 on AF241848. Similar results were obtained by Promoter Scan that identified smaller promoter with coordinates 2665–2905 and score 76.99 that was above the prediction cut-off value of 53.00. This predicted promoter is completely embedded within the pGLM6 DNA insert most active in luciferase assay. NNPP prediction algorithm pointed out two potential promoters, one located within the larger isoform of the first exon of RFP2 (score 0.84) and another preceding the imperfect GGGGA repeat (score 0.90). The latter promoter prediction was endorsed by its independent listing in the output file of the Markov Chain Promoter Finder. Results produced by Promoter 2.0 Prediction Server yielded two marginally acceptable predictions with reliability scores 0.657 and 0.569, both located upstream of the most active DNA fragment studied experimentally (Fig. 3).
Fig. 3.

Computationally predicted promoter elements located within the sequence of the promoter area of the human RFP2 gene. DNA fragment corresponding to insert in pGLM6 most active in luciferase assay is highlighted by blue background. First exon of the human RFP2 gene is in the underlined letters in red color, shorter isoform of this exon is shown in bold. Bold lines depict locations of the computationally predicted promoter elements. Results of the human first exon finder are underlined in blue, results of the promoter scan—in green, and NNPP prediction algorithm results in brown. Nucleotides shown in capital red letters indicate transcription start sites predicted by NNPP. Transcription Start Sites predicted by Markov chain promoter finder and by Promoter 2.0 Prediction Server are shown by green and blue arrows, respectively. Nucleotide positions shown in bold blue letters and in bold green letters are occupied by imperfect repeats forming quadruplexes at non-coding and coding DNA strands, respectively.
Quadruplex formation, nucleosome binding profile, and the profile of the dipole moments on DNA surface in RFP2 promoter
To get a detailed view on sequence-specific characteristics of the hyperactive fragment of RFP2 promoter, the 550-nt sequence of the pGLM6 insert was subjected to analysis with the previously described Quadparser algorithm. Two potential quadruplex forming regions were revealed. One of these regions is located on the non-coding DNA strand and is unusually long. It consists of 35 imperfect GGG-containing repeats that can fold into 32 possible quadruplexes, with a maximum of eight at once (coordinates 223–418 according to the sequence of pGLM6 insert and 2712–2908 according to the sequence of AF241848). Another region is located on the coding DNA strand and occupies nucleotide positions 28–61 according to the sequence of pGLM6 insert, and 2517–2550 according to the sequence of AF241848. The latter region can be folded in three types of quadruplexes, but only one of the folds could be realized at a time (Fig. 3).
Analysis of the electrostatic interactions on the surface of DNA structure formed by nucleotide sequence AF241848 indicated that region with coordinates 2700–2900 is characterized by anomalously high electrostatic fields induced by sequence-dependent dipoles (Fig. 4). Moreover, entire fragment of the RFP2 promoter inserted in pGLM6 region possesses an extremely low affinity to the histone octamer. As could be seen in Fig. 4A, the nucleosome forming potential (NFP) of the pGLM6 insert is lower than mean the NFP value (−1.48 ± 0.20) calculated for the promoters of the set of housekeeping genes earlier [17]. Even the sequences of RFP2 exon 1 and part of its intron 1 are characterized by low NFP values (e.g., −2.27 for nucleotide positions 2988–3038).
Fig. 4.
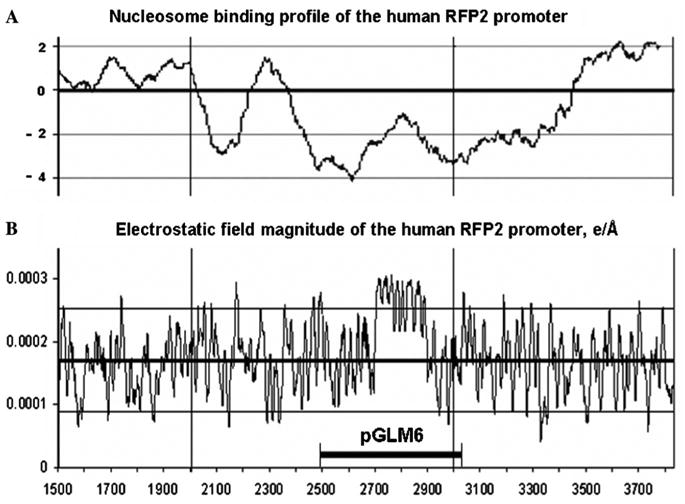
Human RFP2 promoter nucleosome binding profile (A) and the profile of electrostatic field magnitudes induced by sequence-dependent dipoles (B) for free DNA double helix produced by RFP2 promoter nucleotide sequence.
Discussion
This is the first report of the functional analysis of the promoter that drives the expression of human RFP2, a gene previously identified as a tumor suppressor candidate for a number of hematopoietic malignancies [1,3]. At the beginning of this study, the nucleotide sequence of this promoter remained unknown as it was embedded within a gap of unknown length present in the draft version of the human chromosome 13 sequence. To close this gap, sub-cloning of the RFP2 exon 1 containing fragments of cosmid clone LANL116c1 into the vector pUC18 was performed. Surprisingly, even the shortest of such inserts (400–700 bp) were found to be extremely resistant both to PCR amplification primed within vector sequences and to conventional sequencing methods. ‘‘Clumping’’ of the sequence lines has been overcome by the presence of betaine monohydrate allowing the sequencing only the coding DNA strand within the inserts of interest. The reconstructed sequence of the entire promoter and untranslated exons of RFP2 has been deposited in GenBank (Accession No. AF241848). The entire line of the tested luciferase containing constructs has been generated by various means allowing minimization of the original insert contained with the construct pGLM3 (see Results).
The expression activities of the luciferase constructs containing various parts of the putative promoter region of the human RFP2 were assessed after their transient transfection into the HEK293 cell line. In all experiments reported here, the transfection efficiency was normalized by co-transfecting with the plasmid pCMV-Basic that contains the β-galactosidase gene under control of the human cyto-megalovirus immediate early gene promoter. The pGL3-Control vector featuring SV40 promoter and enhancer sequences that provide strong expression of luc+ in many types of mammalian cells has been used to assay baseline luciferase activity [12]. In this assay, the RFP2 promoter was found to possess very high transcriptional activity comparable to that of the combination of the promoter and enhancer of SV40. When the RFP2 promoter has been subjected to minimization, one of its fragments (550 nt in size) has been found to possess activity seven times the strength of the combined SV40 promoter/enhancer.
Most likely, the unusual properties of the 550-nt fragments of the human RFP2 promoter are due to the imperfect GGGA repeat located upstream of its major start of transcription. Similar quadruplex forming motifs are shown to regulate expression activities of a number of human genes [15,16,18–20], many involved in tumorigenesis [15,16,19,20]. Other unusual features of the RFP2 promoter include anomalously high electrostatic fields induced by sequence-dependent dipoles and very low nucleosome forming potential. The magnitude of the electrostatic field on the DNA surface may facilitate DNA binding to transcriptional machinery components [21,22]. On the other hand, availability of DNA sequence for protein binding depends upon its conformation defined by nucelosome positioning. It has been shown recently that the histone eviction plays a causal role in mediating vigorous transcription in vivo and is not merely a consequence of it [23]. Histone octamers less tightly bound to the DNA are easier to evict. DNA affinity to histones depends on the intrinsic flexibility of the DNA affected both by the sequence-dependent conformational preferences of individual base steps and by the nature and location of the exocyclic groups of the DNA bases [24]. By applying automated computer tools for characterization of nucleosome packaging density [13,17] we demonstrated that the hyperactive region of the RFP2 promoter is less attractive to histone octamers than many promoters of housekeeping genes. It is peculiar that the low histone affinity feature extends to RFP2 exon 1 and downstream sequences occupied by putative promoter of the previously described non-coding antisense RNA RFP2OS [1]. It is possible that RFP2OS and RFP2-encoded transcripts are capable of mutual regulation. The local drops in DNA affinity to histones and the formation of DNA quadruplexes may be a part of such regulation and need further investigation.
Sequence analysis did not reveal a TATA or CAAT box motif around the transcription initiation site of human RFP2. Extensive analysis of the RFP2 promoter area yielded a variety of the predictions. The results most closely resembling experimental findings were obtained with the Human First Exon Finder software implementing the discriminant functions into a decision tree [25] and the Promoter Scan that takes advantage of a combination of elements similar to neural networks and genetic algorithms [26]. Other computational tools also predicted promoter-like activities within the sequence of interest, but placed them outside of the fragment being inserted into hyperactive construct pGLM6 (Fig. 3). Nevertheless, presence of these distant elements may also influence expression of the transcripts encoded by RFP2 and/or RFP2OS.
The truncated version of the RFP2 promoter represented by pGLM6 insert could be used for the highly efficient transcription of the various transgenes in the mammalian cells. Possible areas of application include the construction of the plasmids for stable and transient overexpression of the transgenes in the cultured cells and in the model animals, and possibly for the gene therapy of human diseases. As the minimization of RFP2 promoter resulted in increase of its strength, we can anticipate that the mutagenesis may help to improve its performance even further. According to the previous observations published in [1], the RFP2 promoter is ubiquitous as it directs transmission in all mammalian cells, regardless of its tissue origin. It will be interesting to see whether an engineering of its tissue-specific version is possible without the simultaneous drop in the quantity of the produced mRNA.
Acknowledgments
Authors are extremely grateful to Prof. I.A. Zakharov (VIGG, Moscow, Russia) who inspired authors to proceed with this study and to Dr. V. Levitsky and Dr. M. Ponomarenko for their extremely valuable help with the biophysical aspects. Masoumeh Sikaroodi and Mike Estep greatly helped with the proofreading of this manuscript. Dr. J. Huppert helped a lot with quadruplex predictions. Dr. R. Lebovitz helped a lot with filing a patent regarding findings described in this manuscript. A.B. was partially covered by NIH 1R15CA113331-01.
References
- 1.Baranova A, Hammarsund M, Ivanov D, Skoblov M, Sangfelt O, Corcoran M, et al. Distinct organization of the candidate tumor suppressor gene RFP2 in human and mouse: multiple mRNA isoforms in both species- and human-specific antisense transcript RFP2OS. Gene. 2003;321:103–112. doi: 10.1016/j.gene.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 2.van Everdink WJ, Baranova A, Lummen C, Tyazhelova T, Looman MW, Ivanov D, et al. RFP2, c13ORF1, and FAM10A4 are the most likely tumor suppressor gene candidates for B-cell chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2003;146:48–57. doi: 10.1016/s0165-4608(03)00126-2. [DOI] [PubMed] [Google Scholar]
- 3.Kapanadze B, Kashuba V, Baranova A, Rasool O, van Everdink W, Liu Y, et al. A cosmid and cDNA fine physical map of a human chromosome 13q14 region frequently lost in B-cell chronic lymphocytic leukemia and identification of a new putative tumor suppressor gene, Leu5. FEBS Lett. 1998;426:266–270. doi: 10.1016/s0014-5793(98)00357-3. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Corcoran M, Rasool O, Ivanova G, Ibbotson R, Grander D, et al. Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia. Oncogene. 1997;15:2463–2473. doi: 10.1038/sj.onc.1201643. [DOI] [PubMed] [Google Scholar]
- 5.Elnenaei MO, Hamoudi RA, Swansbury J, Gruszka-Westwood AM, Brito-Babapulle V, Matutes E, Catovsky D. Delineation of the minimal region of loss at 13q14 in multiple myeloma. Genes Chromosomes Cancer. 2003;36:99–106. doi: 10.1002/gcc.10140. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov DV, Tyazhelova TV, Lemonnier L, Kononenko N, Pestova AA, Nikitin EA, et al. A new human gene KCNRG encoding potassium channel regulating protein is a cancer suppressor gene candidate located in 13q14.3. FEBS Lett. 2003;539:156–160. doi: 10.1016/s0014-5793(03)00211-4. [DOI] [PubMed] [Google Scholar]
- 7.Kapanadze BI, Brodianskii VM, Baranova AV, Yu Sevat’ianov S, Fedorova ND, Kurskov MM, et al. Cosmid libraries containing DNA from human chromosome 13. Genetika. 1996;32:331–340. [PubMed] [Google Scholar]
- 8.Mytelka DS, Chamberlin MJ. Analysis and suppression of DNA polymerase pauses associated with a trinucleotide consensus. Nucleic Acids Res. 1996;15:2774–2781. doi: 10.1093/nar/24.14.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stepanova M, Tiazhelova T, Skoblov M, Baranova A. A comparative analysis of relative occurrence of transcription factor binding sites in vertebrate genomes and gene promoter areas. Bioinformatics. 2005;21:1789–1796. doi: 10.1093/bioinformatics/bti307. [DOI] [PubMed] [Google Scholar]
- 10.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Groskreutz DJ, Sherf BA, Wood KV, Schenborn ET. Increased expression and convenience with the new pGL3 luciferase reporter vectors. Promega Notes Mag. 1995;50:2–5. [Google Scholar]
- 13.Levitsky VG, Ponomarenko MP, Ponomarenko JV, Frolov AS, Kolchanov NA. Nucleosomal DNA property database. Bioinformatics. 1999;15:582–592. doi: 10.1093/bioinformatics/15.7.582. [DOI] [PubMed] [Google Scholar]
- 14.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J Am Chem Soc. 2005;137:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter, Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 17.Levitsky VG, Podkolodnaya OA, Kolchanov NA, Podkolodn NL. Nucleosome formation potential of eukaryotic DNA: calculation and promoters analysis. Bioinformatics. 2001;17:998–1010. doi: 10.1093/bioinformatics/17.11.998. [DOI] [PubMed] [Google Scholar]
- 18.Catasti P, Chen X, Moyzis RK, Bradbury EM, Gupta G. Structure–function correlations of the insulin-linked polymorphic region. Mol Biol. 1996;264:534–545. doi: 10.1006/jmbi.1996.0659. [DOI] [PubMed] [Google Scholar]
- 19.Cogoi S, Quadrifoglio F, Xodo LE. G-rich oligonucleotide inhibits the binding of a nuclear protein to the Ki-ras promoter and strongly reduces cell growth in human carcinoma pancreatic cells. Biochemistry. 2004;43:2512–2523. doi: 10.1021/bi035754f. [DOI] [PubMed] [Google Scholar]
- 20.Sun D, Pourpak A, Beetz K, Hurley LH. Direct evidence for the formation of g-quadruplex in the proximal promoter region of the RET protooncogene and its targeting with a small molecule to repress RET protooncogene transcription. Clin Cancer Res. 2003;9:A218. [Google Scholar]
- 21.Ahmad S, Sarai A. A moment-based prediction of DNA-binding proteins. J Mol Biol. 2004;341:65–71. doi: 10.1016/j.jmb.2004.05.058. [DOI] [PubMed] [Google Scholar]
- 22.Shanahan HP, Garcia MA, Jones S, Thornton JM. Identifying DNA-binding proteins using structural motifs and the electrostatic potential. Nucleic Acids Res. 2004;32:4732–4741. doi: 10.1093/nar/gkh803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, Herrera-Diaz J, Gross DS. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/ Snf and is not correlated with RNA polymerase II density. Mol Cell Biol. 2005;25:8985–8999. doi: 10.1128/MCB.25.20.8985-8999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Travers A, Drew H. DNA recognition and nucleosome organization. Biopolymers. 1997;44:423–433. doi: 10.1002/(SICI)1097-0282(1997)44:4<423::AID-BIP6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Davuluri RV, Grosse I, Zhang MQ. Computational identification of promoters and first exons in the human genome. Nat Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- 26.Knudsen S. Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]


