Abstract
The epithelial cells of the gastrointestinal tract are exposed to toxins and infectious agents that can adversely affect protein folding in the endoplasmic reticulum (ER) and cause ER stress. The IRE1 genes are implicated in sensing and responding to ER stress signals. We found that epithelial cells of the gastrointestinal tract express IRE1β, a specific isoform of IRE1. BiP protein, a marker of ER stress, was elevated in the colonic mucosa of IRE1β–/– mice, and, when exposed to dextran sodium sulfate (DSS) to induce inflammatory bowel disease, mutant mice developed colitis 3–5 days earlier than did wild-type or IRE1β+/– mice. The inflammation marker ICAM-1 was also expressed earlier in the colonic mucosa of DSS-treated IRE1β–/– mice, indicating that the mutation had its impact early in the inflammatory process, before the onset of mucosal ulceration. These findings are consistent with a model whereby perturbations in ER function, which are normally mitigated by the activity of IRE1β, participate in the development of colitis.
Introduction
The epithelial cell’s role in the development of inflammatory bowel disease (IBD) is poorly understood. Experimental models involving genetic lesions that impact on IBD mainly affect genes controlling immune function and whose protein products are expressed predominantly in nonepithelial cells (reviewed in refs. 1–3). Such mutations reduce anti-inflammatory cytokine production (4) and alter the balance of effector immune cells involved in initiating and sustaining the inflammatory process (5, 6). Fewer genes have been identified that act at the level of the epithelial cell. Trefoil proteins are thought to play a role in the resistance of the mucosa to injury and in the repair of existing mucosal lesions (7), and loss of trefoil protein function exacerbates toxin-induced experimental colitis (8). Prostaglandin production is another epithelial process that, when perturbed, impacts on the outcome on IBD by modifying the production of inflammatory mediators and altering the resistance of the epithelium to injury (9). Perturbation of antigen presentation by epithelial cells may also impact on the development of IBD, and mutations that affect this process predispose to IBD or exacerbate experimental lesions (10). Mutations in the multidrug resistance gene (MDR1A), encoding a transporter expressed in the epithelium and other cells, predisposes to IBD, but it is not certain that the epithelial cell is the site of action of that mutation (11).
The intestinal epithelial cell is the first to encounter many environmental toxins and infectious agents. Recent studies emphasize the interplay between toxic and infectious agents and intracellular routes for secretory protein trafficking and disposition (12). The endoplasmic reticulum (ER), the site of secreted and membrane protein synthesis and assembly, is involved in the response to environmental agents. Ingested environmental toxins like the tunicamycins (the cause of annual rye-grass disease in sheep), inhibit N-linked glycosylation and impair protein folding in the ER (13). Others, exemplified by the Cholera toxin, Pseudomonas exotoxin A, and Ricin toxin, undergo retrograde transport from the endocytic compartment to the ER, whence they are delivered into the cytoplasm through the SEC61 pore (14–16). The enteric pathogen rotavirus replicates in intestinal epithelial cells and the assembled viral particles bud through the ER, activating an organelle-specific stress response (17). Enveloped viruses elaborate surface glycoproteins that are produced in the host-cell’s ER and can stress the organelle (18, 19). Because of its barrier function, the intestinal epithelial cell is likely to be especially exposed to such interactions between the ER and environmental agents.
The environment in the ER is controlled by signaling pathways that respond to the presence of unfolded or misfolded proteins in the lumen of the organelle. The upstream components of these signaling pathways consist of several ER-localized transmembrane proteins with lumenal domains responsive to ER stress signals and cytoplasmic effector domains that transmit the signal to downstream components (reviewed in ref. 20). Yeast have only one known ER stress transducer encoded by IRE1 (21, 22). Yeast Ire1p oligomerizes in response to protein malfolding in the ER (23). Upon oligomerization, the COOH-terminal cytoplasmic effector domain of Ire1p, which has intrinsic protein kinase activity, undergoes trans-autophosphorylation unmasking a second intrinsic endonucleolytic activity resident in the same cytoplasmic effector domain (24, 25). Activated Ire1p cleaves its substrate HAC1 mRNA promoting a sequence-specific splicing event. Unspliced HAC1 mRNA is inefficiently translated, whereas the spliced, activated, form of the mRNA is efficiently translated (26, 27). Increased production of Hac1p, a transcription factor, activates the expression of multiple genes involved in folding and degradation of malfolded proteins in the ER (28–30). Thus IRE1 promotes homeostasis of the yeast ER.
In mammals the ER stress response has diversified considerably. Two IRE1 genes have been identified (IRE1α [ref. 31] and IRE1β [ref. 32]). In addition to their conserved function of regulating gene expression (31, 32), the mammalian IRE1s also link ER stress to activation of JUN NH2-terminal protein kinases (33). Mammals (and other metazoans) have a second type of ER stress transducer, known as PERK (or PEK), that responds to the malfolded protein signal in the ER by phosphorylating the translation initiation factor eIF2α inhibiting protein synthesis (34).
IRE1s and PERK sense the protein folding environment in the ER by the association of their conserved lumenal domains with the ER chaperone BiP. BiP-binding maintains these related transmembrane proteins in an inactive monomeric state, and perturbations of the protein-folding environment in the ER activate signaling by promoting BiP dissociation and IRE1 and PERK oligomerization (35, 36). Activation of this signaling pathway promotes BiP expression and BiP serves as both a marker for ER stress and feeds back negatively on the activity in this pathway (20). The two arms of the mammalian ER stress response coordinately promote the expression of genes required for maintaining a proper folding environment in the ER and prevent overloading the organelle by newly synthesized proteins. Here we report that IRE1β expression is limited to the epithelial cells of the gastrointestinal tract and that the phenotype of mice lacking IRE1β suggests a link between the handling of ER stress by the intestinal epithelial cell and susceptibility to environmental agents that promote colitis.
Methods
Gene targeting.
A positive-negative selection vector, pPNT (37), was used to replace 2.6 kb of genomic DNA (containing exons 12 and 13 that encode the transmembrane domain, amino acid residues 408–497 of mouse IRE1β) with a Neor gene transcribed in the opposite orientation. Long range PCR was performed on a PAC clone containing the IRE1β gene using a sense primer that incorporated a unique KpnI site at the 5′ end (7S, TCT GAG GTA CCT GAC GGC AGC CTG TAT GTC T, corresponding to residues 278–297 in the mouse cDNA sequence) and an antisense primer (6AS, TGG GAG CTC CCT GTA GTG GTG CT, corresponding to cDNA residues 2551–2573). To generate the 5′ homology arm, the greater than 11-kb PCR product was digested by KpnI and HindIII (the HindIII site that defines the 3′ end of the arm is in intron 11). This 4.4-kb fragment was ligated into the KpnI-XbaI sites of pPNT. The 3′ homology arm was obtained by digesting the same 11-kb PCR product by EcoRI and NdeI (the EcoRI site that defines the 5′ end of this fragment is in intron 13). The 3.5-kb EcoRI-NdeI fragment was ligated into the NotI-XhoI sites of pPNT.
129svev-derived W4 embryonic stem (ES) cells were electroporated with this targeting vector and selected for Neor and Ganciclovir resistance. Two hundred resistant clones were picked, and homologous recombination was verified by long-range PCR using the primer pairs: 15S (GAG ACT TTG TGG TTG CCC AGA C, corresponding to cDNA residues 1–22) and UTR3 (GAG ATC AGC AGC CTC TGT TCC ACA, in the 3′ untranslated region of the PGK gene in the pPNT plasmid) to detect recombination at the 5′ end and primer pairs 255R (GCT ACC GGT GGA TGT GGA ATG TG, in the PGK promoter in pPNT) and 6AS to detect recombination at the 3′ end. Two correctly targeted clones, 2G11 and 3F9, were injected into C57BL/6 blastocysts, and chimeric males that transmitted the mutant clone through their germline were obtained from both.
A three-primer PCR strategy was developed to genotype animals. The primer pairs i.1S (GGA GAG ATT GGA GAA GCA CTA G, annealing to a sequence in intron 11 that is at the 3′ end of the 5′ homology fragment) and 14AS (AG CCC TGG ATA AGA GTC TGA AG, in exon 12) give rise to a 450-bp fragment that is present only in the wild-type allele and the primer pairs i.1S and UTR3S give a 231-bp fragment that identifies the mutant allele.
Animal husbandry and induction of DSS colitis.
All experimental procedures involving mice were approved by New York University School of Medicine’s IACUC. The IRE1β mutation was maintained in two genetic backgrounds: inbred 129svev and as a backcross into C57BL/6 (Taconic, Germantown, New York, USA). Mice were maintained in a specific pathogen–free facility. To compare mice with IRE1β+/– and IRE1β–/– genotypes (in either 129svev or C57BL/6 backcross) we used sex-matched siblings derived from matings of IRE1β+/– and IRE1β–/– animals. To compare mice with wild-type and IRE1β–/– genotypes we used sex- and age-matched otherwise isogenic individuals from matings established among wild-type or IRE1β–/– 129svev individuals.
DSS was administered when the mice were between 3 and 6 months of age. Same-sex animals of different IRE1β genotypes were caged together and exposed to dextran sodium sulfate (DSS; mol wt = 40,000; lot number 7359C; ICN Biomedicals, Aurora, Ohio, USA) dissolved in the drinking water (3.5% wt/vol) (38, 39). In experiments not shown here, we determined that water consumption was not influenced by IRE1β genotype (in the presence or absence of DSS). Progress of colitis was monitored by daily examination for rectal bleeding, perianal soiling, lack of grooming, hunched posture, weight loss, and mortality. Although all parameters correlated closely with one another in a given animal, only the last two were easy to quantify objectively, and they provided the basis of our analysis.
Histology and immunohistochemistry.
The colon was removed, emptied of fecal material by gently washing the lumenal contents with a syringe, fixed overnight in 4% paraformaldehyde in PBS, dehydrated in graded ethanol solutions, embedded in paraffin, sectioned longitudinally, stained in hematoxylin and eosin, and mounted in resin. To evaluate ICAM-1 expression, 5-μm sections of tissue frozen in OCT were fixed in cold 4% paraformaldehyde, rinsed in PBS-Tween 0.1%, preincubated in 20% dialyzed serum at 37°C for 2 hours, and endogenous avidin binding proteins were blocked using avidin/biotin blocking kit (Vector Laboratories, Burlingame, California, USA). Sections were incubated for 1 hour at room temperature in a 1/200 dilution of biotinylated anti-mouse ICAM-1 antibody (PharMingen, San Diego, California, USA). Endogenous peroxidase activity was blocked by 0.5% H2O2 in methanol. Sections were then reacted with avidin/peroxidase complex (Vector Laboratories) followed by diaminobenzidine (150 μg/ml, 3% H2O2 in PBS) and counterstained with hematoxylin.
Protein isolation, immunoprecipitation, and immunoblotting.
Rabbit polyclonal antisera against mouse IRE1α, IRE1β, ribophorin I, BiP, and p38 MAP kinase have been previously described (33, 35, 40). Tissues were prepared by scraping the mucosa of the colon, stomach, or small intestine or by mincing tissue in ice cold PBS with a razor blade (in the case of brain, spleen, skeletal muscle, salivary gland, liver, and pancreas). Tissue fragments were centrifuged for 5 minutes at 800 g at 4°C, resuspended in 1 ml “1% triton lysis buffer” (20 mM HEPES [pH 7.5], 150 mM NaCl, 1% Triton-X100, 10% glycerol, 1 mM EDTA, 10 mM tetrasodium pyrophosphate, 100 mM NaF, 17.5 mM β-glycerophosphate, 2 mM phenylmethylsulfonyl fluoride, 8 μg/ml aprotinin, and 4 μg/ml pepstatin) and homogenized on ice in a Dounce homogenizer. Lysates were clarified at 16,000 g for 10 minutes and by preincubation for 1 hour with 10 μl of protein-A Sepharose. Soluble proteins were immunoprecipitated with 1 μl of anti-IRE1β or anti-IRE1α antibody bound to 10 μl of protein A-Sepharose and washed three times in RIPA buffer, and the bound proteins were resolved by 10% SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes as described previously (33, 35). Blots were incubated with diluted rabbit anti-IRE1β (1/10,000), IRE1α (1/5,000), or ribophorin I (1/1,000), and signals were revealed with protein A-HRP (1/5,000) and chemiluminescence (33, 35).
To prepare microsomes, epithelial cells were scraped from the mucosa in ice cold PBS, collected by centrifugation, and resuspended in SI buffer (50 mM Tris [pH 7.9], 1 mM EDTA, 2 mM phenylmethylsulfonyl fluoride, 8 μg/ml aprotinin, and 4 μg/ml pepstatin). Mucosal fragments were placed on ice for 5 minutes and then disrupted by passage through a 29G needle, breaking the cell membrane but leaving the nucleus and internal membranes intact. The extract was adjusted to 0.25 M sucrose and centrifuged at 2,000 g for 5 minutes at 4°C to remove nuclei and debris. The supernatant was recovered, and a membrane fraction was isolated by ultracentrifugation at 150,000 g for 1 hour at 4°C. The membrane pellet was resuspended in 2%-SDS buffer and resolved for immunoblotting by SDS-PAGE as already described here.
Epithelial cell isolation.
Colonic epithelial cells were isolated by a modification of a procedure described previously (41). Briefly, the colon was removed and washed free of fecal material with solution “A” (96 mM NaCl, 27 mM sodium citrate, 1.5 mM KCl, 0.8 mM KH2PO4, 5.6 mM Na2HPO4, 5,000 U/L penicillin, 5 mg/L streptomycin, 0.5 mM DTT, and 2 mM phenylmethylsulfonyl fluoride [pH 7.4]). Square pieces of tissue were placed in 10 ml of solution A at 37°C for 10 minutes with gentle shaking. This removed the mucus, bacteria, and other lumenal contents, which were discarded. The tissue fragments were then incubated in solution “B” (0.1 mM EDTA, 115 mM NaCl, 25 mM NaHCO3, 2.4 mM K2HPO4, 0.4 mM KH2PO4, 5,000 U/l penicillin, 5 mg/l streptomycin, 0.5 mM DTT, 2.5 mM glutamine, and 2 mM phenylmethylsulfonyl fluoride [pH 7.4]) at 37°C for 30 minutes with gentle shaking; the disruption of the mucosa and elution of cells was stopped by adjusting to 1 mM CaCl2. Tissue fragments were removed, and the cells recovered in the suspension were collected by centrifugation and lysed in Triton buffer as described earlier here for immunoprecipitation and immunoblotting. The residual mucosa was scraped off the tissue fragments and processed for immunoprecipitation and immunoblotting as already described here.
To examine the cellular composition of the eluted fraction, an aliquot of the cellular suspension was placed on Alcian blue–coated glass coverslips. Cells were allowed to adhere to the coverslip for 30 minutes at 37°C, washed with PBS, and fixed in cold paraformaldehyde for 10 minutes and stained with a pan-keratin mAb and a secondary antibody (Texas red conjugated goat anti-mouse IgG) counterstained with H33258 (1μg/mL) and visualized by epifluorescent microscopy.
Analysis of CHOP:GFP activity by flow cytometry.
Chinese hamster ovary cells (CHO) with a stably integrated CHOP:GFP reporter consisting of the mouse CHOP gene promoter fused to the green fluorescent protein (GFP) reporter (32) were plated 24 hours before DSS treatment at a density of 105 cells per 2-cm well. DSS was dissolved into culture media (Ham’s F12, 10% FCS) at the indicated concentration, filter sterilized, and added to the cells for the indicated period. GFP activity was measured in single cells by FACScan (Becton Dickinson Immunocytometry Systems, Franklin Lakes, New Jersey, USA).
Results
The murine IRE1β gene was targeted by deleting the two exons containing the transmembrane domain (Figure 1, a and b). The mutant allele produced no protein reactive with a polyclonal antiserum to IRE1β (Figure 1c). This observation, and the absence of a gain-of-function phenotype in the heterozygous mutant state (see later here), indicate that the targeted mutation results in a nonfunctional allele. IRE1β–/– mice constitute the expected 25% of offspring born to IRE1β+/– parents. In a specific pathogen–free facility, IRE1β–/– mice develop normally, are indistinguishable from wild-type and IRE1β+/– littermates, and both sexes are fully fertile as adults. To guide us in the analysis of the mutant mice, we determined the tissue distribution of IRE1β protein.
Figure 1.
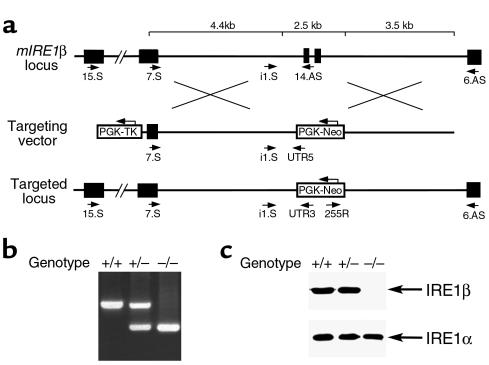
An induced mutation in the murine IRE1β locus. (a) Structure of the wild-type locus in the area encoding the transmembrane domain of IRE1β (top), the targeting vector (middle), and the targeted locus after homologous recombination (bottom). The deleted exons encoding the transmembrane domain are depicted as filled boxes, and intronic sequences are thin lines. The positions of the primers used to distinguish wild-type and mutant alleles are indicated, and the PCR protocols used are described in detail in Methods. (b) PCR analysis of genomic DNA from wild-type, heterozygous, and homozygous mutant mice. (c) Mutant mice do not express proteins reactive with a COOH-terminal anti-IRE1β antiserum. Lysates from colonic mucosa of mice with the indicated genotypes were immunoprecipitated with antisera to IRE1β and IRE1α and immunoblotted with the same antiserum.
Immunoprecipitation, followed by immunoblot of extracts of mouse tissues, revealed IRE1β protein only in the stomach, small intestine, and colon (Figure 2, a and b). No protein was detected in extracts from IRE1β–/– mice, confirming the identity of the IRE1β band in the immunoblot. IRE1α was found in all tissues and served as a positive control for the detection of a functionally related ER resident protein. The signal of both IRE1s was significantly weaker in the small intestine than in the stomach or colon, as was the signal for a third ER membrane protein, PERK (Figure 2b and data not shown). Nonetheless, IRE1β was detectable by immunoblot of lysates prepared from purified microsomes from small intestine (Figure 2b), establishing the highly restricted distribution of IRE1β to the gastrointestinal tract. The broad expression of IRE1α protein is consistent with the ubiquitous distribution of its mRNA (31).
Figure 2.
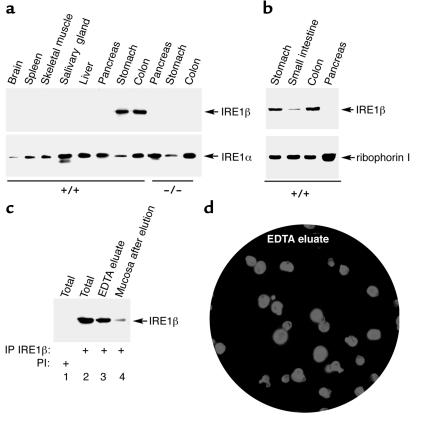
Expression of IRE1β is restricted to the epithelium of the gastrointestinal tract. (a) Immunoprecipitation (IP) followed by immunoblot detection of IRE1β and IRE1α in tissue extracts from wild-type and IRE1β–/– mice. (b) Detection by immunoblotting of IRE1β in microsomes prepared from all three segments of the gastrointestinal mucosa and pancreas. The ER resident protein ribophorin I, detected by immunoblot, serves as a recovery marker. (c) Detection of IRE1β by immunoprecipitation and immunoblotting in lysates prepared from epithelial cells eluted by EDTA treatment from the colon of a wild-type mouse. Lanes 1 and 2 are from lysates prepared from the intact tissue. Lane 3 is from the cells eluted from the tissue fragment in the calcium-free EDTA-containing buffer, and lane 4 is from a lysate prepared from cells that remain associated with the tissue fragment after (partial) depletion of epithelial cells. Lane 1 was immunoprecipitated with preimmune serum (PI), whereas lanes 2–4 were immunoprecipitated with anti-IRE1β immune serum. (d) Immunostaining of cells in an aliquot of the fraction used in lane 3 of c, with mAb’s to keratins. A counterstain with the karyophilic dye H33258 reveals the nuclei of all the cells in the field. Note that almost all the cells in the field stained positive with the keratin antibodies, attesting to their epithelial identity.
The vast majority of IRE1β ESTs are from colon epithelial cells, suggesting that IRE1β is expressed in the epithelial component of the intestine. To test this, we purified epithelial cells from the colonic mucosa by dissociating the tissue in a calcium-free solution containing EDTA (41). IRE1β protein was readily detected in this enriched cellular fraction (Figure 2c). The signal from the lysate prepared from cells that remained associated with the tissue fragment can be attributed to residual epithelial cells that had not been eluted (Figure 2c, lane 4). Staining of the eluted cells with keratin mAb’s confirmed that the cellular fraction expressing IRE1β consisted almost entirely of epithelial cells (Figure 2d). Therefore, we focused our attention on this group of cells.
We had previously mapped the human IRE1β gene to the pericentromeric region of chromosome 16 (32), within a single PAC interval of the polymorphic simple sequence repeat D16s417 (data not shown). This region of the human genome harbors one or more genes implicated in modifying the risk for the development of inflammatory bowel disease (42, 43). This coincidence led us to examine a possible role for mouse IRE1β in the development of experimental inflammatory bowel disease.
The gastrointestinal tract of the IRE1β–/– mice was histologically indistinguishable from that of wild-type or IRE1β+/– mice, and longitudinal follow-up of up to 1 year revealed that IRE1β–/– mice do not spontaneously develop intestinal inflammation. However, when challenged with DSS in the drinking water IRE1β–/– mice develop colitis 3–5 days earlier than do wild-type or IRE1β+/– littermates (the latter two groups have an indistinguishable response to DSS). Genotype-specific differences in progression and severity of colitis were reflected in the time course of weight loss and in the weight nadir, which was 28 ± 4% in the IRE1β–/– mice and 9 ± 5% in IRE1β+/– littermates (Figure 3a). Mortality of the IRE1β–/– mice also accrued approximately 5 days earlier than in wild-type or IRE1β+/– littermates (Figure 3b; data not shown).
Figure 3.
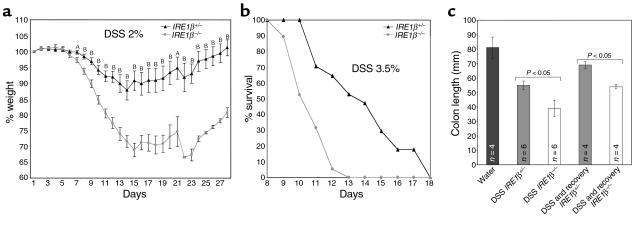
Increased severity of DSS colitis in IRE1β–/– mice. (a) Body weight, expressed as percentage of weight on the day of first exposure to DSS, is plotted against time. Mice were exposed continuously to 2% DSS in the drinking water for days 1–12. Shown are the mean and SEM of a typical experiment carried out on cohort of 23 IRE1β+/– and 23 sibling IRE1β–/– 129svev mice and reproduced three times. AP < 0.05; BP < 0.0005 for the difference in mean value between the two groups (two-tailed t test). (b) Survival of a cohort of 17 age-matched IRE1β+/+ and 19 IRE1β–/– 129svev mice exposed continuously to 3.5% DSS in the drinking water for days 1–12. (c) Comparison of the colon length in untreated mice, and mice of the indicated genotypes treated with 2% DSS for 12 days (DSS) or mice treated with 2% DSS for 12 days and allowed to recover for 15 days (DSS and recovery). Shown are the means and SEM and P values calculated by two-tailed t test.
Increased severity of the disease was observed in the IRE1β–/– mice across a range of DSS doses (2–4%) and in all of five separate direct comparisons of cohorts of age-matched mice with divergent IRE1β genotypes. The effect of genotype was observed in two different strain backgrounds: inbred 129svev (Figures 3 and 4) and a fifth generation backcross into a C57BL/6 background (data not shown). The latter is important, given the role of strain background in susceptibility to DSS colitis (44, 45).
Figure 4.
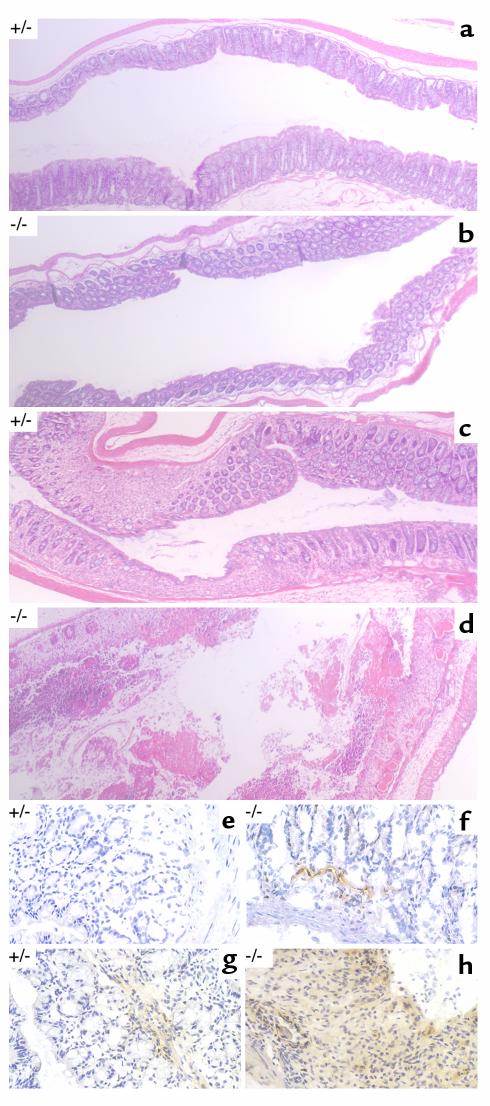
Histopathology of DSS-treated mice. Representative photomicrographs (×50) of paraffin-embedded, hematoxylin and eosin–stained longitudinal sections of the distal colon from untreated mice of the indicated genotypes (a and b) or mice that had been exposed continuously to 2% DSS for 12 days (c and d). Photomicrographs (×400) of longitudinal frozen sections of colon stained with an mAb to murine ICAM-1 revealed by diaminobenzidine (brown precipitate) and lightly counterstained with hematoxylin for orientation. Mice of the indicated genotypes were exposed to 2% DSS for 6 days (e and f) or 10 days (g and h).
Colitis induced by DSS was qualitatively indistinguishable in the mice with different IRE1β genotypes: pathological changes were most obvious in the distal large bowel, with marked shortening of its long axis (39), a change that was more severe in the IRE1β–/– mice (Figure 3c). Histopathological evaluation revealed that the distal colonic mucosa remained intact for the first 7 days of treatment. The only change noted was a mild flattening of the crypts, which was evident in both genotypes. Appearance of inflammatory cells followed, with surface ulceration occurring still later. All of these were more pronounced and occurred 3–5 days earlier in the homozygous mutant mice (Figure 4, a–d). However, the quality of the submucosal inflammatory infiltrate, consisting predominantly of mononuclear cells, and the surface ulceration, consisting of focal lesions interspersed with areas with intact surface epithelium, were indistinguishable in the different genotypes, such that IRE1β–/– mice on day 10 of the protocol resembled many of their wild-type and heterozygous littermates 3–5 days later. Genotype-dependent differences in the extent of inflammation and ulceration were also reflected in the persistence of mucosal lesions into the recovery period in those IRE1β–/– mice that survived the DSS treatment. This compared with a trend toward resolution of the lesions in the wild-type and heterozygous littermates (data not shown).
One of the earliest changes noted in DSS-treated rodents is the expression of ICAM-1 in the vasculature of the submucosa in the distal large bowel (46). ICAM-1 is a cell-surface protein that is induced first on the endothelium of the submucosal blood vessels and whose expression precedes surface ulceration (46). It is thought to play a role in recruiting effector immune cells to the mucosa and in establishing the inflammatory process (47, 48). To determine whether the IRE1β mutation acts early in the sequence of events that lead to chronic colitis, we compared the ICAM-1 staining in DSS treated IRE1β–/– and IRE1β+/– mice. In both genotypes, ICAM-1 staining in the submucosa was first found around blood vessels, and its induction preceded surface ulceration, as described previously (46). However, ICAM-1 staining occurred 3–5 days earlier in the mutant mice (Figure 4, e–h). These differences in ICAM-1 staining indicate that IRE1β acts early in DSS colitis, before the onset of surface ulceration. Later in the process, ICAM-1 staining was observed throughout the mucosa, correlating with the egress of ICAM-1–expressing immune effector cells into the tissue (Figure 4h).
In yeast, IRE1 controls the expression of genes involved in ER function and its absence causes ER stress. To determine whether IRE1β impacts on the level of ER stress, we compared the levels of BiP protein between IRE1β–/– and IRE1β+/+ tissues. BiP protein levels were indistinguishable in the pancreas, liver, and small intestine of IRE1β+/+ and IRE1–/– mice; however, BiP levels were at least threefold higher in lysates from stomach and colon of mutant mice (Figure 5a), tissues that express high levels of IRE1β (Figure 2a). Protein loading is controlled by ribophorin 1, an ER-resident protein that is not induced by ER stress. The elevated BiP content suggests that these tissues are experiencing higher levels of ER stress in the mutant mice. The basal level of stress in the colonic mucosa of the IRE1β–/– mice is also reflected in higher levels of p38 MAP kinase activation, revealed by immunoblot with an antiserum that detects the active form of the kinase. To increase the likelihood that our measurements would reflect events taking place in the epithelium, we restricted the analysis to the early phase of DSS treatment, before the egress of inflammatory cells into the mucosa. Whereas p38 MAP kinase activity increased, as expected, in the mucosa of DSS-treated wild-type mice, activity in this pathway was already elevated in untreated IRE1β–/– mice and remained elevated during DSS exposure (Figure 5b).
Figure 5.
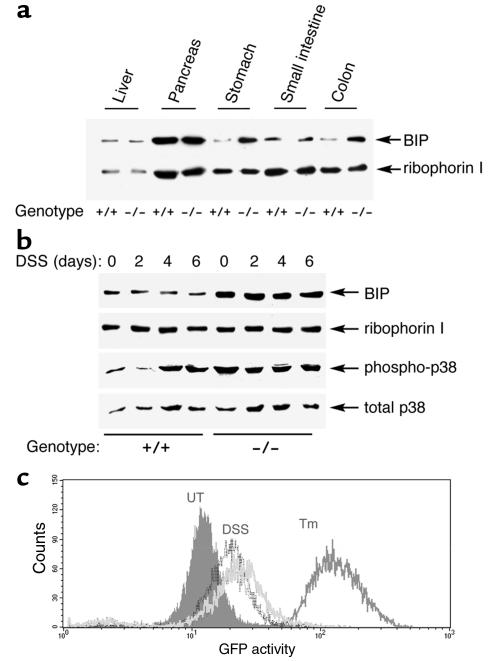
Higher levels of ER stress in gastric and colonic mucosa of IRE1β–/– mice. (a) Immunoblot of BiP and ribophorin I content in tissue lysates of mice with the indicated genotypes. (b) Immunoblot of phosphorylated (active) p38 MAP kinase, total p38 MAP kinase, BiP, and ribophorin I content of colon lysates from mice of the indicated genotype exposed to 3% DSS for the indicated period. (c) Fluorescent activated cell sorting signals of CHO cells expressing an integrated transgene consisting of the CHOP promoter fused to a green fluorescent protein (GFP) reporter. Cells were left untreated (UT), exposed to 2.5 μg/mL tunicamycin for 12 hours (Tm), or cultured in the presence 3.5% DSS for 24 hours (dark gray histogram) or 4.5% DSS for 24 hours (light gray histogram).
In mice BiP can be activated by ER stress in an IRE1-independent manner (33). However, BiP levels were not measurably increased in the mucosa of DSS-treated mice (Figure 5b). To determine whether DSS exposure causes ER stress we turned to a more sensitive system. Rapid onset of ER stress can be detected by the activation of a marker gene CHOP (also known as GADD153) (49). We exposed a CHO cell line containing a sensitive integrated CHOP:GFP reporter gene to DSS and measured GFP expression by fluorescent-activated cell sorting. DSS treatment increased the fluorescent signal (Figure 5c). Activation of the CHOP:GFP reporter by DSS was reproducible, although significantly weaker than that elicited by tunicamycin, an agent that rapidly causes high levels of ER stress. We conclude that DSS treatment likely causes ER stress, reflected in the activation of a target gene of the ER stress response.
Discussion
Mammals have two IRE1 genes: a ubiquitously expressed and essential α isoform, and a β isoform that is restricted in its expression to the epithelial component of the gastrointestinal tract. Homozygous loss-of-function mutations in mouse IRE1α cause early embryonic lethality consistent with its broad and early expression (31, 33). This study has uncovered a role for IRE1β in promoting resistance to DSS-induced colitis. Mice lacking IRE1β develop colitis of increased severity and with shorter latency compared with wild-type and IRE1β+/– heterozygote animals. The selective expression of IRE1β to the epithelial component of the gastrointestinal tract suggests that its activity in promoting resistance to DSS-induced colitis is exerted at the level of the epithelial cell. Epithelial expression is constitutive, consistent with evidence that signaling in the IRE1β pathway is activated post-translationally (23, 35, 36). These features set IRE1β apart from many other genes known to impact on the development of spontaneous or experimental colitis in mice, in that the latter are active predominantly in immune effector cells that initiate and modulate the inflammatory process, rather than the epithelial cells that are its target (50–52).
IRE1 proteins are internal stress receptors, responding to changes in the folding environment in the ER. It is possible that some feature related to the secretory function of the intestinal epithelium (e.g., mucin production) may have favored the elaboration of a tissue-specific IRE1 isoform that is required for maintaining a specialized environment in the epithelial cell ER. The elevated level of BiP in the gastric and colonic mucosa of the IRE1β–/– mice is consistent with this possibility. The periodical exposure of the gastrointestinal tract to infectious agents or toxins that impair ER function or produce ER stress signals may have served as an additional stimulus to the evolution of a tissue-specific isoform of IRE1. Yeast IRE1 is the ancestor of RNase L, a component of the innate immune system (20). In this vein, it is intriguing to consider that a specialized isoform of IRE1 evolved in the gastrointestinal to couple intracellular ER-stress signals to immune effects.
The mechanism by which DSS ingestion induces colitis is not known. The topological features of the gastrointestinal tract dictate that epithelial cells are the first to be exposed to the lumenal content and suggest that the epithelium is also likely to have a primary role in the development of the inflammatory process. Toxicity from ER stress (or any other cause) resulting in death of epithelial cells is unlikely, however, to account for initiation of disease in DSS-treated animals, as the onset of inflammation precedes the development of ulcerative lesions (39, 46). Furthermore, even though the colonic bacterial flora can influence the course of the disease, DSS colitis also develops in germ-free mice (53). These observations suggest that a simple breach of the barrier function of the gastrointestinal epithelium is unlikely to be a primary instigator inflammation. It is more likely that DSS acts on the epithelial cells to set in motion a process that leads to inflammation and ulceration. ICAM-1 expression in the vasculature of the submucosa appears to be an early marker for this proposed chain of events in which the epithelium signals to the endothelial cells to trigger inflammation (46). We propose that the acquisition of ICAM-1 staining 3–5 days earlier in DSS–treated IRE1β–/– mice reflects increased production of such signals by the stressed epithelium. The observations that DSS treatment activates an ER stress-induced gene and that IRE1β is part of the apparatus responsive to ER stress support a model whereby a DSS-induced perturbation in ER function, which is partially mitigated by the activity of IRE1β in wild-type mice, plays an early role in the development of inflammation.
In yeast, IRE1 mitigates ER stress by activating genes that reduce the impact of protein malfolding (28–30). Our findings of higher levels of BiP and more activity of p38 MAP kinase in IRE1β–/– colonic mucosa suggest that mutant epithelial cells are subject to more ER stress, in both the basal state and presumably also after exposure to DSS. Higher levels of ER stress might feed into the inflammatory response by modifying the expression of cell-surface proteins or secreted factors that interact with the immune system. For example, ER stress might activate the expression of proinflammatory genes utilizing pathways that are IRE1β independent. Hypothetical candidates might include genes encoding MHC-like MICA proteins, the human forms of which are known to be inducible by stress (54, 55) and activate intraepithelial lymphocytes (54, 56). Alternatively, IRE1β may play a role in the expression of anti-inflammatory genes, directly or indirectly by promoting a proper environment in the ER. Either of these would be impaired in the IRE1β–/– mice.
The observation that IRE1β maps to a region of the human genome that harbors IBD1, a major modifier locus for the development of inflammatory bowel disease, suggests that polymorphism in the human gene may account for some of the variance in the risk for this disease. Regardless of the ultimate disposition of IRE1β as a candidate for the putative IBD1 gene, the findings here suggest that events in the epithelial cell’s ER may modify the course of chronic colitis. Our working model for this relationship predicts that IRE1β–/– mice will have altered sensitivity to certain naturally occurring infectious or toxic agents that attack the gastrointestinal epithelial cells and that identification of genes activated by IRE1β will advance our understanding of inflammation in the gastrointestinal tract. Both questions can be approached experimentally.
Acknowledgments
We thank L.G. Axelson, S. Murthy, S. Brant, and J. Lafaille for insight and suggestions; A. Auerbach and the staff at New York University School of Medicine Transgenic Facility for assistance in production of chimeric animals; and G. Kreibich and L. Hendershot for antisera to ribophorin I and BiP. Supported by grants from the NIH (DK-47119 and CA-60945). A. Bertolotti is a recipient of an Human Frontier Science Program award; I. Novoa was supported in part by a Basque Government Fellowship; and D. Ron is a Scholar of the Leukemia and Lymphoma Society of America.
References
- 1.Sartor RB. Cytokine regulation of experimental intestinal inflammation in genetically engineered and T-lymphocyte reconstituted rodents. Aliment Pharmacol Ther. 1996;10(Suppl. 2):36–42. doi: 10.1046/j.1365-2036.1996.22164018.x. [DOI] [PubMed] [Google Scholar]
- 2.Strober W, et al. Reciprocal IFN-gamma and TGF-beta responses regulate the occurrence of mucosal inflammation. Immunol Today. 1997;18:61–64. doi: 10.1016/s0167-5699(97)01000-1. [DOI] [PubMed] [Google Scholar]
- 3.Bhan AK, Mizoguchi E, Smith RN, Mizoguchi A. Colitis in transgenic and knockout animals as models of human inflammatory bowel disease. Immunol Rev. 1999;169:195–207. doi: 10.1111/j.1600-065x.1999.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 4.Berg DJ, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadlack B, et al. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 6.Mombaerts P, et al. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell. 1993;75:274–282. doi: 10.1016/0092-8674(93)80069-q. [DOI] [PubMed] [Google Scholar]
- 7.Sands BE, Podolsky DK. The trefoil peptide family. Annu Rev Physiol. 1996;58:253–273. doi: 10.1146/annurev.ph.58.030196.001345. [DOI] [PubMed] [Google Scholar]
- 8.Mashimo H, Wu DC, Podolsky DK, Fishman MC. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science. 1996;274:262–265. doi: 10.1126/science.274.5285.262. [DOI] [PubMed] [Google Scholar]
- 9.Morteau O, et al. Impaired mucosal defense to acute colonic injury in mice lacking cyclooxygenase-1 or cyclooxygenase-2. J Clin Invest. 2000;105:469–478. doi: 10.1172/JCI6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saubermann LJ, et al. Activation of natural killer T cells by alpha-galactosylceramide in the presence of CD1d provides protection against colitis in mice. Gastroenterology. 2000;119:119–128. doi: 10.1053/gast.2000.9114. [DOI] [PubMed] [Google Scholar]
- 11.Panwala CM, Jones JC, Viney JL. A novel model of inflammatory bowel disease: mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J Immunol. 1998;161:5733–5744. [PubMed] [Google Scholar]
- 12.Falkow S, Isberg RR, Portnoy DA. The interaction of bacteria with mammalian cells. Annu Rev Cell Biol. 1992;8:333–363. doi: 10.1146/annurev.cb.08.110192.002001. [DOI] [PubMed] [Google Scholar]
- 13.Jago M, Payne A, Peterson J, Bagust T. Inhibition of glycosylation by corynetoxin, the causative agent of annual ryegrass toxicity: a comparison with tunicamycin. Chem Biol Interact. 1983;45:223–234. doi: 10.1016/0009-2797(83)90070-4. [DOI] [PubMed] [Google Scholar]
- 14.Jackson ME, et al. The KDEL retrieval system is exploited by Pseudomonas exotoxin A, but not by Shiga-like toxin-1, during retrograde transport from the Golgi complex to the endoplasmic reticulum. J Cell Sci. 1999;112:467–475. doi: 10.1242/jcs.112.4.467. [DOI] [PubMed] [Google Scholar]
- 15.Orlandi PA. Protein-disulfide isomerase-mediated reduction of the A subunit of cholera toxin in a human intestinal cell line. J Biol Chem. 1997;272:4591–4599. [PubMed] [Google Scholar]
- 16.Simpson JC, Dascher C, Roberts LM, Lord JM, Balch WE. Ricin cytotoxicity is sensitive to recycling between the endoplasmic reticulum and the Golgi complex. J Biol Chem. 1995;270:20078–20083. doi: 10.1074/jbc.270.34.20078. [DOI] [PubMed] [Google Scholar]
- 17.Xu A, Bellamy AR, Taylor JA. BiP (GRP78) and endoplasmin (GRP94) are induced following rotavirus infection and bind transiently to an endoplasmic reticulum-localized virion component. J Virol. 1998;72:9865–9872. doi: 10.1128/jvi.72.12.9865-9872.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peluso RW, Lamb RA, Choppin PW. Infection with paramyxoviruses stimulates synthesis of cellular polypeptides that are also stimulated in cells transformed by Rous sarcoma virus or deprived of glucose. Proc Natl Acad Sci USA. 1978;75:6120–6124. doi: 10.1073/pnas.75.12.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoeckle MY, et al. 78-kilodalton glucose-regulated protein is induced in Rous sarcoma virus-transformed cells independently of glucose deprivation. Mol Cell Biol. 1988;8:2675–2680. doi: 10.1128/mcb.8.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 21.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 22.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 23.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 24.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 25.Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 26.Chapman RE, Walter P. Translational attenuation mediated by an mRNA intron. Curr Biol. 1997;7:850–859. doi: 10.1016/s0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- 27.Kawahara T, Yanagi H, Yura T, Mori K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol Biol Cell. 1997;8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casagrande R, et al. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol Cell. 2000;5:729–735. doi: 10.1016/s1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- 29.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 30.Travers KJ, et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 31.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang XZ, et al. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urano F, et al. Coupling of stress in the endoplasmic reticulum to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 34.Harding H, Zhang Y, Ron D. Translation and protein folding are coupled by an endoplasmic reticulum resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 35.Bertolotti A, Zhang Y, Hendershot L, Harding H, Ron D. Dynamic interaction of BiP and the ER stress transducers in the unfolded protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 36.Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress-response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 37.Tybulewicz V, Crawford C, Jackson P, Bronson R, Mulligan R. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1553–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 38.Okayasu I, et al. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 39.Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69:238–249. [PubMed] [Google Scholar]
- 40.Wang X-Z, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP-kinase. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 41.Sundaram U, West AB. Effect of chronic inflammation on electrolyte transport in rabbit ileal villus and crypt cells. Am J Physiol. 1997;272:G732–G741. doi: 10.1152/ajpgi.1997.272.4.G732. [DOI] [PubMed] [Google Scholar]
- 42.Hugot JP, et al. Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature. 1996;379:821–823. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 43.Brant SR, et al. American families with Crohn’s disease have strong evidence for linkage to chromosome 16 but not chromosome 12. Gastroenterology. 1998;115:1056–1061. doi: 10.1016/s0016-5085(98)70073-3. [DOI] [PubMed] [Google Scholar]
- 44.Mahler M, et al. Differential susceptibility of inbred mouse strains to dextran sulfate sodium-induced colitis. Am J Physiol. 1998;274:G544–G551. doi: 10.1152/ajpgi.1998.274.3.G544. [DOI] [PubMed] [Google Scholar]
- 45.Mahler M, et al. Genetic analysis of susceptibility to dextran sulfate sodium-induced colitis in mice. Genomics. 1999;55:147–156. doi: 10.1006/geno.1998.5636. [DOI] [PubMed] [Google Scholar]
- 46.Breider MA, Eppinger M, Gough A. Intercellular adhesion molecule-1 expression in dextran sodium sulfate-induced colitis in rats. Vet Pathol. 1997;34:598–604. doi: 10.1177/030098589703400608. [DOI] [PubMed] [Google Scholar]
- 47.Bennett CF, et al. An ICAM-1 antisense oligonucleotide prevents and reverses dextran sulfate sodium-induced colitis in mice. J Pharmacol Exp Ther. 1997;280:988–1000. [PubMed] [Google Scholar]
- 48.Bendjelloul F, et al. Intercellular adhesion molecule-1 (ICAM-1) deficiency protects mice against severe forms of experimentally induced colitis. Clin Exp Immunol. 2000;119:57–63. doi: 10.1046/j.1365-2249.2000.01090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X-Z, et al. Signals from the stressed endoplasmic reticulum induce C/EBP homologous protein (CHOP/GADD153) Mol Cell Biol. 1996;16:4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brandtzaeg, P., Halstensen, T., and Kett, K. 1992. Immunopathology of inflammatory bowel disease. In Inflammatory bowel disease. R. MacDermott and W. Stenson, editors. Elsevier Science. New York, New York, USA. 95–136.
- 51.Strober W, Ehrhardt RO. Chronic intestinal inflammation: an unexpected outcome in cytokine or T cell receptor mutant mice. Cell. 1993;75:203–205. doi: 10.1016/0092-8674(93)80062-j. [DOI] [PubMed] [Google Scholar]
- 52.Elson CO, Sartor RB, Tennyson GS, Riddell RH. Experimental models of inflammatory bowel disease. Gastroenterology. 1995;109:1344–1367. doi: 10.1016/0016-5085(95)90599-5. [DOI] [PubMed] [Google Scholar]
- 53.Bylund-Fellenius A-C, Landström E, Axelsson L-G, Midtvedt T. Experimental colitis induced by dextran sulphate in normal and germfree mice. Microbial Ecology in Health and Disease. 1994;7:205–215. [Google Scholar]
- 54.Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 55.Groh V, et al. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Griffith E, Ramsburg E, Hayday A. Recognition by human gut gamma delta cells of stress inducible major histocompatibility molecules on enterocytes. Gut. 1998;43:166–167. doi: 10.1136/gut.43.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]


